Abstract
A novel antiinfective approach is to exploit stresses already imposed on invading organisms by the in vivo environment. Fe metabolism is a key vulnerability of infecting bacteria because organisms require Fe for growth, and it is critical in the pathogenesis of infections. Furthermore, humans have evolved potent Fe-withholding mechanisms that can block acute infection, prevent biofilm formation leading to chronic infection, and starve bacteria that succeed in infecting the host. Here we investigate a “Trojan horse” strategy that uses the transition metal gallium to disrupt bacterial Fe metabolism and exploit the Fe stress of in vivo environments. Due to its chemical similarity to Fe, Ga can substitute for Fe in many biologic systems and inhibit Fe-dependent processes. We found that Ga inhibits Pseudomonas aeruginosa growth and biofilm formation and kills planktonic and biofilm bacteria in vitro. Ga works in part by decreasing bacterial Fe uptake and by interfering with Fe signaling by the transcriptional regulator pvdS. We also show that Ga is effective in 2 murine lung infection models. These data, along with the fact that Ga is FDA approved (for i.v. administration) and there is the dearth of new antibiotics in development, make Ga a potentially promising new therapeutic for P. aeruginosa infections.
Introduction
Recently much attention has been focused on the need for new antimicrobial agents. Heavy antibiotic use and person-to-person spread of bacteria have greatly increased antibiotic resistance due to genetic mutation, and this problem is continually increasing in severity (1). The bacterium Pseudomonas aeruginosa is a prime example: 30% of clinical isolates from ICU or nursing home patients are now resistant to 3 or more drugs, and a similar situation exists for other organisms (2). Despite these trends, only 1 new antibacterial drug with a completely novel mechanism of action has been introduced in the past 3 decades (linezolid), and very few new antibiotics are in advanced development (1).
New agents are also needed because conventional antibiotics generally work poorly in chronic infections, even when the bacteria are susceptible when tested ex vivo. A key factor accounting for this is that the infecting organisms live in biofilms, surface-associated bacterial communities encased in a polymeric matrix (3, 4). Physiological changes inherent to biofilm growth make bacteria far more resistant to killing by the immune system and antibiotics than cells in the free-living (planktonic) state (5). Examples of biofilm infections include the airway infections in cystic fibrosis (CF) patients, chronic wound and sinus infections, endocarditis, and medical device infections, among others (3, 4).
A novel approach to combating infection is to exploit stresses already imposed on organisms by the in vivo environment or host defenses. Fe metabolism is a major vulnerability for infecting bacteria for 2 main reasons. First, in almost all pathogens, Fe is essential for growth and the functioning of key enzymes, such as those involved in DNA synthesis, electron transport, and oxidative stress defense (6). Second, free Fe levels are extremely low in vivo (approximately 10–18 M) due to multiple host defenses that sequester Fe, and this is a potent deterrent to both acute and chronic infection (6). The importance of Fe limitation in blocking acute infection has been established using at least 18 different bacterial species. In these studies, increasing the amount of available Fe markedly reduced resistance to acute infections (6, 7). For example, a single injection of Fe decreased the lethal dose of a P. aeruginosa strain (in a murine acute infection model) from more than 104 organisms to fewer than 10 (8).
Work by a number of laboratories also links Fe metabolism to the pathogenesis of chronic infections, in that high Fe levels (i.e., more Fe than is required for growth) promote biofilm development. High levels of Fe are required for the formation of cell clusters early in biofilm development and for the maturation of biofilms into 3-dimensional structures (9–12). Fe-withholding defenses may also be protective during infection, as bacteria taken from infection sites show gene expression profiles of Fe starvation (13–15).
The fact that host defenses severely limit available Fe and the importance of Fe in infection suggest that invading organisms may be vulnerable to interventions that further disrupt Fe acquisition or metabolism. Unfortunately, exploiting this vulnerability has proved to be difficult. Fe chelation therapy has been tried, but some organisms use Fe complexed to chelators, and secondary infections occurred (16). Targeting bacterial Fe acquisition is also problematic because most pathogens have many redundant uptake systems; P. aeruginosa has more than 30 genes encoding Fe receptors (17). This redundancy reduces the likelihood that Fe uptake systems could be effectively blocked.
Here we explore an alternative approach that uses the transition metal Ga as a “Trojan horse” to disrupt bacterial Fe metabolism. Ga has an ionic radius nearly identical to that of Fe, and many biologic systems are unable to distinguish Ga3+ from Fe3+ (18). Ga can disrupt Fe-dependent processes because, unlike Fe3+, Ga3+ cannot be reduced, and sequential oxidation and reduction are critical for many of Fe’s biological functions (18). Furthermore, Ga is already FDA approved, and large doses are given (i.v.) to treat hypercalcemia of malignancy (19).
