Abstract
The HIV-1 co-receptor CCR5 has been thought a relevant target for small interfering RNA (siRNA)-based therapeutics. However, recent findings suggest that siRNA can stimulate innate cytokine responses in mammals. All siRNA agents tested were able to down-regulate the expression of CCR5, albeit with different efficiency (51–74% down-regulation), block HIV-induced syncytia formation between HIV-1 BaL-infected and uninfected CD4+ cells or block single-round HIV-1 infection as measured by a luciferase reporter assay (46–83% inhibition). Conversely, siRNA directed against CCR5 did not affect replication of a vesicular stomatitis virus (VSV) pseudotyped virus, suggesting that inhibition of HIV replication was specific to CCR5 down-regulation. However, two of four siRNA tested were able to induce the production of interleukin (IL) IL-6 (sixfold induction) and IL-8 (ninefold induction) but no interferon (IFN)-α, IFN-β, IFN-γ, tumour necrosis factor (TNF)-α, monocyte chemoattractant protein (MCP)-1, macrophage inflammatory protein (MIP)-1α, MIP-1β, RANTES, IL-1β, IL-10 or IL-12p70 cytokine induction was noted. In the absence of detectable IFN-α, IL-6 or IL-8 may represent markers of non-specific effects triggered by siRNA.
Keywords: CCR5, HIV, off-target effects, RNA interference
Introduction
RNA-mediated interference (RNAi) is a post-transcriptional gene-silencing phenomenon in which double-stranded RNA (dsRNA) may trigger the degradation of homologous mRNA in the cytoplasm of a cell. RNAi has become a consolidated tool for the study of gene function [1]. RNAi has been used to target the expression of viral genes to inhibit HIV-1 infection or to identify cellular genes that regulate virus infection and replication [2–8]. However, therapeutic approaches may be limited by the ability of HIV to escape to RNA interference [9,10], the efficient delivery of siRNA into the target cells or the specificity of the silencing effect.
It was believed that short interfering RNAs [< 30 base pairs (bp), siRNAs] were unable to trigger a similar cellular immune response [11] to that observed for longer double-stranded RNAs (dsRNAs) [12]. However, a number of studies have reported non-specific effects induced [13] or not [14] by siRNAs related to the interferon response. Several possible sensors for siRNAs in cells have been described, including Toll-like receptor 3 (TLR3) in macrophages [15], TLR7 and TLR8 in plasmacytoid dendritic cells [16], and more recently the RNA helicase RIG-I in T98G cells [17]. Nevertheless, the underlying mechanism triggering non-specific siRNA-induced responses is not well understood, and more importantly, a protocol for detection of these undesirable effects is not well established.
Materials and methods
Cells
U87-CD4+ CCR5+ cells (NIH, AIDS Reagent Program, Bethesda, MD, USA) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% of fetal bovine serum (FBS) in the presence of 0·5 mg/ml of geneticine (Invitrogen, Madrid, Spain) and 2 µg/ml of puromycin (Sigma-Aldrich, Madrid, Spain). Uninfected and HIV-1 chronically infected MOLT-4/CCR5+ cells were generated as described previously [18] and cultured in RPMI supplemented with 10% FBS.
siRNAs
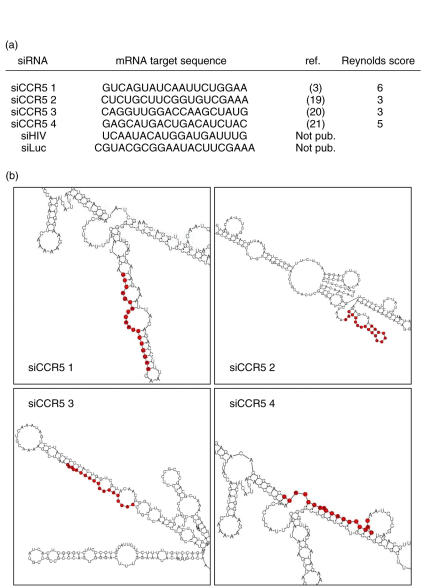
The sequences of CCR5-targeting siRNAs (siCCR5s) and control siRNAs used are shown in Fig. 1. Sequences are based on previously published papers [3,19–21]. siCCR5#1, siLuc and siHIV were purchased from Dharmacon (Lafayette, CO, USA). SiCCR5#2, siCCR5#3 and siCCR5#4 were provided by Qiagen (Crawley, UK). siRIG-I was purchased from Ambion (Huntingdon, UK). All siRNA were resuspended in RNAase free, HEPES-base buffer.
