Abstract
The relationship between the occurrence of cryoglobulins and hepatitis C virus (HCV) productive infection in peripheral blood and bone marrow-derived lymphocytes was explored. HCV minus strand RNA, the viral replicative intermediate, was searched for by a polyA+ tract strand-specific Tth-based reverse transcriptase–polymerase chain reaction (RT–PCR) in lymphoid cells of 46 patients with acute and chronic infection. The HCV minus strand was demonstrated in RNA extracted from six (13%) and five (11%) peripheral blood and bone marrow-derived lymphocytes, respectively. The HCV replicating form in lymphoid cells was associated strictly with mixed cryoglobulinaemia (MCG), in that it was found in six of 13 (46%) MCG patients, including two with B cell non-Hodgkin's lymphoma (NHL). No traces of HCV-negative strand RNA were found in four patients with acute hepatitis C, in 15 with chronic active hepatitis without extrahepatic disorders, in seven with monoclonal gammopathy of undetermined significance, and in seven with B-NHL without MCG. These results emphasize the direct role of the virus in the pathogenesis of MCG and support the contention that HCV is not specifically lymphotropic, its entry and replication in lymphoid cells being determined largely by selective interactions.
Keywords: cryoglobulinaemia, HCV RNA minus strand, mononuclear cells, non-Hodgkin's lymphoma, Tth-based RT–PCR
Introduction
Hepatitis C virus (HCV) is an enveloped RNA virus of Hepacivirus genus in the Flaviviridae family [1]. Although permissive cell culture has been described recently [2], it is still difficult to characterize the virus replication cycle. Its genome carries a single positive-strand RNA containing a single open reading frame encoding for a long viral polyprotein of 3010–3040 amino acids, flanked at both ends by highly conserved untranslated regions (UTR) [3].
RNA replication takes place in distinct compartments of the cell cytoplasm and requires viral (NS3–NS5) and host proteins [4]. During replication, HCV genomic RNA is transcribed into a complementary RNA strand, which subsequently constitutes a template for the synthesis of new viral genome [4]. The presence of the minus strand is essential to prove active viral replication in tissues and cells. Detection of plus-strand RNA merely implies contamination of circulating virions. HCV RNA negative strand is detected commonly in liver biopsies, and the ratio of replicative intermediate versus genomic RNA is estimated to be in the range of 1 : 10–1 : 100 [5,6].
The existence of extrahepatic reservoirs of HCV replication remains highly controversial [7]. Due to the weak amount of negative strand RNA, a sensitive and specific system should be employed to demonstrate the HCV replicative cycle in the cells. Using primer-specific reverse transcription–polymerase chain reaction (RT–PCR) plus RNA may act as a template for the synthesis of ‘false’ negative strand molecules [8,9]. Non-specific priming may include self and random priming, due to the complex secondary structure of the virus [8] and contaminating cellular nucleic acids [10]. Attempts have been made in the last few years to overcome these drawbacks, including chemical blocking of the free 3′ end of RNA [10], tagged primers directed against 5′ end [8] or core [11] regions of HCV genome and the use of rTth, a thermostable enzyme that has both reverse transcriptase and DNA polymerase activity [12].
Although these assays have greatly improved the specificity of HCV minus strand RNA detection, they have not definitively eliminated non-specific priming events associated with false detection of incorrect strand. Takyar et al. [13] described a novel procedure with a high degree of discrimination, in that false detection of negative strand does not occur even in the presence of 1010 molecules of the positive strand. By this method, with some modifications, we provide evidence for the occurrence of HCV replication in bone marrow (BM) and peripheral blood (PB) mononuclear cells (MC) of HCV-infected patients with MCG.
Materials and methods
Study population
Samples were obtained from patients enrolled in a prospective natural-history study of HCV infection starting from January 2002. Patients were included if they were: (a) HCV RNA positive, regardless of their HCV serostatus; (b) HBsAg and anti-HIV negative; and (c) not receiving anti-viral therapy at the beginning of the study.
Baseline evaluation included disease history, current signs and symptoms and previous medications. Physical examination and laboratory values were recorded. Serum cryoglobulins were determined as described elsewhere [14], and the monoclonal component in the serum or in the cryoprecipitate was characterized by immunofixation (Paragon, Beckman, Fullerton, CA, USA).