We tested this approach using the opportunistic pathogen P. aeruginosa because it causes deadly acute infections and it avidly forms biofilms leading to intractable chronic infections. We found that Ga inhibits P. aeruginosa growth, prevents biofilm formation, and has bactericidal activity. Furthermore, Ga is effective in 2 different experimental animal infections that model acute lethal pneumonia and an airway biofilm infection. Our work also sheds light on Ga’s mechanism of action, showing that Ga is taken up by P. aeruginosa, that it decreases bacterial Fe uptake, and that it works in part by repressing the Fe-responsive transcriptional regulator pvdS.
Results
Ga inhibits growth and kills planktonic bacteria.
As a first step in studying the antimicrobial action of Ga, we examined its effects on growth of the laboratory P. aeruginosa strain PA01 in batch cultures. For consistency, experiments were performed with Ga(NO3)3 (the FDA-approved formulation) in biofilm medium (see Methods) at 37°C, unless specific assays required other conditions. Ga inhibited the growth of PA01 in a dose-dependent manner at concentrations greater than 1 μM (Figure 1A). Ga’s growth-inhibitory action was somewhat more pronounced at room temperature (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI30783DS1), and NaNO3 (a control for the effect of nitrate) had no effect on growth. Bactericidal assays showed that higher levels of Ga also killed log- (data not shown) and stationary-phase bacteria (Figure 1B).
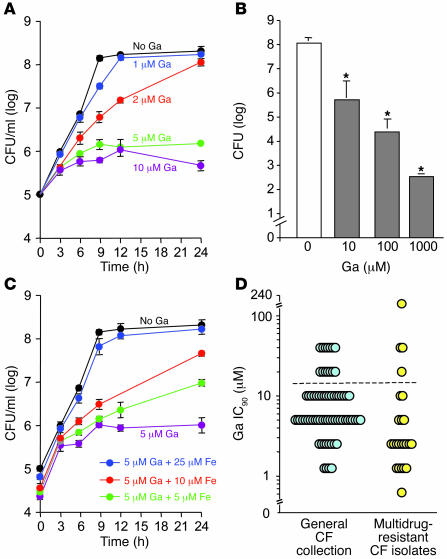
Figure 1. Effect of Ga on P. aeruginosa growth.
(A) Ga(NO3)3 inhibits P. aeruginosa growth in a concentration-dependent manner. Experiments were performed in biofilm medium at 37°C, and data are the mean of 4 experiments; error bars indicate SEM. (B) Ga kills P. aeruginosa. Bacteria were grown in TSB to stationary phase and incubated with indicated concentrations of Ga(NO3)3 for 24 hours. Cells were then pelleted and washed 3 times to remove Ga, and viable bacteria were enumerated by plate counting. Data are the mean of 3 experiments; error bars indicate SEM; *P < 0.001 versus the untreated control. (C) The addition of Fe (FeCl3) diminishes Ga’s growth-inhibitory effects. Data are the mean of 3 experiments; error bars indicate SEM. (D) Ga sensitivity of P. aeruginosa isolates from chronically infected CF patients. Clinical isolates and PA01 bacteria were grown for 24 hours at 37°C in BM2 succinate medium with 0.1% casamino amino acids with increasing (2-fold) concentrations of Ga(NO3)3. The IC90 of PA01 (determined by measuring OD600) was 10 μM Ga in this medium. Isolates represented by points lying above the dotted line were more resistant than PA01. Data are mean of 2 measurements for each isolate.
To begin investigating whether the antimicrobial action of Ga was mediated by effects on Fe-dependent processes, we tested Ga in the presence of increasing concentrations of Fe (FeCl3). Fe addition reversed Ga’s growth-inhibitory effects; however, a higher molar ratio of Fe as compared to Ga (approximately 5:1) was required to restore growth to control levels (Figure 1C).
Multidrug-resistant CF isolates are sensitive to Ga.
During chronic infections, bacteria undergo genetic change as they adapt to selective pressures within the host (20). Because Fe is severely limited in vivo, Fe uptake mechanisms may evolve during infection, and this could affect Ga’s action. To investigate this, we tested 3 collections of CF isolates for sensitivity to the growth-inhibitory effects of Ga.