Fig. 1.
Small interfering RNA (siRNAs) sequences and predicted secondary structures. (a) mRNA target sequences of siRNAs, source references and Reynolds score [22] for siRNA targeting CCR5. (b) Predicted CCR5 mRNA secondary structure. siCCR5-targeted regions are indicated in red.
CCR5 mRNA secondary structure prediction was performed using the MWG Biotech online siRNA design tool (siMAX design tool, http://www.mwg-biotech.com) using a default temperature of 37°C. Reynolds scores for siCCR5s were calculated as described previously [22]. In vitro-transcribed dsRNA (> 700 bp) was used as control.
Transfections
Cells (1–1·5 × 106) were seeded in 24-well plates. siRNAs were transfected in a concentration of 70–100 nM. The Block-iT fluorescent oligo (Invitrogen) was used as a control of transfection as detected by flow cytometry. Transfections were performed with lipofectamine 2000, during 4–5 h in serum-free medium (Optimem, Invitrogen) following the manufacturer's instructions; 48 h after transfection, cells were stained for surface CCR5, co-cultured with HIV infected cells or infected with HIV. Supernatants were stored at −20°C until cytokine quantification assays.
Flow cytometry
Transfected cells were detached gently with trypsin-free ethylenediamine tetraacetic acid (EDTA) solution (Versene, 1 : 5000; Invitrogen) and stained with the following monoclonal antibodies as described previously [23]:CD4–peridinin–chlorophyll complex protein (PerCP) and CCR5–phycoerythrin (PE) (BD Biosciences, Madrid, Spain).
Quantitative real-time polymerase chain reaction (PCR)
Total cellular RNA was purified from transfected cells by RNeasy mini kit (Qiagen) as recommended by the manufacturer, including a DNAse I treatment step. Relative levels of RIG-I (DDX58) and TLR3 mRNA were measured by two-step quantitative reverse transcription (RT)–PCR and normalized by the comparative carboxy terminal (Ct) method to glyceraldehyde-3-phosphate-dehydrogenase (GADPH) mRNA expression. In all experiments, a sample treated in the absence of RT enzyme was used as a negative control. Primers and DNA probes were purchased from Applied Biosystems (assay-on-demand), selecting assays whose probes span an exon junction to avoid detecting genomic DNA, and reactions analysed on an ABI PRISM 7000 (Applied Biosystems, Madrid, Spain).
Viral stocks
The CCR5 using (R5) HIV-1 strain BaL was grown in U87-CD4+ CCR5+ cells. The HIV-1 NL4-3 expression plasmid lacking functional envelope (pNL4-3.Luc.R–E–, NIH AIDS Reagent Program), together with plasmid expressing HIV-1 R5 envelope or envelope protein G of the vesicular stomatitis virus (VSV-G) were used to produce viral stocks in 293T cells as described previously [6]. Cell-free viral stock was estimated using an enzyme-linked immunoassay for antigen HIV-p24 detection (Innotest™ HIV-Ag; Innogenetics, Barcelona, Spain) and titrated for use in 24-well plate infection assays.
Co-cultures of infected and uninfected cells
Transfected cells were co-cultured with HIV-1 (BaL) persistently infected MOLT/CCR5+ cells (MOLT-BaL) in a 10 : 1 ratio, in the absence or presence of HIV-1 inhibitors as described previously [18]: CCR5 antagonist TAK779 [24] (NIH AIDS Reagent Program) and 1 µg/ml of the RT inhibitor zidovudine (AZT; Sigma, Madrid, Spain). Co-cultures were incubated at 37°C for 24 h, fixed and evaluated for syncytium formation by phase-contrast microscopy.