Patients were divided as follows: 15 patients with chronic active liver disease (CALD), without signs of extrahepatic disorders; 18 patients with CALD associating type II MCG in 11 and a monoclonal gammopathy of undetermined significance (MGUS) in seven (IgGk in six and IgGë in one); nine patients with B cell non-Hodgkin's lymphoma (B-NHL, two of them developed in MCG patients); and four patients with acute hepatitis C. Criteria for the diagnosis of acute hepatitis C were: (a) HCV RNA-positive without anti-HCV seroconversion; (b) negative markers for hepatitis A virus (HAV), Epstein–Barr virus (EBV) [IgM anti-Epstein–Barr virus nuclear antigen (EBNA) and anti-virus capsid antigen (VCA)], and cytomegalovirus (CMV) (IgM anti-CMV); (c) negative markers for autoimmune diseases [anti-nuclear antibody (ANA), anti-mitochondrial antibody (AMA), anti-liver–kidney microsome (LKM)-1]; (d) alanine aminotransferase (ALT) levels at least 10 times higher than the upper limit of normal values; and (e) no administration of potential hepatotoxic drugs; (f) histology compatible with acute liver damage.
All patients underwent liver biopsy. Total activity score and degree of fibrosis were evaluated according to Ishak et al. [15]. The study was approved by the Institutional Ethical Committee, and written informed consent was obtained from all patients. HCV RNA levels in cells and sera were quantified by signal amplification, using branched DNA in a sandwich hybridization assay (Quantiplex HCV RNA, version 3·0; Chiron Corp., Emeryville, CA, USA). The main clinical, histological and virological features of these patients are summarized in Table 1.
Table 1.
Epidemiological, virological, laboratory and histological parameters in 46 patients with acute and chronic hepatitis C virus (HCV) infection.
| Epidemiology | Virology | Laboratory | Liver histology | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotypes | |||||||||||
| Patients | No. | Sex (F/M) | Age (years) | Serum HCV RNA (IU/ml) | 1 | 2 | 3 | ALT (IU/l) | Diagnosis (CAH/C/MCh) | Activity index (mean ± s.d.) | Stage (mean ± s.d.) |
| Chronic infection | |||||||||||
| Without lymphoproliferation | 15 | 6/9 | 62 ± 14 | 1463 568 ± 1 685 452 | 8 | 6 | 1 | 184 ± 80 | 13/2/0 | 7·2 ± 2·2 | 2·4 ± 0·5 |
| With lymphoproliferation | |||||||||||
| MCG | 11 | 8/3 | 61 ± 10 | 988 450 ± 1 050 116 | 4 | 7 | 0 | 70 ± 44 | 10/1/0 | 4·6 ± 2·3 | 2·1 ± 0·8 |
| MGUS | 7 | 4/3 | 63 ± 9 | 1246 350 ± 1 126 440 | 4 | 2 | 1 | 88 ± 51 | 5/2/0 | 5·0 ± 2·1 | 3·0 ± 1·2 |
| B-NHL with MCG | 2 | 1/1 | 69 ± 6 | 1288 410 ± 1 450 910 | 1 | 1 | 0 | 45 ± 11 | 1/0/1 | 3·2 ± 0·5 | 2·2 ± 0·3 |
| B-NHL without MCG | 7 | 3/4 | 68 ± 11 | 1446 896 ± 1 160 110 | 4 | 2 | 1 | 40 ± 42 | 2/0/5 | 3·6 ± 1·0 | 2·3 ± 0·9 |
| Acute infection | 4 | 1/3 | 45 ± 18 | 326 404 ± 415 670 | 1 | 2 | 1 | 624 ± 827 | – | – | – |
CAH: chronic active hepatitis; C: cirrhosis; MCh: minimal changes; MCG: mixed cryoglobulinemia; MGUS: monoclonal gammopathy of undetermined significance; B-NHL: B-cell non-Hodgkin's lymphoma.
Purification of RNA
Cell RNA was purified as described elsewhere [16]. All steps including centrifugation and lymphomonocytes separation were performed at 37°C. Syringes and solutions were prewormed. Briefly, peripheral blood mononuclear cells (PBMC) and bone marrow mononuclear cells (BMMC) were separated by density gradient centrifugation on a suitable medium (Pharmacia, Uppsala, Sweden). The mononuclear cell layer was washed in RPMI-1640 medium supplemented with 1% HEPES. Cells were resuspended at a final concentration of 106/ml in RPMI-1640 and processed with the BioRobot EZ1 and EZ1 RNA cell kits (Qiagen, Valencia, CA, USA). This is a silica-based RNA purification assay which allows nucleic acids to be isolated through their binding to the silica surface in the presence of chaotropic salts. DNA was removed by RNase-free DNase I. The quality of extracted RNA was controlled by the UIA small nuclear RNP gene. To this end, reverse transcription with anti-sense primer and PCR amplification of the uroporphyrin isomerase (UI) region were performed as described previously [17].