In a general collection of 71 CF isolates (Figure 1D), only 13% were more resistant to Ga than the laboratory strain PA01. We also tested a separate collection of 23 CF isolates resistant to 3 or more antibiotics and again found relatively few (17%) that were more resistant than PA01 (Figure 1D). Finally, we examined whether Ga sensitivity changes during infection by testing paired clonally related isolates from 20 patients that were collected at an early and late stage of infection (isolates were collected 5–20 years apart; ref. 20). The Ga sensitivity of the majority of isolates remained constant or decreased during the course of infection (data not shown).
Of note, the degree of Ga resistance found was generally modest. Except for 1 outlier (of 115 strains tested), the most resistant clinical isolate had an inhibitory concentration only 4-fold higher than that of PA01. These data suggest that neither the bacterial adaptations associated with CF infection nor resistance to conventional antibiotics are likely to compromise Ga’s antimicrobial activity.
Low concentrations of Ga prevent biofilm formation.
The link between Fe and biofilm formation (9–12) and the fact that Ga can disrupt Fe-dependent processes (21) led us to investigate the effect of Ga on P. aeruginosa biofilm development. Studying Ga’s effects on biofilms was also of interest because low concentrations of a number of antibiotics promote biofilm formation, perhaps because biofilm growth can be induced as a consequence of stress (22–25). These findings have raised the worrisome possibility that antibiotic concentrations that fail to completely kill may actually promote chronic infection.
To determine whether Ga affects biofilm formation, we grew biofilms in the presence and absence of a low concentration of Ga(NO3)3 that modestly slowed planktonic growth (Figure 2A and Supplemental Figure 1). In control medium (no Ga), biofilms developed in a typical manner. Bacteria attached to the growth surface, formed cell clusters, and then matured into tower-shaped biofilm structures (Figure 2A). The addition of Ga completely blocked biofilm formation; attached bacteria remained as separated individual cells even after 5 days (Figure 2A). As with Ga’s growth-inhibitory effects, a higher molar ratio of Fe than Ga (approximately 10:1) was required to reverse the effect of Ga on biofilm formation (Figure 2B).
Figure 2. Ga prevents P. aeruginosa biofilm formation.
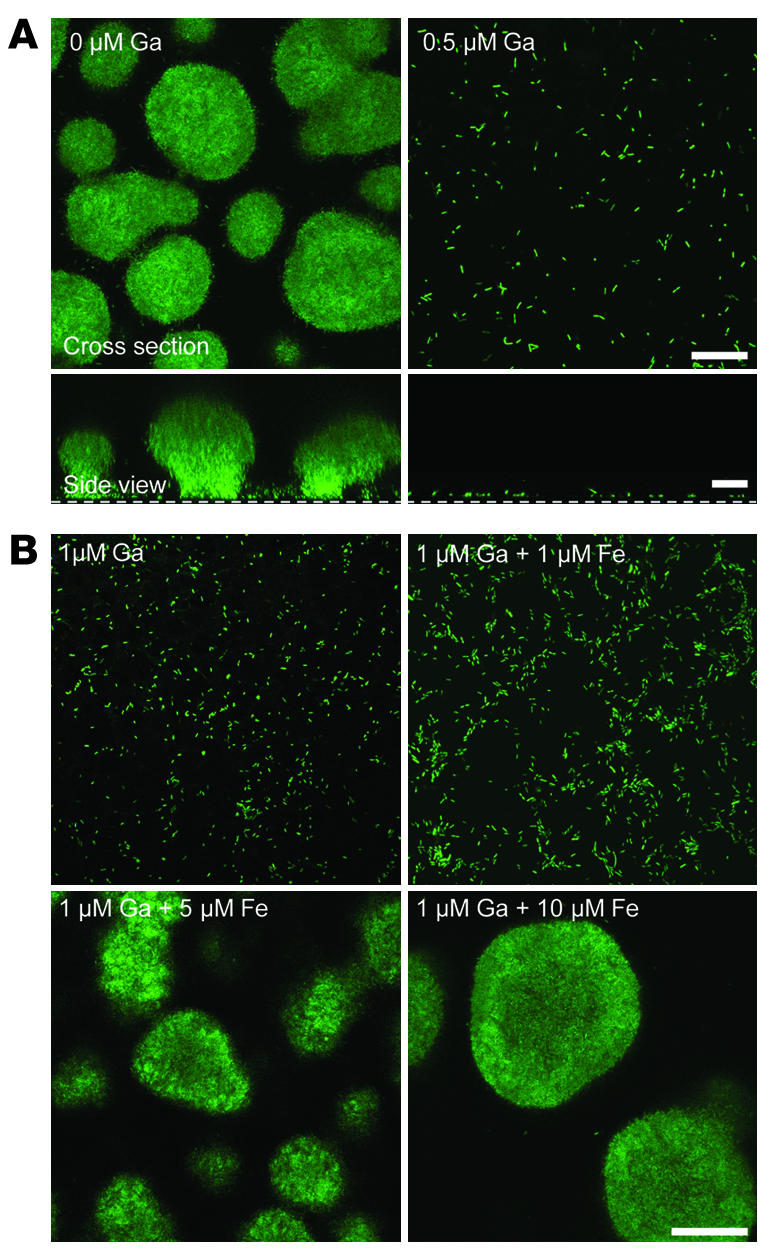
(A) Confocal microscopic images of GFP-expressing P. aeruginosa in flow cells perfused with biofilm medium without (left) and with (right) Ga, 5 days after inoculation. Experiments were performed at 25°C using 0.5 μg/ml Ga(NO3)3; this concentration modestly inhibited growth at 25°C (see Supplemental Figure 1). Higher concentrations [1, 5, and 10 μM Ga(NO3)3] also inhibited biofilm formation. Top images are top-down views (x-y plane); bottom images are side views (x-z plane); the dotted line represents the biofilm growth surface; scale bars: 50 μm. Results are representative of 6 experiments. (B) Biofilm flow cells were perfused with increasing concentrations of FeCl3 in the presence of 1 μM Ga(NO3)3. Approximately 10 μM FeCl3 was required to completely reverse the antibiofilm effect of 1 μM Ga(NO3)3. Images are top-down views (x-y plane); scale bar: 50 μm. Results are representative of 3 experiments.