HIV infection and replication
Cells were infected 48 h after siRNA transfection with 300 pg/ml of HIV-1 pseudotyped with R5 envelope (HIVenv) or 10 pg/ml of HIV-1 pseudotyped with VSV (HIVvsv). After 3 days, cells were detached with trypsin treatment (Invitrogen), pelleted and kept frozen (−80°C). These viruses encode for luciferase gene instead of envelope protein, and levels of viral replication were determined by luciferase assay using the Bright-Glo™ luciferase assay system (Promega, Madrid, Spain). HIV inhibitors were used as controls when indicated. The R5 HIV-1 strain BaL was used for replication assays in the presence or absence of interleukin (IL) IL-6 and IL-8 (Peprotech, London, UK). Viral replication was quantified after 3 days by determining extracellular p24 in supernatants as described previously [25].
Cytokine quantification
Inflammatory cytokines were measured by flow cytometry using a cytometric bead array (CBA, Human Inflammation Kit, BD Biosciences) for the quantification of IL-8, IL-1β, IL-6, IL-10, TNF and IL-12p70 (20–5000 pg/ml), as described previously [26]. Human Chemokine Kit II was also used for the detection of IL-8, RANTES, monocyte chemoattractant protein (MCP)-1, macrophage inflammatory protein (MIP)-1α, MIP-1β or MCP-1. For the detection of interferon (IFN)-α, IFN-β and IFN-γ, conventional quantitative sandwich enzyme immunoassays (ELISA) were used: human IFN-α ELISA kit (10–500 pg/ml or 40–5000 pg/ml, R&D Systems Minneapolis, MN, USA), human IFN-β ELISA kit (250–10 000 pg/ml, R&D Systems) and human IFN-gamma BD OptEIA ELISA set (12·5 pg/ml detection limit, BD Biosciences).
Statistical analysis
Results are given as the mean ± s.e.m. of at least three experiments.
Results
siRNA induce down-regulation of CCR5
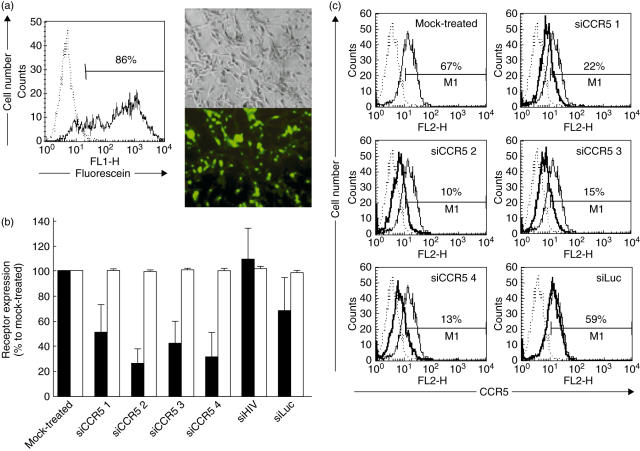
siRNA may be transfected into CCR5+, CD4+ U87 cells with an efficiency of 86% (Fig. 2a). All four siRNAs targeting CCR5 [3,19–21] were able to reduce CCR5 expression but with different efficiency (Fig. 2b,c). Relative levels of surface expression of CCR5 showed 51–74% down-regulation, compared to mock-transfected cells or cells treated with non-related siRNAs (siHIV and siLuc). The order of potency of the tested siRNAs was siCCR5#2 > siCCR5#4 > siCCR5#3 > siCCR5#1. Conversely, none of the siRNAs reduced the expression of CD4. CCR5 down-regulating efficiency did not correlate with the expected efficiency as calculated by Reynolds design criteria [22], which pointed to siCCR5#3 as the most effective (Fig. 1a). The predicted mRNA secondary structure (Fig. 1b) did not support any preference to siCCR5#2 as the most potent siRNA. siCCR5#2 partially targets a hairpin structure, which has been suggested as a negative selection factor for siRNA sequences [27,28].
Fig. 2.
