Abstract
Because primary immunodeficiencies (PID) are rare diseases, transnational studies are essential to maximize the scientific outcome and lead to improved diagnosis and therapy. Immunologists in Europe have united to determine the prevalence of PID in Europe and to establish and evaluate harmonized guidelines for the diagnosis and treatment of PID as well as to improve the awareness of PID in Europe. In order to achieve this aim we have developed an internet-based database for clinical and research data on patients with PID. This database forms the platform for studies of demographics, the development of new diagnostic and therapeutic strategies and the identification of novel disease-associated genes. The database is completely secure, while providing access to researchers via a standard browser using password and encrypted log-in sessions and conforms to all European and national ethics and data protection guidelines. So far 2386 patients have been documented by 35 documenting centres in 20 countries. Common variable immunodeficiency (CVID) is the most common entity, accounting for almost 30% of all entries. First statistical analyses on the quality of life of patients show the advantages of immunoglobulin replacement therapy, at the same time revealing a mean diagnostic delay of over 4 years. First studies on specific questions on selected PID are now under way. The platform of this database can be used for any type of medical condition.
Keywords: clinical studies, ESID, online database, primary immunodeficiency, registry
Introduction
Primary immunodeficiencies (PID) are rare diseases caused by inherent defects of the immune system. Advances in medical research have led to the identification of 206 different subtypes of PID, the prevalence of which varies considerably depending on the type of disease [1].
The prevalence of PIDs in the general population has not been identified clearly. Estimates on the minimum prevalence of PIDs are based on the information compiled from a number of national registries on cases of PIDs, established in countries such as Norway [2], the United States [3], Australia [4], Spain [5], Switzerland [6] and Sweden [7]. The birth prevalence is estimated at one per 10 000 individuals for significant PIDs, with a diagnostic delay of over 5 years due to the rarity and complexity of symptoms associated with the diseases [8–10].
PID are, by definition, genetic defects. Of the 206 PIDs, about 110 gene defects have been identified to date. A given genotype, however, can result in a variable clinical phenotype depending on many heterogeneous factors, such as environmental effects. Thus, the relationship between genetic and clinical phenotype is not linear; instead it is a combination of molecular defects regulated by endogenous and exogenous factors, which leads to a complex set of phenotypic features [11–13]. In order to establish genotype–phenotype correlations, this overall complexity can be analysed by combining the molecular information with the clinical phenotype information on PIDs. It is therefore necessary to collect large amounts of data regularly over a long period of time.
However, most medical centres have only a few patients with a specific subtype of PID, which leads to difficulties in diagnosis and therapy management. Only through international collaboration can sufficient patient numbers be made available for adequate research. Thus European immunologists formed a network of clinicians and researchers, the European Society for Immunodeficiencies (ESID; http://www.esid.org) to support research on the cause, mechanism and therapy in immunodeficiencies. This in turn led to the development of a registry for PID in 1994 in Sweden, which accumulated data on 9707 patients from 26 European countries [14]. This was a first step in creating a basis for European-wide research on PID, in particular for clinical studies. However, the database had several shortcomings: it provided a single-point registration only and was of no benefit to the attending physician. The database was maintained centrally in Stockholm, and all participating centres had to send in their data by post or fax to the co-ordinator, who then transferred the data into the registry itself. In particular, it was not possible to carry out follow-up documentation on patients which is vital for research. Data analysis could be requested, but there was no direct access to the database by the documenting centres.
Given these shortcomings, ESID has now developed a new state-of-the-art database for primary immunodeficiencies which is presented here. The ESID database is a secure, internet-based patient registry, which combines both clinical and laboratory data of PID patients. It is stored on secure servers within the firewall of the University of Freiburg, Germany. The database allows the documentation of patients with a PID into an online system via a standard web browser, while at the same time conforming to standard ethics and data protection requirements. It provides a common data set for 206 different PIDs. This core data set includes data on diagnosis, therapy, laboratory data and secondary outcome measures. First analyses of the data are presented here and demonstrate the potential available in health services research. It will lead to improved diagnosis, classification, prognosis and therapy by enabling the exchange of information among centres, assuring detailed long-term documentation and facilitating large genetic and therapeutic projects.