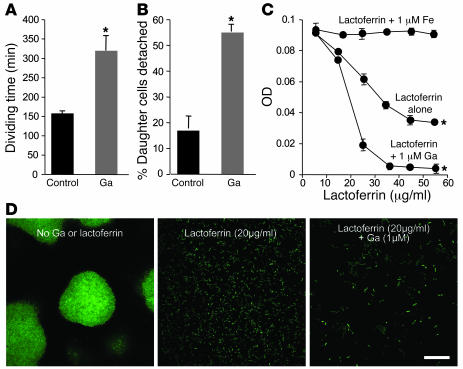
The antibiofilm effect of Ga appeared to be similar to that seen in previous work with the Fe-binding protein lactoferrin (9). Fe sequestration by lactoferrin was found to stimulate bacterial surface motility, thereby preventing the formation of cell clusters. It also increased the rate at which bacteria detached from the growth surface and slowed the growth of attached cells somewhat. To determine whether Ga acted in a similar manner, we performed time-lapse microscopy of P. aeruginosa growing in biofilm reactors in the presence and absence of Ga. Ga slowed the growth of attached bacteria and increased the rate at which cells detached (Figure 3, A and B), as was seen with lactoferrin (9). However, Ga did not stimulate bacterial surface motility in biofilm reactors or in motility assays (Supplemental Figure 2). These data suggest that Ga and lactoferrin inhibit biofilm formation, at least in part, by different mechanisms.
Figure 3. Ga inhibits growth of biofilm-forming cells, stimulates detachment, and its activity is enhanced by lactoferrin.
(A–B) The dividing time (A) and detachment rate (B) of attached bacteria were increased by 1 μM Ga(NO3)3. Time-lapse images of GFP-labeled P. aeruginosa in biofilm reactors from 4 separate experiments were collected in the presence and absence of 1 μM Ga(NO3)3. Dividing times of attached bacteria were measured by counting the number of frames between 50 cell divisions. The percentage of daughter cells detaching from the surface was determined by observing 100 daughter cells and averaging the results; *P < 0.001 versus the untreated control. (C) Ga potentiates the growth-inhibitory effects of lactoferrin. 104 planktonic P. aeruginosa were inoculated in biofilm medium with various concentrations of lactoferrin alone or lactoferrin mixed with Fe or Ga. Cultures were then grown for 24 hours, and the OD600 was measured. Fe (1 μM) blocked the growth-inhibitory effects of lactoferrin, but Ga potentiated it. Results are the mean of 3 separate experiments; error bars are SEM; *P < 0.001 versus the untreated control. (D) Ga potentiates the antibiofilm effects of lactoferrin. Confocal microscopic images of GFP-labeled P. aeruginosa grown in flow cells with biofilm medium alone and medium containing lactoferrin (20 μg/ml) or lactoferrin (20 μg/ml) plus Ga(NO3)3 (1 μM). Images are top views (x-y plane) obtained after 5 days of growth; scale bar: 50 μm. Results are representative of 3 separate experiments. See Supplemental Figure 3 for counts of attached bacteria.