HCV minus strand RNA was purified from total RNA using the Oligotex Direct mRNA isolation system purchased from Qiagen [18]. By this procedure, pure polyA+ RNA was isolated directly from cells, utilizing rigorous denaturing lysis conditions to generate an immediate RNase-free environment for the isolation of intact mRNA. The RNA samples were hybridized with a dT oligomer coupled covalently to the surface of polystyrene-latex particles. In this context, the polyA+ tail found toward the 5′ end of the HCV minus strand provided a useful tool for the separation from RNA molecules without polyA+ tails that were easily washed away.
The majority of RNA molecules are tRNAs and rRNAs, whereas mRNA accounts for only 1–5% of the total cellular RNA. In this respect, it is highly desirable to reduce the amount of rRNAs and tRNAs in a total RNA preparation, in order to increase the relative amount of polyA+ containing RNA molecules. Thus, particle-bound polyA+ RNA including HCV minus strand RNA was washed free of contaminating RNA, eluted and processed for RT–PCR using sense primer in the RT step.
The RT–PCR was performed using rTth essentially as described by Lanford et al. [9], with minor modifications. cDNA was generated in 20 µl of a reaction mixture containing 50 pM of primer (sense: 17–32; 5′-GAGGGCGACACTCCACCA-3′), 1× RT buffer (Perkin-Elmer, Inc., Foster City, CA, USA), 1 mM MnCl2, 200 µM of each deoxynucleoside triphosphate, 5 units of rTth (Perkin-Elmer, Inc.). After 20 min at 70°C, Mn2+ was chelated with 8 µl of 10× Ethylene glycol tetraacetic acid (EGTA) buffer (Perkin-Elmer, Inc.), then 50 pM primer (anti-sense: 254–274; 5′-TCGCGACCCAACACTACTC-3′) was added, the volume was adjusted to 100 µl, and the MgCl2 concentration was adjusted to 2·2 mM. The mixture was subjected to 45 cycles of 94°C for 1 min, 62°C for 1 min and 74°C for 1 min, followed by 72°C for 7 min.
Two µl of the first-round product, 259-base pairs (bp) in length in the 5′-UTR, were used to perform a nested PCR with inner primers (sense: 98–117; GAGTGTCGTGCAGCCTCCAG-3′ and anti-sense: 239–258; CTCGGCTAGCAGTCTCGCGG-3′) designed to target a 160-bp segment, using 94°C for 20 s, 55°C for 30 s and 72°C for 30 s, followed by a 72°C extension period. The products of the reactions were analysed by gel electrophoresis and sequenced in both directions by using an ABI 377 automated DNA sequencer (Applied Biosystems, Foster City, CA, USA) and the Dye Dideoxy TM terminator cycle sequencing kit (Applied Biosystems).
Production of synthetic 5′ UTR HCV RNA
Total RNA was extracted from 10 mg of liver biopsy tissue of each HCV chronic carrier, as described elsewhere [16]. cDNA using Moloney murine leukaemia virus (M-MLV) reverse transcriptase with an anti-sense primer (1679–1700: 5′-ATGCGCTCTGGGCACCCGGAC-3′) was synthesized and amplified by a heminested PCR reaction involving 30 cycles at 94°C for 20 s, 50°C for 1 min and 72°C for 1 min, followed by a final elongation step at 72°C.
Sense (1–18: 5′-GCCAGCCCCTGTTGGGGG-3′) and anti-sense (409–426: 5′-AGTTCCCCGGGTGGCGGTG-3′) primers were used in the first round, whereas sense (1–15: 5′-GCCAGCCCCCTGATG-3′) and anti-sense (353–371: 5′-TTTTCTTTGAGGTTTAGG-3′) were used in the second round, as reported elsewhere [19].
The PCR product was cloned directly into topoisomerase (TOPO) TA dual promoter vector and a recombinant was digested with the restriction enzyme KpnI to orientate the insert. The orientation was confirmed by DNA sequencing. Positive sense RNA was synthesized from 1 µg of HindIII linearized recombinant plasmid by transcription from the T7 RNA polymerase promoter. Similarly, synthetic negative sense RNA was produced from the XhoI SP6 promoter. The synthetic transcripts were purified extensively to remove plasmid template. Concentrations of both positive and negative strands were determined spectrophotometrically at a 260 nm wavelength and their concentrations equalized.
Results
The efficacy of HCV minus strand RNA capture through a polyA+ isolation procedure designed to purify polyadenylated mRNA was assessed with a 371-nt region corresponding to the 5′-UTR HCV genome amplified from a chronically HCV-infected patient; 5′-UTR amplicon was cloned and synthetic positive and negative polarity RNA transcripts were produced. Transcripts were subjected to the polyA+ isolation procedure and different numbers of HCV genomic equivalent (Eq) molecules were added in the RT step with sense and antisense primers.
When using sense primer in the RT reaction, the amount of HCV minus strand RNA was unchanged after the polyA+ purification step. As shown in Fig. 1a, 104 genomic molecules of minus strand could be demonstrated, with or without non-polyadenylated RNA removal. Plus-strand HCV RNA originated by non-specific priming of sense primer was demonstrated, starting from samples containing 1010 genomic molecules of HCV minus strand RNA.