Methods
System structure
The central servers of the ESID Online Database are hosted within the secure server network of the IT Centre of the University Hospital Freiburg, Germany and run within the validated firewall of the hospital. The complete system is being developed and maintained continuously by the Centre for web-based Research and Patient Databases (CwebRD), which is a core facility at the same institution. The ESID database is accessible via standard browsers from the ESID web site (http://www.esid.org). The complete online database system is a complex platform based on the Java 2 Platform Enterprise Edition (J2EE), which can manage easily large amounts of data. The individual components of the system enable data storage, data entry, reporting and the integration of pre-existing data sources within an enterprise business-to-business integration (B2B).
Administration and participating centres
The database is administered by the head of the ESID Registry and his team at the University Hospital Freiburg, Germany. Centres specializing in immunology wishing to participate in the project must sign an agreement with ESID and send in applications for a password in order to receive the necessary database access information. The ESID database complies with the European legislation on good clinical practice (Clinical Trial Directive 2001/20/EC). At present, there is no standard procedure for obtaining data protection and ethical approval at the European level. This means that every centre has to obtain these approvals according to its local laws and regulations, which have to follow the EC directive 95/46/EC on data protection. In addition, patients must give their informed consent before their data are entered into the database.
For accessing the database, users receive a personal user name and password. Every centre has access only to their own patients' data. For cross-centre studies, the respective centres must agree explicitly to share their data with other centres. Patient-identifying information is always removed prior to sharing.
Currently 75 documenting centres in 28 countries have obtained passwords for the online documentation. Two existing registries have been integrated into the database by electronic import: the Italian National PID Registry Associazione Italiana Ematologia Oncologia Pediatrica: (AIEOP); co-ordinator Dr A. Plebani [15]); and the pan-European common variable immunodeficiency (CVID) Registry compiled by Professor Lennart Hammarström at the Karolinska Institute in Huddinge, Sweden [14].
Database structure
The ESID database for PID consists of 206 disease-specific registries, which are grouped within eight main categories and 68 subcategories. The categorization is based on the latest classification defined by the IUIS (International Union of Immunological Societies) in 2005 [1]. All disease-specific registries share a common core data set. This core data set was designed to bring together basic data for all PID patients in order to make general evaluations possible. It contains information on diagnosis, therapy and quality of life as well as the most important laboratory data for PID (blood and lymphocyte count, immunoglobulin (Ig) levels and data on important cell surface markers).
In addition to the core data set, 28 extended disease-specific data sets have been implemented. These include, among others, X-linked agammaglobulinaemia, CVID, FOXP3 deficiency, Nijmegen breakage syndrome and DiGeorge syndrome. The additional information provided by these extended data sets comprises data on aetiology, genetics, infection history, additional diagnoses, collected samples, biopsies and surgery events, as well as clinical and additional investigation analyses.
Additional extended data sets are being developed continuously in order to provide platforms for a detailed documentation of a given PID. The extended data sets contain the relevant information for a particular disease, and are defined by steering committees comprised of specialists in the respective disease. These committees are nominated by the ESID board.
The diagnostic criteria defined by ESID for the different PIDs have been included in the extended disease-specific data sets as a query box, with the aim of standardizing and clarifying diagnosis. Another feature available in the ESID database is the Health Survey Form 36 (SF-36, http://www.qualitymetric.com), which is used for disease-specific studies on the quality of life of these PID patients. For evaluation purposes, it is important to receive standardized data for sections such as infection history, additional diagnoses and surgery. Therefore, the ESID database offers an ICD-10 search tool which ensures that data is documented according to the World Health Organization (WHO) 10th International Classification of Diseases (ICD-10) [16], using the codes and text as provided by the WHO.