Ga augments the antimicrobial action of lactoferrin in vitro.
Lactoferrin is abundant on mucosal surfaces and is secreted by neutrophils at sites of infection (26). Thus, in clinical use, Ga would likely be applied to sites where high concentrations of lactoferrin are present. Because lactoferrin works by binding Fe, the activity of Ga and lactoferrin may be enhanced in combination, as Fe limitation produced by lactoferrin could increase Ga’s effects (see Figure 1C and Figure 2B). An antagonistic interaction is also possible if Ga liberated lactoferrin-sequestered Fe.
To distinguish between these possibilities, we examined the growth-inhibitory and antibiofilm actions of lactoferrin in the presence of Ga. Whereas Fe blocked the growth-inhibitory action of lactoferrin, Ga addition increased it (Figure 3C). Ga also enhanced the antibiofilm activity of lactoferrin (Figure 3D and Supplemental Figure 3). These findings are consistent with previous work showing that lactoferrin has a higher affinity for Fe than for Ga (27) and with our finding that Ga’s activity is enhanced in low Fe conditions (Figure 1C and Figure 2B). If these findings reflect the situation in vivo, Ga is not likely to interfere with the host defense functions of lactoferrin and may in fact augment them.
Ga kills established P. aeruginosa biofilms.
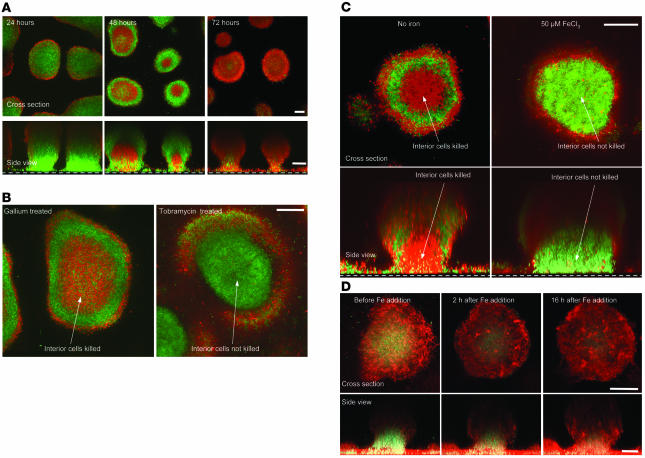
To determine whether Ga kills established biofilms in vitro, we grew biofilms for 3 days (with no Ga present) and then switched to medium containing 10, 100, and 1,000 μM Ga. Bacterial viability was assayed using a live/dead stain. Ga killed biofilm bacteria in a dose-dependent manner, and by 72 hours the bulk of biofilm bacteria treated with 100 and 1,000 μM Ga were killed (Figure 4A and Supplemental Figure 4). Ga killed biofilm bacteria with an unexpected pattern, as cells located in the center of biofilm structures were particularly sensitive to Ga (Figure 4, A and B). This is of interest because some antibiotics, including tobramycin (Figure 4B) and ciprofloxacin, are relatively ineffective at killing cells in the interior of biofilms (28, 29).
Figure 4. Ga kills established biofilms.
(A) Biofilms were grown using GFP-labeled P. aeruginosa for 3 days in the absence of Ga; treated with 100 μM Ga(NO3)3 for the indicated time periods; and stained with 30 μM propidium iodide, which labels dead cells red. Top images are top-down views (x-y plane); bottom images are side views (x-z plane); the dotted line represents the biofilm growth surface; scale bars: 50 μm. Results are representative of 4 separate experiments. (B) High-magnification image of the biofilm killing pattern of Ga and tobramycin. Three-day-old P. aeruginosa biofilms (grown without Ga) were treated with Ga(NO3)3 (100 μM) or tobramycin (1 μg/ml) for 48 hours and stained as described in A. Ga preferentially killed bacteria in the central regions, whereas tobramycin preferentially killed bacteria located in the biofilm periphery. Images are top-down views (x-y plane); scale bar: 50 μm. Results are representative of 3 separate experiments. (C) Fe addition eliminates the central biofilm killing pattern of Ga. GFP-labeled P. aeruginosa biofilms were grown in flow cells for 3 days with or without FeCl3 (50 μM), treated with 100 μM Ga(NO3)3 for 48 hours, and then stained as described in A. Fe addition eliminated the central killing pattern of Ga. Top images are top-down views (x-y plane); bottom images are side views (x-z plane); the dotted line represents the biofilm growth surface; scale bar: 50 μm. Results are representative of 3 separate experiments. (D) Evidence for Fe starvation in the central biofilm regions. P. aeruginosa constitutively expressing red florescent protein and containing the pvdA-gfp(S) reporter (which fluoresces green during Fe limitation; see Supplemental Figure 4) were grown in biofilms for 3 days. In the absence of added Fe, the central biofilm regions appeared green, indicating Fe limitation. Fe addition reduced the size of the central Fe-starved (green) region within 2 hours. Images are top-down views (x-y plane); scale bar: 50 μm. All images in D were taken with identical confocal microscope settings, and results are representative of 3 separate experiments.