Different small interfering RNAs (siRNAs) inhibited HIV-1 co-receptor CCR5 with different efficiency. (a) Transfection efficiency of U87-CD4+ CCR5+ cells, using a RNA oligonucleotide labelled with fluorescein and followed by fluorescence microscopy and flow cytometry. (b) Relative surface expression of CCR5 (black bars) and CD4 (white bars) in U87-CD4+ CCR5+ cells 48 h after treatment with siRNAs. CCR5 and CD4 were quantified by flow cytometry and expressed as percentage of mock-treated control. Mean ± s.e.m. of four independent experiments is shown. (c) siCCR5s inhibited surface levels of CCR5 quantified by flow cytometry. Non-stained cells (dotted line), CCR5 levels of mock-treated cells (narrow line) and CCR5 levels of siRNA-treated cells (thick line) are shown. One representative experiment is shown.
siRNA targeting CCR5 blocked HIV replication
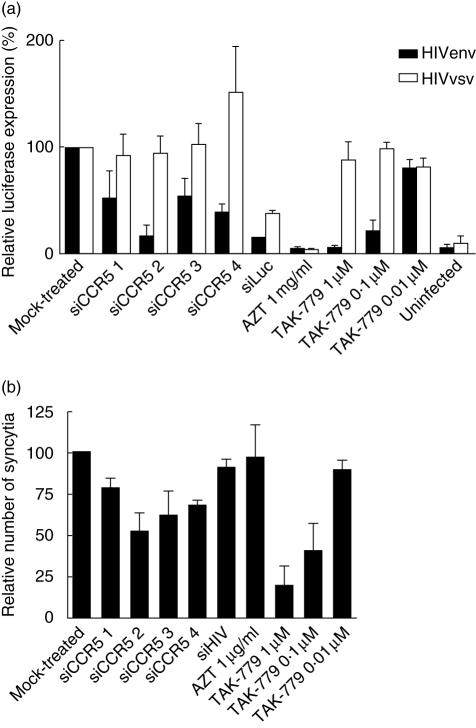
The effect of siRNA in HIV infection is shown in Fig. 3. All siRNAs targeting CCR5 blocked HIV replication ina single cycle replication assay (Fig. 3a). The potency of siRNA at blocking HIV replication was siCCR5#2 > siCCR5#4 > siCCR5#3 > siCCR5#1. The siRNA targeting the luciferase gene was able to block luciferase formation and all controls (the reverse transcriptase inhibitor AZT and the fusion inhibitor TAK-779) blocked HIV replication, as expected. None of the siRNA tested blocked the replication of a VSV-G/HIV chimera. However, siCCR5#4-tranfected cells showed higher virus replication levels (150 ± 44%).
Fig. 3.
Small interfering RNAs (siRNAs) against CCR5 specifically inhibited HIV-1 pathogenesis and replication. (a) Drug- and siRNA-treated cells were infected with envelope protein (env) pseudotyped HIV (HIVenv, black bars) and vesicular stomatitis virus (VSV)-G pseudotyped HIV (HIVvsv, white bars), and replication was detected as luciferase expression. Data were normalized to mock-treated samples and expressed as relative luciferase expression. Mean ± s.e.m of at least three independent experiments. Each experiment was run at least in duplicate. (b) Relative number of syncytia after co-culture of siRNA-treated cells with HIV-1 persistently infected cells (Molt-BaL), normalized to the co-culture with mock-treated cells. Mean ± s.e.m of three independent experiments is shown. Each experiment was run in triplicate, and at least three fields were counted for each triplicate.
All siRNAs targeting CCR5 reduced syncytium formation when compared to mock-treated cells (Fig. 3b). The order of potency was siCCR5#2 > siCCR5#3 > siCCR5#4 > siCCR5#1. Fusion inhibitor TAK-779 showed dose-dependent activity. As expected, AZT or siHIV, targeting post-entry events of viral cycle, did not have an effect on fusion events between infected and uninfected cells. Taken together, these results suggest that the inhibitory effect of siRNAs targeting CCR5 was specific for HIV envelope-dependent fusion, i.e. CCR5-dependent.