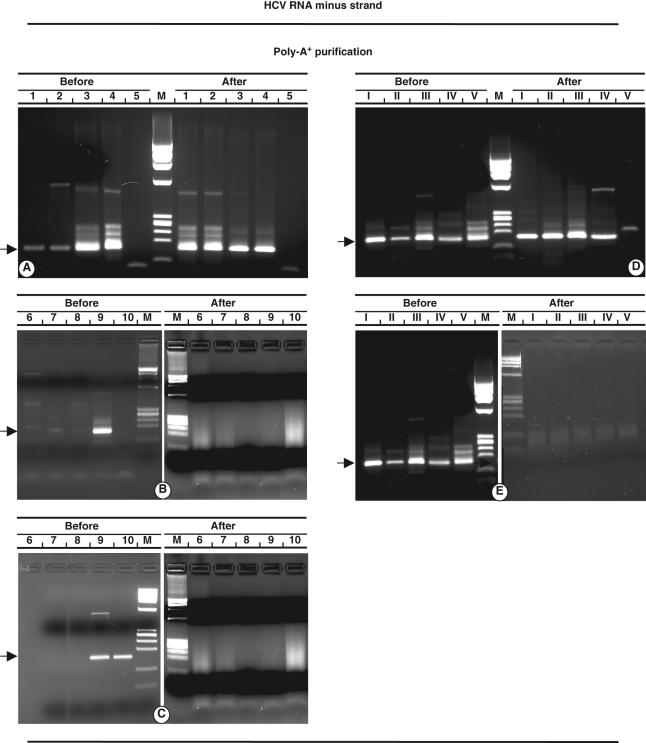
Fig. 1.
Analyses before and after polyA+ purification procedure. Detection of hepatitis C virus (HCV) minus strand RNA (a), which shows serial dilutions of synthetic transcript by using sense primer in the RT reaction; (b) demonstration of HCV plus strand RNA as the result of non-specific priming; (c) anti-sense primer detects HCV minus strand RNA as mispriming effect. (d, e) Specific detection of HCV minus strand RNA from different mixtures of HCV plus and minus strand RNA added to 300 ng cellular RNA is depicted. Sense (d) and anti-sense (e) primers were used to detect minus and plus RNA strands in reverse transcription (RT) reaction. Arabic numbers in each lane represent copies HCV RNA (log) added to RT reaction. Roman numerals represent different ratio of positive/negative strands: I, 108/107; II, 107/106; III, 106/105; IV, 105/104; V, 104/103. M: (174 RF DNA/HaeIII. The arrows indicate 160 base pairs amplified product.
Detection of HCV minus strand RNA in biological samples is strictly dependent upon the concentration of plus strand. It can be assumed that any sample containing more than 106 copies of positive strand has the potential to generate a false minus strand product. Purification of polyA+ containing RNA molecules provided an effective procedure, in that it resulted in the absence of the signal of the plus strand samples, including those containing 1010 genomic molecules (Fig. 1b).
When using anti-sense primers in the RT step, detection of minus strand as the result of non-specific priming was demonstrated from samples containing 109 genomic molecules HCV RNA. The polyA+ purification procedure was effective in abolishing the mispriming signal, in that HCV minus strand RNA was never shown (Fig. 1c). Efficient removal of plus strand RNA was defined by detection of the amplified product in samples containing 1010 positive strand HCV RNA.
The effect of cellular RNA on minus strand-specific detection was investigated after polyA+ purification. Cellular RNA was isolated from an uninfected liver biopsy to which HCV minus strand RNA and different amounts of HCV plus strand RNA were added. After polyA+ purification, samples were processed for RT–PCR. Minus strand detection was demonstrated in all samples (Fig. 1d), whereas plus strand was found in samples before polyA+ purification (Fig. 1e). Thus, in the presence of cellular RNA there was no loss of minus strand and no effect on HCV plus strand RNA removal following the polyA+ purification procedure.
Fifteen patients with CALD without extrahepatic manifestations, 11 with type II MCG, seven with MGUS, nine with B-NHL and four with acute hepatitis C formed the cohort of HCV carriers whose epidemiological, clinical, biological and histological findings are reported in Table 1. The median age of B-NHL patients was higher than that of other groups. With regard to sex distribution, females prevailed among MCG patients.
The total liver necroinflammatory grade ranged between 3 and 12, with a higher median in patients with CALD without extrahepatic manifestations which differed significantly from that found in MCG and B-NHL patients (P < 0·05). Fibrosis ranged from stages 2–5, and no significant difference was found between patient groups. Cirrhosis occurred almost equally. Minimal changes (MCh), including non-specific portal inflammatory cells, intraparenchymal cytolytic foci and hyperplasia of sinusoidal cells, were found in the B-NHL group only.