Although the mechanisms producing antimicrobial resistance of biofilms are incompletely understood, a leading hypothesis posits that internal populations resist killing because the metabolic activity of overlying cells limits the nutrient reaching them. This produces resistance because nutrient limitation generally decreases the effectiveness of antibiotics (30). However, our data with planktonic bacteria (Figure 1C and Figure 2B) indicate that the limitation of at least one nutrient, Fe, enhances the antimicrobial activity of Ga. This led us to hypothesize that internal biofilm populations may be more susceptible to Ga, in part, because of Fe limitation in these regions.
We tested this hypothesis in 2 ways. First, we added Fe during Ga treatment and found that central killing did not occur (Figure 4C). Second, to localize regions within biofilms where Fe limitation occurs, we used a P. aeruginosa strain containing an Fe-responsive promoter (transcribed when Fe levels are low; see Supplemental Figure 5) linked to a short-half-life GFP reporter. The highest level of fluorescence intensity was seen in the central biofilm regions, and the addition of Fe decreased reporter expression within 2 hours (Figure 4D). A similar result was seen using a long-half-life reporter (Supplemental Figure 6). These data indicate that the promoter used is Fe responsive when grown in biofilms and suggest that cells in the internal regions experience Fe limitation. Taken together, these data are consistent with the hypothesis that internal populations are Ga sensitive because of relative Fe starvation.
Ga is taken up by P. aeruginosa, and mutations in major iron uptake systems do not produce Ga resistance.
Fe acquisition by some bacterial systems involves Fe reduction (31). Because Ga cannot be reduced, it was not clear whether Ga would be taken up by P. aeruginosa or whether it exerted its effects from the outside of cells, for example by blocking Fe uptake systems. To test this, we incubated bacteria with radioactive Ga (67Ga), washed them repeatedly in the chelator EDTA to remove noninternalized metal, and measured Ga uptake using a gamma counter. As shown in Figure 5A, WT P. aeruginosa took up Ga in a concentration-dependent manner.
Figure 5. The uptake and antimicrobial activity of Ga does not depend on major P. aeruginosa Fe-uptake systems.
(A) P. aeruginosa takes up Ga. WT P. aeruginosa takes up Ga in concentration-dependent manner. Isogenic strains lacking functional pyoverdine, pyochelin, pyoverdine and pyochelin, and ferric uptake systems also took up Ga. Values are mean of 4 experiments and are not statistically different from each other (P = 0.17–0.37). (B–E) P. aeruginosa strains lacking functional pyoverdine, pyochelin, pyoverdine and pyochelin, and ferric citrate uptake systems showed sensitivity to Ga’s growth-inhibitory effect that was similar to that of WT P. aeruginosa. Arrows show the final culture density obtained when WT P. aeruginosa was treated with the same concentration of Ga(NO3)3 (10 μM). Results are the mean of 3 separate experiments; error bars are SEM. (F) Fe-uptake mutants are sensitive to the antibiofilm effects of Ga. GFP-labeled bacteria were grown in flow cells perfused with biofilm medium without (top) and with (bottom) 1 μM Ga(NO3)3. Inactivation of the pyoverdine system produced biofilms with a flat structure, as described previously (11). Images were obtained after 5 days and are top-down views (x-y plane); scale bar: 50 μm. Results are representative of 3 experiments.