Cytokine production by cells transfected with siRNAs
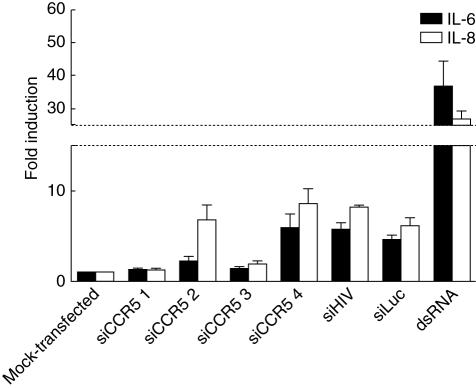
Recent findings [16,29] suggest that siRNAs may induce a type I IFN response. Thus, we evaluated the presence of a number of cytokines in the supernatants of cells treated with siRNAs. CBAs were performed to detect differences in cytokine secretion compared to mock-transfected cells. No IFN-α, IFN-β, IFN-γ, TNF-α, MCP-1, MIP-1α, MIP-1β, RANTES, IL-1β, IL-10 or IL-12p70 cytokine induction was noted (data not shown). However, production of the inflammatory cytokines IL-6 and IL-8 was up-regulated depending on the siRNA used (Fig. 4). Levels of IL-6 and IL-8 in supernatants of mock-transfected cells were 99·6 ± 19·6 and 660·0 ± 52·8 pg/ml, respectively. siCCR5#4 induced 6 ± twofold and 9 ± twofold the production of both IL-6 and IL-8 (600·6 ± 124·8 and 5632·1 ± 333·2 pg/ml, respectively) similarly to control siRNAs (siLuc and siHIV), whereas siCCR5#2 stimulated mainly IL-8 (7 ± twofold; 4528·0 ± 187·4 pg/ml) and siCCR5#1 and #3 did not show significant changes (< twofold increase). Long (> 700 bp) double-stranded RNA (dsRNA) at 16 ng/ml induced a strong secretion of IL-6 and IL-8 (37 ± 8 and 27 ± twofold; 3652·2 ± 494·6 and 13639·9 ± 516·1 pg/ml, respectively). In addition, low levels of RANTES and MIP-1α were detected when cells were treated with long dsRNA (data not shown). IL-6 and IL-8 secretion were not associated with the functionality of siRNAs, as control siRNAs (siLuc and siHIV) could also induce cytokine up-regulation.
Fig. 4.
Small interfering RNA (siRNAs) stimulated cytokine secretion. Interleukin (IL) IL-6 (black bars) and IL-8 (white bars) in the supernatants of siRNA-treated cells were quantified by cytometric bead array (CBA). Data are expressed as the induction of secretion relative to mock-transfected cells. Mean ± s.e.m of at least three experiments are shown.
Of note, we tested that exogenous IL-6 or IL-8 alone or in combination did not have an effect on CCR5 expression, HIV replication or the observed siRNA anti-viral effects (data not shown).
Double-stranded RNA(dsRNA) induced activation of RIG-I
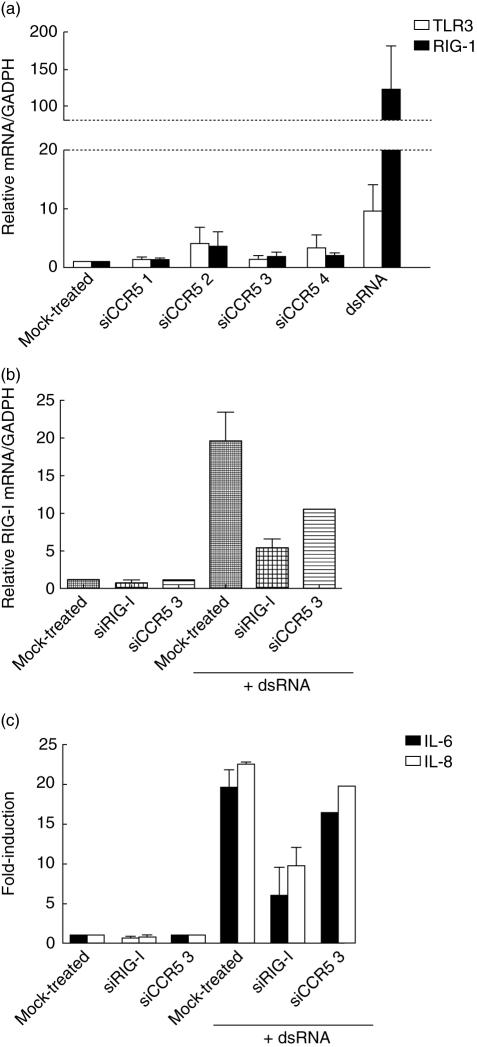
Incubation of cells with siRNAs in the absence of the transfection reagent did not induce RNA uptake and IL-6/IL-8 secretion (data not shown). This observation suggests that an immune response may be triggered by an intracellular dsRNA sensor [30]. The RNA helicase RIG-I is a cytoplasmic sensor of dsRNA [31] and has been associated recently with non-specific effects triggered by siRNAs [17]. TLR3 has also been suggested as a sensor for siRNA [15] and is located in the cell surface and endosomes [32]. Thus, we quantified mRNA of RIG-I and TLR3 in siRNA-treated cells. mRNA of RIG-I was increased when cells were transfected with dsRNA (124·1 ± 58·6-fold) (Fig. 5a). However, we were not able to detect RIG-I mRNA up-regulation in cells treated with siRNAs (< fivefold-induction). Similarly, none of the siRNAs showed higher mRNA levels of TLR3, but dsRNA only increased 9·6 ± 4·4 times its levels normalized to GADPH, suggesting that detection of dsRNA may be mediated by RIG-I. In addition, treatment of U87-CD4+ CCR5+ cells with an siRNA against RIG-I, prior to transfection with dsRNA, prevented RIG-I mRNA up-regulation (Fig. 5b) and impaired IL-6/IL-8 induction (Fig. 5c), providing evidence that RIG-I acts as a dsRNA sensor.