In chronic HCV carriers, ALT levels were significantly higher in patients without extrahepatic manifestations compared with MCG, MGUS or B-NHL patients (P < 0·05). All patients were viraemic and comparable mean serum HCV RNA levels were demonstrated in chronically infected patients, whereas lower levels were found in acutely infected patients. A slight prevalence of genotype 2 was found in MCG patients, but no distinct profile was found in other groups. Three patients without extrahepatic manifestations, four with MCG and one with MGUS, had received blood transfusions 20–25 years previously. In the remaining patients, the source of HCV infection was not identified. All denied a history of intravenous drug abuse.
BMMC and PBMC were examined for the presence of HCV minus strand RNA before and after the polyA+ purification procedure (Table 2). Using an anti-sense primer in the RT step, all samples were shown to be positive for HCV plus strand HCV RNA. With a sense primer, HCV minus strand RNA was detected in PBMC from two of 15 (13%) patients without extrahepatic manifestations, six of 11 (54%) MCG patients, two of seven (28%) MGUS, two of two B-NHL with MCG, one of seven (14%) B-NHL without MCG and none of patients with acute infection.
Table 2.
Hepatitis C virus (HCV) negative polarity strand RNA in peripheral blood and bone marrow in different groups of HCV-infected patients.
| HCV negative strand RNA | ||||
|---|---|---|---|---|
| Poly A+ separation procedure | ||||
| PBMC | BMMC | |||
| Patients | Before (%) | After | Before (%) | After |
| Chronic infection | ||||
| Without lymphoproliferation | 2/15 (13) | 0 | n.a. | n.a. |
| With lymphoproliferation | ||||
| MCG | 6/11 (54) | 4 (36) | 6 (54) | 3 (27) |
| MGUS | 2/7 (28) | 0 | 1 (14) | 0 |
| B-NHL with MCG | 2/2 (100) | 2 (100) | 2 (100) | 2 (100) |
| B-NHL without MCG | 1/7 (14) | 0 | 0 | 0 |
| Acute infection | 0/4 | 0 | n.a. | n.a. |
n.a.: Not available.
In BMMC, plus and minus strand RNA were examined only in chronically-infected patients with extrahepatic disorders. HCV minus strand RNA was demonstrated in six (54%) MCG patients, one (14%) MGUS patient, and the two patients with B-NHL and MCG. After polyA+ purification, HCV minus strand RNA was detected in PBMC of four (36%) patients with MCG and two with B-NHL and MCG. Only three (27%) MCG patients and the two with B-NHL and MCG were shown to contain HCV minus strand RNA in bone marrow-derived cells.
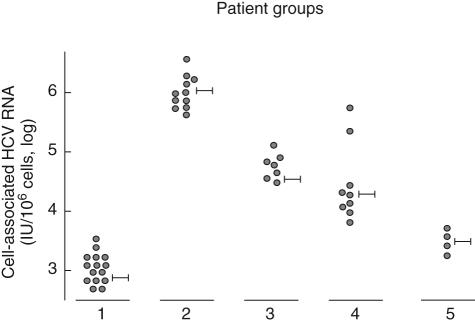
Quantitative analysis of HCV RNA was also performed in cell preparations used for the detection of HCV minus strand RNA. Results depicted in Fig. 2 show that in patients without extrahepatic manifestations mean levels of cell-associated HCV RNA were significantly lower than those found in MCG patients (1847 ± 890 versus 992 650 ± 345 669 HCV RNA IU/106 cells) (P < 0·01). Intermediate levels were found in MGUS (34 912 ± 40 448 HCV RNA IU/106 cells) and in B-NHL without MCG (21 460 ± 25 400 HCV RNA IU/106 cells). Lower levels of cell-associated HCV RNA were demonstrated in acute patients (2991 ± 3230 HCV RNA IU/106 cells).
Fig. 2.
Levels of cell-associated hepatitis C virus (HCV) RNA in different patient groups. Group 1: patients without extrahepatic disorders; group 2: with mixed cryoglobulinaemia (MCG); group 3: with monoclonal gammopathy of undetermined significance (MGUS); group 4: with B cell non-Hodgkin's lymphoma (B-NHL); group 5: with acute hepatitis.