CF isolates commonly develop mutations in biosynthetic genes of the siderophore pyoverdine, perhaps because pyoverdine is a ligand for the immune system (20). If Ga uptake required the pyoverdine system, its activity could be compromised in CF. However, we found that inactivation of the pyoverdine system had only a modest effect on Ga uptake (Figure 5A) and actually somewhat increased the sensitivity of P. aeruginosa to Ga’s growth-inhibitory and antibiofilm effects (Figure 5, B and F). We also tested mutants with the siderophore pyochelin and ferric citrate systems inactivated and a double mutant with both the pyoverdine and pyochelin systems inactivated. All took up Ga, and all were sensitive to Ga’s antibiofilm and growth-inhibitory actions (Figure 5, A and C–F). These data indicate that Ga’s activity is not dependent upon any one of the Fe uptake systems we examined and that inactivation of these systems is not likely to produce Ga resistance.
Ga disrupts Fe metabolism in P. aeruginosa.
In experiments described above (Figure 1C and Figure 2B), we found that higher molar ratios of Fe to Ga were required to reverse Ga’s growth-inhibitory and antibiofilm effects. One explanation is that Ga impaired bacterial Fe uptake. We measured Fe uptake by P. aeruginosa in the presence and absence of Ga and found that Ga inhibited the uptake of Fe in a concentration-dependent manner (Figure 6A). Surprisingly, the converse was not true; Fe had little effect on Ga uptake by P. aeruginosa (Figure 6B). These data suggest that the dynamics of Ga and Fe uptake are likely complex and not solely determined by a simple competition for uptake systems.
Figure 6. Ga blocks Fe uptake by P. aeruginosa.
(A) WT bacteria were grown to mid-log phase in 10% TSB medium with increasing concentrations of Ga and 2 μM 59Fe. Ga inhibited Fe uptake by P. aeruginosa in a concentration-dependent manner. Values are mean of 4 experiments. Error bars are SEM; *P < 0.001 versus the untreated control. (B) Fe addition does not block Ga uptake by P. aeruginosa. WT bacteria were grown to mid-log phase in 10% TSB medium with increasing concentrations of Fe and 2 μM 67Ga. Ga had little effect on Fe uptake by P. aeruginosa. Values are mean of 4 experiments and are not statistically different from each other (P = 0.33–0.68); error bars are SEM. (C) Ga represses pyoverdine production normally induced by Fe starvation. WT P. aeruginosa was grown in BM2 medium with indicated concentrations of FeCl3 in the presence and absence of Ga (2 μM). Ga treatment blocked the increase in pyoverdine production normally induced by Fe starvation. Ga also blocked pyoverdine gene expression induced by Fe starvation (Supplemental Figure 7).
To further investigate Ga’s mechanism of action, we examined the effect of Ga on the gene expression profile of P. aeruginosa. Microarray analysis was performed using batch cultures in biofilm medium to the middle of exponential phase in the presence and absence of a subinhibitory concentration of Ga (1 μM; see Figure 1A). Since the Ga-treated bacteria grew at nearly the same rate as untreated cells, we thought these conditions would increase our chances of detecting expression changes directly caused by Ga and would minimize nonspecific toxic effects.
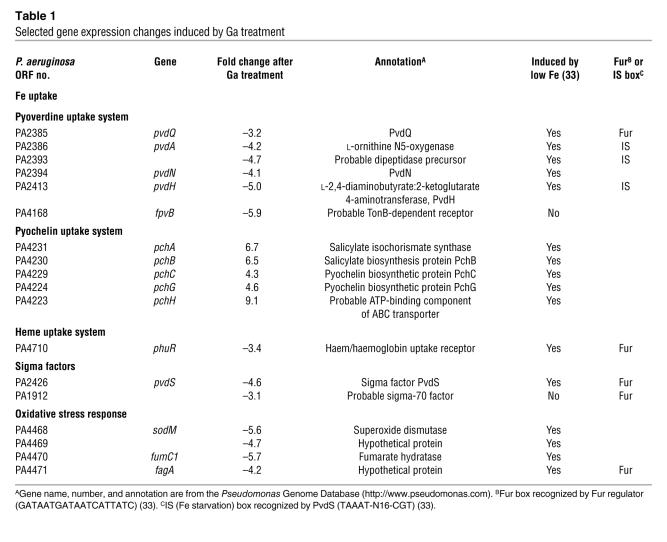
Using a P value threshold of 0.01 and a 3-fold expression change cutoff, we found 27 genes to be induced by Ga treatment and 47 genes repressed (Table 1 and Supplemental Table 1). Several findings were of note. First, Ga treatment altered the expression of genes involved in carbon utilization and reduced expression of ribosomal genes (Supplemental Table 1). These changes may reflect secondary stress effects, despite the fact that the concentration of Ga used had a minimal effect on growth. Second, we found that Ga exposure did not induce the expression of invasive virulence factors. This is significant because previous work has shown that Fe limitation can induce elastase, exotoxin A, rhamnolipids, and other factors that increase the invasiveness and toxicity of P. aeruginosa (32–34).