Genetic mutation documentation
The database offers a Mutation Browser, which is a multi-component system for the deposition of genetic mutation data. This is being developed in co-operation with the Institute of Medical Technology (IMT) in Tampere, Finland. The IMT maintains the ‘IDbases’, an electronic library for genetic mutations in PID patients. Gene mutations entered in the ESID database are stored and validated at the IDbases after which the data is transferred automatically back to the ESID database. The exchanged data is coded and contains no personal patient information. Thus, the data received in Finland are anonymous. This tool works with both genomic DNA and cDNA and includes information related to the number of affected alleles, where applicable. It is currently being extended to 105 of the diseases in the database.
Documentation
The database is designed to be used for long-term documentation. It offers the possibility to add any number of visit dates for a given patient. Users are asked to update their patients' data at least once a year. First-time data entry takes 10–15 min per patient on average for the red fields of the core data set, depending on the availability of the patient data, longer if additional fields are documented. Regular updates take considerably less time, usually under 5 min per patient.
One of the objectives when developing the system was to make it user-friendly. Other than in previous databases, where patient data were recorded on case report forms and entered into a database centrally, the ESID online database uses online technology which makes it possible for every registered physician to enter data directly into the database. The only requirements are a standard internet browser and internet connection, in combination with valid access codes. In addition, it is possible for the user to enter personal data of his patient on a secure server. Thereby, the database can be used easily while seeing a patient. Centres without online access may use hard copy entry forms for documentation.
Data quality
The quality of the data is dependent on the users who enter the data. ESID signs contracts only with known primary immunodeficiency centres. Password application users have to be validated by their head of department who countersigns their application. These users are usually physicians and their documenting assistants, who are trained in the documentation of medical data. ESID database administrators offer specific training on the ESID database to help users. Data monitoring can be carried out both by the database administration and by a person appointed in each centre. Data quality is ensured only if this monitoring takes place regularly. The database has inbuilt quality assurance features; for instance, mandatory fields and year of birth cannot be some date in the future. Further, if a patient has been diagnosed incorrectly and thus entered into the wrong subregistry, the patient data can be transferred to the correct subregistry. Similarly, if the diagnosis of a PID is redefined by identification of a genetic mutation, patient data can be transferred to the appropriate subregistry. Users are aware that this database is to be used for long-term documentation and that data have to be updated regularly. Regular updating of data is encouraged by a small financial compensation of 10 EUR per patient.
Analysis of data and studies
The ESID database offers a platform for different types of studies on primary immunodeficiencies. European researchers have the opportunity to learn from other patients with a specific diagnosis throughout Europe and can contact appropriate documenting centres. The database has been constructed so that users can access data automatically only on patients from their centre. This is both for security and data protection reasons. If a user would like to access data from another centre he has to apply to that centre for permission. This centre, in turn, has to contact the database administrators to request that the inquiring user be allowed access to their data. The data from the database can then be analysed as requested, but only by the database administrators. Specific queries can be sent to them for analysis on patients for whom the user has permission to access. A few specific predefined queries are available in the database (main registry with predefined queries) and are directly accessible to users. We are still in the process of developing the technology further so that a user can directly interrogate the database him- or herself. This will be available in 2008.
Contact details can be obtained from the ESID Registry working party, if the other centre agrees. Furthermore, co-operations between centres and multi-centre studies can be initiated on the basis of information retrieved from the database. First studies are currently running on CVID, X-linked agammaglobulinaemia, DiGeorge syndrome and Nijmegen breakage syndrome. Studies on CD3 antigen gamma subunit deficiency (CD3G), winged-helix nude deficiency (FOXN1) and Wiskott–Aldrich syndrome (WAS) have already been submitted and will be launched in 2007.
The database fields which are considered relevant to such a study are defined in the respective steering committees and highlighted on the user interface. This is achieved by using a colour-coding scheme: the fields are tinted in a pre-assigned colour and are thus clearly recognizable for the user.