Fig. 5.
Double-stranded RNA (dsRNA) stimulates a non-specific immune response. (a) Relative amount of TLR3 (white bars) and RIG-I (black bars) mRNA normalized to mRNA of glyceraldehyde-3-phosphate-dehydrogenase (GADPH) after treatment with small interfering RNA (siRNAs) and dsRNA. (b) Relative quantification by reverse transcription–polymerase chain reaction (RT–PCR) of mRNA RIG-I levels in U87-CD4+ CCR5+ cells, mock-transfected, treated with an siRNA targeting RIG-I (siRIG-I) or siRNA targeting CCR5 (siCCR5 3), before transfection of dsRNA. (c) Induction of interleukin (IL) IL-6 and IL-8 secretion by dsRNA in cells pretreated with siRNA targeting RIG-I (siRIG-I), targeting CCR5 (siCCR5 3) or lipofectamine-treated (mock-treated). Mean ± s.e.m. of two independent experiments.
Discussion
The application of screening assays for non-specific activation of both innate and acquired immunity will be necessary for further development of RNAi as a therapeutic tool [33]. Here, we provide evidence of non-specific, unwanted effects of a series of siRNAs that have been tested as candidate anti-HIV agents and shown to specifically block HIV co-receptor CCR5 expression. Independently of their potency as inhibitors of HIV replication and apparent specificity for the HIV co-receptor, two of four siRNAs targeting CCR5 were able to induce the production of IL-6, IL-8 or both. This effect might have been overlooked by the evaluation of interferon production or 2′,5′-oligoadenylate synthetase 1 (OAS1) up-regulation that are commonly used as reference for non-specific activation of innate immune responses. Regardless of the potency of IL-6 or IL-8 production, these cytokines may act as relevant surrogate markers of the specificity of siRNAs.
CCR5 is an important therapeutic target for anti-HIV intervention and a number of independent laboratories have shown the effect of siRNAs targeting CCR5, making these siRNA of interest to evaluate possible non-specific, off-target effects. The CCR5 siRNA that were tested herein did not obey Reynolds' specificity criteria (low G/C content, a bias towards low internal stability at the sense 3′-terminus, lack of inverted repeats and sense strand base preferences at positions 3, 10, 13 and 19) [22] with regard to potency towards CCR5 down-regulation or anti-HIV inhibition. Furthermore, siRNA-specificity criteria was irrelevant to the ability of siRNA to induce the secretion of IL-6 (siCCR5#2) or IL-6 and IL-8 (siCCR5#4). The selection of specific siRNA targeting any particular gene may require a detailed structure activity relationship that also takes into account an array of markers of the innate immune response.
The observation that production of IL-6 or IL-8 did not have an effect on the replication of HIV after acute infection or the inhibition by siRNAs could indicate that, in our model, cytokine production was not sufficient to affect those parameters evaluated herein. U87-CD4+ cells are IL-6 receptor (CD126)-negative and IL-8 receptor (CDw128)-negative (data not shown). Conversely, B lymphocytes express CD126, and IL-6 has been shown to affect HIV replication and disease progression [34–38]. T cells and monocytes, which are both the main target of HIV infection, express CDw128, suggesting that non-specific effects induced by siRNA could alter the course of infection. Our results highlight the potential and simplicity of the use of siRNA to target cellular or viral genes. However, they caution on the generalized simplification of selecting homologous target sequences that could induce non-specific immune responses.