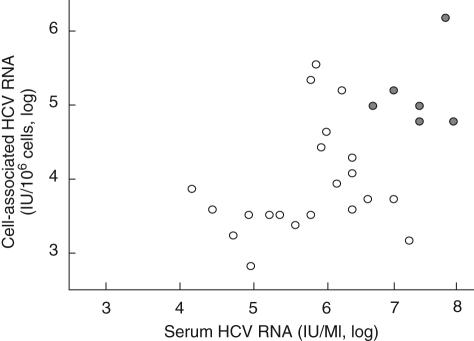
With respect to the occurrence of HCV minus strand RNA, a direct correlation was demonstrated with serum and cell-associated viral load. Compared with those with HCV minus strand RNA negative, positive patients had significantly higher levels of serum and cell-associated HCV RNA (994 943 ± 1478 551 versus 3637 601 ± 2316 523 IU/ml, P < 0·01; 37 900 ± 54 888 versus 881 267 ± 1634 321 IU/106 cells, P < 0·05, respectively) (Fig. 3).
Fig. 3.
Levels of hepatitis C virus (HCV) RNA in peripheral blood mononuclear cells with ( ) and without (○) HCV minus strand RNA are function of serum HCV RNA levels in chronically HCV-infected patients with extrahepatic disorders.
) and without (○) HCV minus strand RNA are function of serum HCV RNA levels in chronically HCV-infected patients with extrahepatic disorders.
From a clinical point of view, patients with productive HCV infection in BMMC and PBMC showed distinct clinical features, namely more frequent flare-up of cutaneous vasculitis, involvement of renal function and peripheral neuropathy compared with negatives (4 ± 2 versus 0·8 ± 0·6 episodes/month; four of six versus one of 14 patients; three of six versus two of 14 patients, respectively).
Discussion
The current study was undertaken to determine whether PBMC and/or BMMC support HCV productive infection. By applying a highly specific and sensitive method for HCV minus strand RNA detection, we provide evidence of virus replication in the PBMC of 13% and in the BMMC of 11% of HCV-infected patients. Our data also indicate that carriage of the HCV replicative form was related to the clinical picture, in that HCV replication was shown in cells of patients with MCG, but not in the other study groups. No evidence of HCV minus strand RNA was provided in both acutely or chronically HCV-infected patients without MCG. These results strongly support the concept of a direct relation between productive infection of lymphoid cells and cryoglobulinaemia. Because lymphocytes from MCG patients have high amounts of HCV particles on their surface, it can be argued that HCV enrichment may favour fusion events and virus entry [20].
The recent description of the complete virus life cycle in cell culture suggests that HCV has a mechanism of entry similar to that of Pestiviruses [21]. Virus entry requires a fusion event between the viral and cellular membranes. HCV binding to cell receptor(s) induces conformational changes in the envelope glycoproteins, allowing membrane fusion [22]. The surface of HCV particles consists of tightly packed glycoprotein oligomers, resembling the features of classical Flaviviruses which consist of 90 envelope protein dimmers [23]. In the Dengue virus, for example, the E dimers are closely packed, making viral membrane inaccessible and fusion impossible. When fusion is initiated by the low pH, conformational changes are induced and the surface area of the viral membrane is exposed [24]. Similarly, HCV entry appears to be pH-dependent, requiring an acidified intracellular compartment [25]. Low pH activates the entry of cell-surface bound HCV particles in a time and temperature-dependent manner [26].
We can speculate that HCV enrichment on lymphoid cells from MCG patients may lead to dramatic activation of low pH-induced conformational changes to E1–E2 heterodimers, resulting in a remarkable extension of the exposing area for the fusion step. HCV may be primed for cell entry by receptor-binding molecules such as CD81 [27], scavenger receptor class B type I [28], low density lipoprotein [29] or as-yet-unidentified molecules. Factors responsible for HCV enrichment in lymphoid cells from MCG patients are unknown, but may be related to the higher density [30,31] and/or genetic polymorphism of receptor genes [19].
The evidence of HCV minus strand RNA in a distinct cohort of patients enables the division of HCV-infected patients into two subgroups: the first corresponding to a compartment model, which assumes that viral replication occurs only in the liver; the other compatible with a second extrahepatic replication compartment, in addition to the liver [32]. Our results suggest that cryoglobulins imply lymphoid compartmentalization of HCV productive infection.
Although it has been estimated that infected lymphocytes account for about 3% of the viral load in the circulation [33–35], they have a major pathogenetic role acting as an effective circulating reservoir of HCV infection. In this study, replicating HCV was shown in PBMC and/or BMMC of MCG-associated B-NHL patients, whereas no traces of HCV minus strand RNA were detected in cells of HCV-positive B-NHL patients without MCG.
In light of our own findings, which have shown the lack of productive HCV infection in B-NHL cells [36,37], the present results raise intriguing questions as to the role of HCV in lymphomagenesis. The availability of PBMC and BMMC permitted the examination of the minus strand of HCV RNA, providing insights into the replication ability of HCV genome carried by MCG patients. These data lead to the conclusion that HCV infects lymphoid cells, which become capable of producing cryoglobulins; the up-regulation of HCV genome expression in lymphoid cells probably requires host cell factors able to concentrate viral particles on their surface.