Results
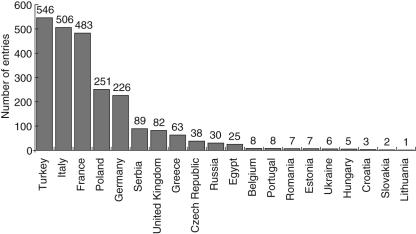
The database was launched in June 2004 and now 2386 patients have been documented by 35 documenting centres in 20 countries. The distribution of the patients by country is shown in Fig. 1. Another 29 centres have signed agreements with ESID but they have not yet been able to begin documentation, mainly because of the necessity to obtain ethics and data protection approval as well as signed consent statements from their patients. The number of entries in the database is growing constantly. Regularly updated numbers are available on the ESID website (http://www.esid.org/statistics.php).
Fig. 1.
Distribution of patients in the database by country. The numbers above the columns are absolute patient numbers.
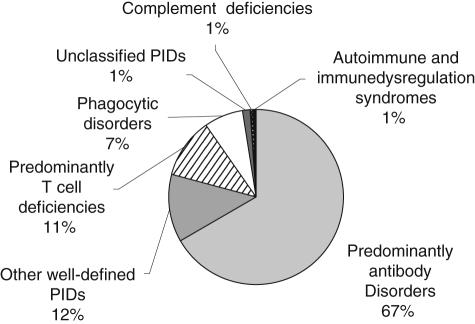
Entries have been made into 78 of the 206 disease-specific registries, some of which represent very rare cases throughout Europe. The distribution of entries among the main PID categories is displayed in Fig. 2. The breakdown of distribution by disease is located in Table 1.
Fig. 2.
Distribution of entries in the European Society for Immunodeficiencies (ESID) online database by main primary immunodeficiencies (PID) category.
Table 1.
Main diseases (by genotype) in the European Society for Immunodeficiencies (ESID) online database.
| Disease | Number | Percentage |
|---|---|---|
| Common variable immunodeficiency (CVID) | 676 | 31·6 |
| Agammaglobulinaemia, X-linked (BTK) | 235 | 11·0 |
| Immunoglobulin A deficiency | 197 | 9·2 |
| Isolated IgG subclass deficiency | 155 | 7·2 |
| Transient hypogammaglobulinaemia of infancy | 144 | 6·7 |
| Ataxia telangiectasia (ATM) | 106 | 5·0 |
| DiGeorge syndrome | 88 | 4·1 |
| Chronic granulomatous disease, X-linked (CYBB) | 72 | 3·4 |
| Wiskott–Aldrich syndrome with mutations in WASP | 66 | 3·1 |
| CSR defects and HIGM syndromes with unknown genetic cause | 52 | 2·4 |
| Hyper IgE syndrome | 39 | 1·8 |
| Other hypogammaglobulinaemias | 38 | 1·8 |
| Nijmegen breakage syndrome (NBS1) | 36 | 1·7 |
| Wiskott–Aldrich syndrome with unknown genetic cause | 34 | 1·6 |
| CGD with unknown genetic cause | 34 | 1·6 |
| Unclassified immunodeficiencies | 34 | 1·6 |
| T-B + SCID with unknown genetic cause | 30 | 1·4 |
| CD40 antigen ligand deficiency (CD154) | 28 | 1·3 |
| Other unclassified T cell disorders | 27 | 1·3 |
| Severe combined immunodeficiency, X-linked (SCIDX1) | 26 | 1·2 |
| Agammaglobulinaemias with unknown genetic cause | 23 | 1·1 |
| Total number of patients | 2140 | 100·0 |
Of all entries, 66·8% fall within the major PID category of predominantly antibody disorders, which includes CVID. CVID forms the largest cohort in the database with 676 entries, which accounts for 28·3% of all documented cases. Of these, 81 entries were imported electronically from the preceding CVID database run by Professor Hammarström. There are still 305 entries in that database which have not been imported because informed consent has not yet been collected from these patients.