Acknowledgments
This work was supported in part by the European TRIoH Consortium (LSHB-CT-2003–503480) and the Spanish MEC (BFU2006-00966) and FIPSE (36549-6) and FIS (PI060624).
References
- 1.Novina CD, Sharp PA. The RNAi revolution. Nature. 2004;430:161–4. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]
- 2.Berkhout B, Haasnoot J. The interplay between virus infection and the cellular RNA interference machinery. FEBS Lett. 2006;580:2896–902. doi: 10.1016/j.febslet.2006.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez MA, Gutierrez A, Armand-Ugon M, et al. Suppression of chemokine receptor expression by RNA interference allows for inhibition of HIV-1 replication. AIDS. 2002;16:2385–90. doi: 10.1097/00002030-200212060-00002. [DOI] [PubMed] [Google Scholar]
- 4.Pauls E, Este JA. RNA interference as a tool for target validation. Drug Discov Today Technol. 2004;1:135–40. doi: 10.1016/j.ddtec.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Martinez MA, Clotet B, Este JA. RNA interference of HIV replication. Trends Immunol. 2002;23:559–61. doi: 10.1016/s1471-4906(02)02328-1. [DOI] [PubMed] [Google Scholar]
- 6.Pauls E, Senserrich J, Clotet B, Este JA. Inhibition of HIV-1 replication by RNA interference of p53 expression. J Leukoc Biol. 2006;80:659–67. doi: 10.1189/jlb.0306189. [DOI] [PubMed] [Google Scholar]
- 7.Jacque JM, Stevenson M. The inner-nuclear-envelope protein emerin regulates HIV-1 infectivity. Nature. 2006;441:641–5. doi: 10.1038/nature04682. [DOI] [PubMed] [Google Scholar]
- 8.Li Z, Xiong Y, Peng Y, et al. Specific inhibition of HIV-1 replication by short hairpin RNAs targeting human cyclin T1 without inducing apoptosis. FEBS Lett. 2005;579:3100–6. doi: 10.1016/j.febslet.2005.04.074. [DOI] [PubMed] [Google Scholar]
- 9.Das AT, Brummelkamp TR, Westerhout EM, et al. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J Virol. 2004;78:2601–5. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabariegos R, Gimenez-Barcons M, Tapia N, Clotet B, Martinez MA. Sequence homology required by human immunodeficiency virus type 1 to escape from short interfering RNAs. J Virol. 2006;80:571–7. doi: 10.1128/JVI.80.2.571-577.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–8. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 12.Minks MA, West DK, Benvin S, Baglioni C. Structural requirements of double-stranded RNA for the activation of 2′,5′-oligo(A) polymerase and protein kinase of interferon-treated HeLa cells. J Biol Chem. 1979;254:10180–3. [PubMed] [Google Scholar]
- 13.Kim DH, Longo M, Han Y, Lundberg P, Cantin E, Rossi JJ. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat Biotechnol. 2004;22:321–5. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- 14.Semizarov D, Frost L, Sarthy A, Kroeger P, Halbert DN, Fesik SW. Specificity of short interfering RNA determined through gene expression signatures. Proc Natl Acad Sci USA. 2003;100:6347–52. doi: 10.1073/pnas.1131959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kariko K, Bhuyan P, Capodici J, Weissman D. Small interfering RNAs mediate sequence-independent gene suppression and induce immune activation by signaling through Toll-like receptor 3. J Immunol. 2004;172:6545–9. doi: 10.4049/jimmunol.172.11.6545. [DOI] [PubMed] [Google Scholar]
- 16.Hornung V, Guenthner-Biller M, Bourquin C, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11:263–70. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 17.Marques JT, Devosse T, Wang D, et al. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat Biotechnol. 2006;24:559–65. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- 18.Bosch B, Blanco J, Pauls E, et al. Inhibition of coreceptor-independent cell-to-cell human immunodeficiency virus type 1 transmission by a CD4-immunoglobulin G2 fusion protein. Antimicrob Agents Chemother. 2005;49:4296–304. doi: 10.1128/AAC.49.10.4296-4304.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arteaga HJ, Hinkula J, van Dijk-Hard I, et al. Choosing CCR5 or Rev siRNA in HIV-1. Nat Biotechnol. 2003;21:230–1. doi: 10.1038/nbt0303-230. [DOI] [PubMed] [Google Scholar]