With different technical approaches recent studies have emphasized the role of productive infection of HCV in lymphoid cells and lymph nodes [38]. None of them, however, defined the relationship between HCV and cryoglobulins.
The occurrence of a subset of HCV-infected patients carrying phenotypically distinct lymphoid cells implicates the existence of high affinity receptor molecules capable of mediating HCV trafficking, fusion and cell entry. Their biological and functional characterization and their relation to cryoglobulin production will require further studies.
Acknowledgments
We are indebted to Mr Vittorio Padolecchia for his invaluable technical assistance. This study was supported in part by grants from: Associazione Italiana per la Ricerca sul Cancro (AIRC, Milan, Italy) − Regional Grants 2005; University of Bari, Italy; Fondazione Banco di Napoli, Naples, Italy and Fondazione Cassa di Risparmio di Puglia, Bari, Italy.
References
- 1.Choo Q-L, Richman KH, Han JH, et al. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–5. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindenbach BD, Evans MJ, Syder AJ, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–6. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 3.Reed KE, Rice CM. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr Top Microbiol Immunol. 2000;242:55–84. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]
- 4.Lohmann VS, Hoffmann U, Herian F, et al. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J Virol. 2003;77:3007–19. doi: 10.1128/JVI.77.5.3007-3019.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindenbach BD, Rice CM. Flaviviridae: the viruses and their replication. In: Knipe DM, Howlwy PM, editors. Field's virology. 4. Vol. 1. Philadelphia, PA: Lippincott-Raven; 2001. pp. 991–1041. [Google Scholar]
- 6.Kuhn RJ, Zhang W, Rossmann MG, et al. Structure of Dengue virus: implications for Flavivirus organization, maturation and fusion. Cell. 2002;108:717–25. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gowans EJ. The distribution of markers of HCV infection throughout the body. Semin Liver Dis. 2000;20:11–28. doi: 10.1055/s-2000-9503. [DOI] [PubMed] [Google Scholar]
- 8.Lanford RE, Sureau C, Jacob JR, et al. Demonstration of in vitro infection of chimpanzee hepatocytes with HCV using strand-specific RT/PCR. Virology. 1994;202:606–14. doi: 10.1006/viro.1994.1381. [DOI] [PubMed] [Google Scholar]
- 9.Lanford RE, Chavez D, Chisari FV, et al. Lack of detection of negative-strand hepatitis C virus RNA in peripheral blood mononuclear cells and other extrahepatic tissues by the highly strand-specific rTth reverse transcriptase PCR. J Virol. 1995;69:8079–83. doi: 10.1128/jvi.69.12.8079-8083.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunji T, Kato N, Hijikata M, et al. Specific detection of positive and negative stranded hepatitis C viral RNA using chemical RNA modification. Arch Virol. 1994;134:293–302. doi: 10.1007/BF01310568. [DOI] [PubMed] [Google Scholar]
- 11.Lerat H, Berby F, Trabaud MA, et al. Specific detection of hepatitis C virus minus strand in hematopoietic cells. J Clin Invest. 1996;97:845–51. doi: 10.1172/JCI118485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanford RE, Chavez D. Strand-specific rTth RT-PCR for analysis of HCV replication. In: Lau JYN, editor. Hepatitis C protocols. London: Humana Press; 1998. pp. 471–81. [Google Scholar]
- 13.Takyar ST, Li DS, Wang Y-H, et al. Specific detection of minus-strand hepatitis C virus RNA by reverse-transcription polymerase chain reaction on PolyA+-purified RNA. Hepatology. 2000;32:382–7. doi: 10.1053/jhep.2000.9094. [DOI] [PubMed] [Google Scholar]
- 14.Dammacco F, Sansonno D, Piccoli C, et al. The cryoglobulins: an overview. Eur J Clin Invest. 2001;31:628–38. doi: 10.1046/j.1365-2362.2001.00824.x. [DOI] [PubMed] [Google Scholar]
- 15.Ishak K, Baptista A, Bianchi L, et al. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22:696–9. doi: 10.1016/0168-8278(95)80226-6. [DOI] [PubMed] [Google Scholar]
- 16.Sansonno D, Tucci FA, De Re V, et al. HCV-associated B cell clonalities in the liver do not carry the t(14;18) chromosomal translocation. Hepatology. 2005;42:1019–27. doi: 10.1002/hep.20887. [DOI] [PubMed] [Google Scholar]
- 17.Sansonno D, Lauletta G, De Re V, et al. Intrahepatic B cell clonal expansions and extrahepatic manifestations of chronic HCV infection. Eur J Immunol. 2004;34:126–36. doi: 10.1002/eji.200324328. [DOI] [PubMed] [Google Scholar]
- 18.Oligotex Handbook. Technical manual. 2. Valencia, CA: Qiagen; 2002. [Google Scholar]