Sex and age distribution
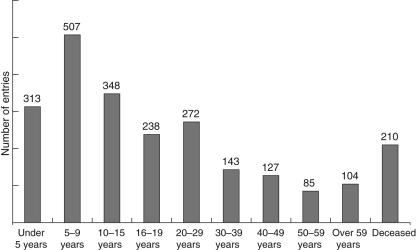
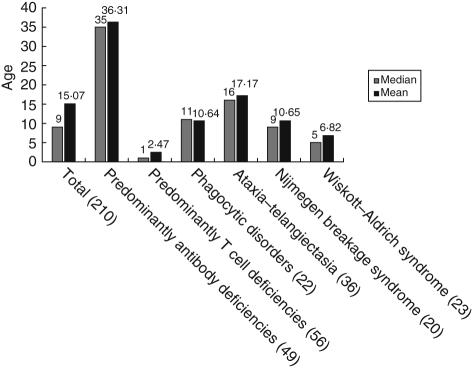
The 2386 patients listed in the ESID database include 1539 male and 847 female patients; 53·6% of the live patients are under 16 years of age. The complete age distribution is given in Fig. 3. Among the registered patients, 210 are deceased. The average age of death of these patients varies considerably, depending on the type of PID. An overview is given in Fig. 4.
Fig. 3.
Age distribution of patients registered in the European Society for Immunodeficiencies (ESID) online database.
Fig. 4.
Average age at death of patients from selected subcategories and diseases from the European Society for Immunodeficiencies (ESID) online database. The absolute number of patients is given in brackets.
Treatment
Therapy is recorded for each patient and includes different kinds of treatment, documenting starting- and end-dates for each treatment. Immunoglobulin replacement, being one of the main treatments in PID, has been documented for 1122 (47·0%) of the patients. Of those patients, 957 (85·3%) undergo intravenous Ig-replacement therapy, 146 (13·0%) are being treated with subcutaneous Ig products, while there are still four patients (0·36%) who receive intramuscular Ig-replacement. The route of Ig replacement was not detailed in the remaining 15 patients. A total of 141 (5·9%) patients have undergone bone marrow transplantation.
Quality of life
A first evaluation of patients' quality of life in relation to immunoglobulin replacement showed that for CVID patients not undergoing Ig replacement, the average number of days missed at work or school during a 1-year period was 23·3 and 19·8 days had to be spent in hospital. In contrast, CVID patients receiving Ig replacement missed an average of only 6·1 days at school or work and had a lower hospital stay of 12·5 days. Days in hospital do not include visits to the out-patient clinic and day clinic. A total of 343 data sets were evaluated for ‘days in hospital’ evaluation, with 313 patients on Ig replacement, 30 without; 171 data sets were evaluated for ‘days missed’, with 133 patients on Ig replacement and 38 without.
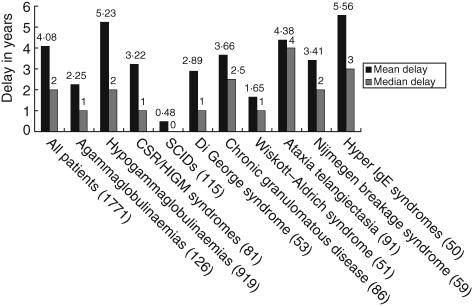
Diagnostic delay
The mean diagnostic delay between the onset of symptoms and the date of diagnosis calculated from the database is currently 4·08 years (median delay: 2 years). In the case of antibody deficiencies, the mean diagnostic delay is 4·75 years (median: 2 years). Further numbers are given in Fig. 5. These figures compare well with previous studies on diagnostic delay. For example, Seymour et al. found a mean delay of 4·4 years (median: 2 years) in a cohort of 87 antibody-deficient patients studied between 1989 and 2002 in a British hospital [8,17].
Fig. 5.
Median and mean diagnostic delay for patients with data entered in the relevant fields from selected subcategories and diseases from the European Society for Immunodeficiencies (ESID) online database. Numbers of patients are given in brackets. Definition of fields in the database: ‘Date of diagnosis’: the date on which all ESID criteria for the respective diagnosis have been fulfilled and the diagnosis was verified by a physician ‘Onset of symptoms’: the date of first severe infection or characteristic manifestation of the primary immunodeficiency.