- 20.Qin XF, An DS, Chen IS, Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci USA. 2003;100:183–8. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song E, Lee SK, Dykxhoorn DM, et al. Sustained small interfering RNA-mediated human immunodeficiency virus type 1 inhibition in primary macrophages. J Virol. 2003;77:7174–81. doi: 10.1128/JVI.77.13.7174-7181.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–30. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 23.Bosch B, Clotet-Codina I, Blanco J, et al. Inhibition of human immunodeficiency virus type 1 infection in macrophages by an alpha-v integrin blocking antibody. Antiviral Res. 2006;69:173–80. doi: 10.1016/j.antiviral.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Este JA. Virus entry as a target for anti-HIV intervention. Curr Med Chem. 2003;10:1617–32. doi: 10.2174/0929867033457098. [DOI] [PubMed] [Google Scholar]
- 25.Armand-Ugon M, Clotet-Codina I, Tintori C, et al. The anti-HIV activity of ADS-J1 targets the HIV-1 gp120. Virology. 2005;343:141–9. doi: 10.1016/j.virol.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 26.Coma G, Pena R, Blanco J, et al. Treatment of monocytes with interleukin (IL) -12 plus IL-18 stimulates survival, differentiation and the production of CXC chemokine ligands (CXCL) 8, CXCL9 and CXCL10. Clin Exp Immunol. 2006;145:535–44. doi: 10.1111/j.1365-2249.2006.03145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schubert S, Grunweller A, Erdmann VA, Kurreck J. Local RNA target structure influences siRNA efficacy: systematic analysis of intentionally designed binding regions. J Mol Biol. 2005;348:883–93. doi: 10.1016/j.jmb.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 28.Luo KQ, Chang DC. The gene-silencing efficiency of siRNA is strongly dependent on the local structure of mRNA at the targeted region. Biochem Biophys Res Commun. 2004;318:303–10. doi: 10.1016/j.bbrc.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 29.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–62. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 30.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 31.Yoneyama M, Kikuchi M, Natsukawa T, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–7. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 32.Dunne A, O'Neill LA. Adaptor usage and Toll-like receptor signaling specificity. FEBS Lett. 2005;579:3330–5. doi: 10.1016/j.febslet.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 33.Marques JT, Williams BR. Activation of the mammalian immune system by siRNAs. Nat Biotechnol. 2005;23:1399–405. doi: 10.1038/nbt1161. [DOI] [PubMed] [Google Scholar]
- 34.Breen EC, Rezai AR, Nakajima K, et al. Infection with HIV is associated with elevated Il−6 levels and production. J Immunol. 1990;144:480–4. [PubMed] [Google Scholar]
- 35.Lane BR, Lore K, Bock PJ, et al. Interleukin-8 stimulates human immunodeficiency virus type 1 replication and is a potential new target for antiretroviral therapy. J Virol. 2001;75:8195–202. doi: 10.1128/JVI.75.17.8195-8202.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee ES, Zhou H, Henderson AJ. Endothelial cells enhance human immunodeficiency virus type 1 replication in macrophages through a C/EBP-dependent mechanism. J Virol. 2001;75:9703–12. doi: 10.1128/JVI.75.20.9703-9712.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poli G, Bressler P, Kinter A, et al. Interleukin 6 induces human immunodeficiency virus expression in infected monocytic cells alone and in synergy with tumor necrosis factor alpha by transcriptional and post-transcriptional mechanisms. J Exp Med. 1990;172:151–8. doi: 10.1084/jem.172.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scala G, Ruocco MR, Ambrosino C, et al. The expression of the interleukin-6 gene is induced by the human-immunodeficiency-virus-1 Tat protein. J Exp Med. 1994;179:961–71. doi: 10.1084/jem.179.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]