- 19.Laporte J, Bain C, Maurel P, et al. Differential distribution and internal translation efficiency of hepatitis C virus quasispecies present in dendritic and liver cells. Blood. 2003;101:52–7. doi: 10.1182/blood-2002-03-0818. [DOI] [PubMed] [Google Scholar]
- 20.Sansonno D, Lauletta G, Montrone M, et al. Virological analysis and phenotypic characterization of peripheral blood lymphocytes of hepatitis C virus-infected patients with and without mixed cryoglobulinaemia. Clin Exp Immunol. 2006;143:288–96. doi: 10.1111/j.1365-2249.2005.02987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeisel MB, Baumert TF. Production of infectious hepatitis C virus in tissue culture: a breakthrough for basic and applied research. J Hepatol. 2006;44:436–9. doi: 10.1016/j.jhep.2005.11.031. [DOI] [PubMed] [Google Scholar]
- 22.Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1–E2 envelope protein complexes. J Exp Med. 2003;197:633–42. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinz FX, Allison SL. Structures and mechanisms in flavivirus fusion. Adv Virus Res. 2000;55:231–69. doi: 10.1016/S0065-3527(00)55005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhn RJ, Zhang W, Rossmann MG, et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–25. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu M, Zhang J, Flint M, et al. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc Natl Acad Sci USA. 2003;100:7271–6. doi: 10.1073/pnas.0832180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tscherne DM, Jones CT, Evans MJ, et al. Time and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J Virol. 2006;80:1734–41. doi: 10.1128/JVI.80.4.1734-1741.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peracca R, Falugi F, Galli G, et al. Structure–function analysis of hepatitis C virus envelope–CD81 binding. J Virol. 2000;74:4824–30. doi: 10.1128/jvi.74.10.4824-4830.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scarselli E, Ansuini H, Cerino R, et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017–25. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agnello V, Abel G, Elfahal M, et al. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci USA. 1999;96:12766–71. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuckerman E, Kessel A, Slobodin G, et al. Antiviral treatment down-regulates peripheral B-cell CD81 expression and CD5 expansion in chronic hepatitis C virus infection. J Virol. 2003;77:10432–6. doi: 10.1128/JVI.77.19.10432-10436.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kronenberger B, Sarrazin C, Hofmann WP, et al. Mutations in the putative HCV-E2 CD81 binding regions and correlation with cell surface CD81 expression. J Viral Hepatol. 2004;11:310–8. doi: 10.1111/j.1365-2893.2004.00508.x. [DOI] [PubMed] [Google Scholar]
- 32.Dahari H, Feliu A, Garcia-Retortillo M, et al. Second hepatitis C replication compartment by viral dynamics during liver transplantation. J Hepatol. 2005;42:491–8. doi: 10.1016/j.jhep.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 33.Lerat H, Berby F, Trabaud MA, et al. Specific detection of hepatitis C virus minus strand RNA in hematopoietic cells. J Clin Invest. 1996;97:845–51. doi: 10.1172/JCI118485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sansonno D, Iacobelli AR, Cornacchiulo V, et al. Detection of hepatitis C virus (HCV) proteins by immunofluorescence and HCV RNA genomic sequences by non-isotopic in situ hybridization in bone marrow and peripheral blood mononuclear cells of chronically HCV-infected patients. Clin Exp Immunol. 1996;103:414–21. doi: 10.1111/j.1365-2249.1996.tb08296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sansonno D, Lotesoriere C, Cornacchiulo V, et al. Hepatitis C virus infection involves CD34(+) hematopoietic progenitor cells in hepatitis C virus chronic carriers. Blood. 1998;92:3328–37. [PubMed] [Google Scholar]
- 36.De Vita S, Sansonno D, Dolcetti R, et al. Hepatitis C virus within a malignant lymphoma lesion in the course of type II mixed cryoglobulinemia. Blood. 1995;86:1887–92. [PubMed] [Google Scholar]
- 37.Sansonno D, De Vita S, Cornacchiulo V, et al. Detection and distribution of hepatitis C virus-related proteins in lymph nodes of patients with type II mixed cryoglobulinemia and neoplastic or non-neoplastic lymphoproliferation. Blood. 1996;88:4638–45. [PubMed] [Google Scholar]
- 38.Pal S, Sullivan DG, Kim S, et al. Productive replication of hepatitis C virus in perihepatic lymph nodes in vivo: implications of HCV lymphotropism. Gastroenterology. 2006;130:1107–16. doi: 10.1053/j.gastro.2005.12.039. [DOI] [PubMed] [Google Scholar]