Discussion
Comparison of the ESID online database with the old ESID Registry reveals that the average number of patients registered per year is about 1000 (old registry: 908 patients per year over a 10-year period; ESID online database: 954 patients per year over a 2-year period). This indicates that the online technology used for the new database has not led to an acceleration in the documentation of cases. This may be explained by the fact that for many centres, irrespective of the method used, documentation means additional work on top of patient care, so that physicians must often document in their free time.
Another reason for the moderate speed of documentation may be data protection and ethical requirements. We expect patient numbers to increase faster once more documenting centres have passed the obstacle of obtaining the necessary approvals. This is a huge but necessary bureaucratic step, which costs the documenting centres a great deal of time and effort. Therefore, we call for a standardized procedure within the European Union whereby databases operating in the EU should have to obtain only one positive data protection and ethics statement, so that it should be sufficient for all participating centres to inform their local data protection authorities of their participation in a standardized format.
On the other hand, the switch from a paper-based one-time registration to internet-based follow-up documentation has already been well accepted. In the second year of the database, participating centres documented follow-up data for 539 of the 919 patients registered during the first year, which equals 59%. Herein we see the potential of the database in the long term.
First analyses on data in the database indicate that not all fields are completed as requested. For example, analysis on Ig substitution therapy of PID patients revealed that for 15 of 1122 patients (1·3%) the route of substitution had not been documented.
This shows that intensive monitoring is essential to ensure an adequate quality of data in the database. Data monitoring can take place at both the level of each documenting centre and by the administrators of the database, whereby it is the responsibility of each centre to ensure that the data it enters are complete. Documenting centres are informed of incomplete data sets discovered during monitoring by the database administrators.
However, features such as the ICD-10 tool and the ESID diagnostic criteria as well as constant quality checks by the database administration will ensure a high level of data quality.
There is still a bias in the documentation due to the restricted numbers of institutions documenting patients; for example, complement deficiencies are under-represented in the current online database and account for only 0·6% of the cases, whereas in the former ESID registry with 9707 PID patients they account for about 6% of PID. This bias will be reduced with the inclusion of additional centres and the increased documentation of patients.
With the growing cohorts in the ESID database it will be interesting to observe the further development of data, such as diagnostic delay. Hereby, it will be possible to study the success of ESID's effort to improve diagnosis at the European level [18,19]. For the evaluation of treatment studies using the quality of life (QoL) data will be of importance. In addition to the QoL questions which have been evaluated recently, the SF-36 questionnaire has now been integrated into the database.
Features which are planned to be implemented in the near future include percentile curves as well as tools for pedigree drawing and reporting. Finally, we would like to point out that this type of database could be used for any type of medical condition.
Acknowledgments
We are grateful to all individuals involved in the work in the documenting centers. The complete list of documenting centres can be viewed at (http://www.esid.org/centres.php). This work was supported by EU grant no SP23-CT-2005–006411 and by PPTA Europe (http://www.plasmatherapeutics.org) sponsorship of ESID.
References
- 1.Notarangelo L, Casanova JL, Conley ME, et al. Primary immunodeficiency diseases: an update from the International Union of Immunological Societies Primary Immunodeficiency Diseases Classification Committee Meeting in Budapest, 2005. J Allergy Clin Immunol. 2006;117:883–96. doi: 10.1016/j.jaci.2005.12.1347. [DOI] [PubMed] [Google Scholar]
- 2.Stray-Pedersen A, Abrahamsen TG, Froland SS. Primary immunodeficiency diseases in Norway. J Clin Immunol. 2000;20:477–85. doi: 10.1023/a:1026416017763. [DOI] [PubMed] [Google Scholar]
- 3.Winkelstein JA, Marino MC, Johnston RB, Jr, et al. Chronic granulomatous disease. Report on a national registry of 368 patients. Medicine (Balt) 2000;79:155–69. doi: 10.1097/00005792-200005000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Baumgart KW, Britton WJ, Kemp A, French M, Roberton D. The spectrum of primary immunodeficiency disorders in Australia. J Allergy Clin Immunol. 1997;100:415–23. doi: 10.1016/s0091-6749(97)70257-4. [DOI] [PubMed] [Google Scholar]
- 5.Matamoros Flori N, Mila Llambi J, Espanol Boren T, Raga Borja S, Fontan Casariego G. Primary immunodeficiency syndrome in Spain: first report of the National Registry in Children and Adults. J Clin Immunol. 1997;17:333–9. doi: 10.1023/a:1027382916924. [DOI] [PubMed] [Google Scholar]
- 6.Ryser O, Morell A, Hitzig WH. Primary immunodeficiencies in Switzerland: first report of the national registry in adults and children. J Clin Immunol. 1988;8:479–85. doi: 10.1007/BF00916954. [DOI] [PubMed] [Google Scholar]
- 7.Fasth A. Primary immunodeficiency disorders in Sweden: cases among children, 1974–79. J Clin Immunol. 1982;2:86–92. doi: 10.1007/BF00916891. [DOI] [PubMed] [Google Scholar]
- 8.Seymour B, Miles J, Haeney M. Primary antibody deficiency and diagnostic delay. J Clin Pathol. 2005;58:546–7. doi: 10.1136/jcp.2004.016204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thickett KM, Kumararatne DS, Banerjee AK, Dudley R, Stableforth DE. Common variable immune deficiency: respiratory manifestations, pulmonary function and high-resolution CT scan findings. QJM. 2002;95:655–62. doi: 10.1093/qjmed/95.10.655. [DOI] [PubMed] [Google Scholar]
- 10.Smith CIE, Ochs HD, Puck JM. Genetically determined Immunodeficiency diseases: A perspective. In: Ochs HD, Smith CIE, Puck JM, editors. Primary immunodeficiency diseases. A molecular and genetic approach. Oxford: Oxford University Press; 1999. pp. 3–12. [Google Scholar]
- 11.Buckley R. Variable phenotypic expression of mutations in genes of the immune system. J Clin Immunol. 2005;115:2974–6. doi: 10.1172/JCI26956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janssen R, van Wengen A, Hoeve MA, et al. The same IkappaBalpha mutation in two related individuals leads to completely different clinical syndromes. J Exp Med. 2004;200:559–68. doi: 10.1084/jem.20040773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maraschio P. Genetic heterogeneity for a Nijmegen breakage-like syndrome. Clin Genet. 2003;63:283–90. doi: 10.1034/j.1399-0004.2003.00054.x. [DOI] [PubMed] [Google Scholar]
- 14.Abedi M. Report from the ESID Registry of Primary Immunodeficiencies. The Source. 2003;February/March:8–9. [Google Scholar]
- 15.Plebani A, Soresina A, Rondelli R, et al. Italian Pediatric Group for XLA-AIEOP. Clinical, immunological, and molecular analysis in a large cohort of patients with X-linked agammaglobulinemia: an Italian multicenter study. Clin Immunol. 2002;104:221–30. doi: 10.1006/clim.2002.5241. [DOI] [PubMed] [Google Scholar]
- 16.World Health Organization (WHO) ICD-10 codes, terms and text. Geneva: World Health Organization; 1992–94. [Google Scholar]
- 17.Blore J, Haeney MR. Primary antibody deficiency and diagnostic delay. BMJ. 1989;298:516–7. doi: 10.1136/bmj.298.6672.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Vries E. Clinical Working Party of the European Society for Immunodeficiencies (ESID) patient-centred screening for primary immunodeficiency: a multi-stage diagnostic protocol designed for non-immunologists. Clin Exp Immunol. 2006;145:204–14. doi: 10.1111/j.1365-2249.2006.03138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sewell WA, Khan S, Dore PC. Early indicators of immunodeficiency in adults and children: protocols for screening for primary immunological defects. Clin Exp Immunol. 2006;145:201–3. doi: 10.1111/j.1365-2249.2006.03126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]