Table 1 .
Selected gene expression changes induced by Ga treatment
Third, Ga treatment (in the absence of Fe limitation) induced some expression changes characteristic of Fe starvation and others that typify the Fe-replete state (Table 1). For example, Ga treatment induced the expression of pyochelin biosynthetic genes, known to be induced by Fe limitation (33). In contrast, Ga repressed the transcriptional regulator pvdS and genes under its control, which are among the most highly induced by Fe starvation (33). The pvdS regulator controls genes involved in the synthesis of pyoverdine, the pyoverdine receptor, and other Fe-responsive genes (33).
To determine whether Fe limitation could overcome Ga-induced repression of pvdS-controlled functions, we exposed P. aeruginosa to Ga in a defined medium and progressively lowered the Fe concentration. Ga exposure abolished the increased pyoverdine biosynthesis (Figure 6C) and pyoverdine gene expression (Supplemental Figure 7) that normally occurs during Fe limitation. The mechanism by which Ga represses pvdS-controlled functions is not known. Previous work showed that pyochelin biosynthesis genes and the pvdS regulon are coordinately controlled by the master Fe regulator Fur (33). Thus, Ga appears to disrupt typical patterns of Fe signaling.
Repression of pvdS contributes to Ga’s antimicrobial activity.
Our findings that Ga decreased bacterial Fe uptake and repressed pvdS (which controls genes involved in Fe acquisition), coupled with the fact that Ga’s activity is enhanced when Fe is low (Figure 1C, Figure 2B, and Figure 3, C and D), led us to hypothesize that repression of pvdS may contribute to Ga’s antimicrobial effects.
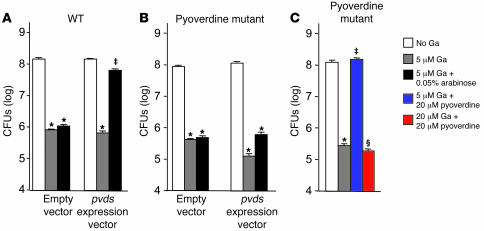
To test this, we constructed a P. aeruginosa strain with pvdS controlled by an arabinose-inducible promoter so that we could override pvdS repression during Ga treatment. We confirmed that this construct inducibly expressed pvdS-controlled genes by measuring pyoverdine production and the fluorescence of a pvdA-gfp reporter (Supplemental Figure 8, A and B). As shown in Figure 7A, overriding the Ga-induced repression of pvdS protected P. aeruginosa from Ga’s growth-inhibitory effects. These data suggest that the antimicrobial activity of Ga is mediated in part by repression of some gene(s) in the pvdS regulon.
Figure 7. Repression of pyoverdine expression via the transcriptional regulator pvdS contributes to Ga’s antimicrobial activity.
(A) Overriding pvdS repression protects P. aeruginosa from the growth-inhibitory effects of Ga. P. aeruginosa containing the arabinose-inducible pvdS construct or an empty vector were grown in biofilm medium with or without 5 μM Ga and 0.05% arabinose (arabinose was present from the beginning of the experiment). Results are the mean of 3 experiments; error bars are SEM; *P < 0.001 versus untreated control; ‡P < 0.001 versus 5 μM Ga. Consistent with this result, inactivation of pvdS somewhat increased Ga sensitivity (see Supplemental Figure 11). (B) The protective effect of pvdS expression depends on pyoverdine. The pvdA mutant containing the inducible pvdS construct or the empty vector was grown in biofilm medium with or without 5 μM Ga, 0.05% arabinose, or 20 μM of pyoverdine. The protective effect of overriding pvdS repression during Ga treatment that was seen in WT cells (A) was lost in the pvdA mutant. (C) Pyoverdine protects P. aeruginosa from the growth-inhibitory effects of Ga. The pvdA mutant was grown in biofilm medium with or without 5 μM Ga, 20 μM Ga, and 20 μM of pyoverdine. Exogenous addition of pyoverdine protected bacteria from the growth-inhibitory effects of 5 μM Ga. Higher levels of Ga (20 μM) overcome the protective effects of pyoverdine. Results are the mean of 3 experiments; error bars are SEM; *P < 0.001 versus untreated control; ‡P < 0.001 versus 5 μM Ga; ΧP < 0.001 versus 5 μM Ga plus 20 μM pyoverdine.
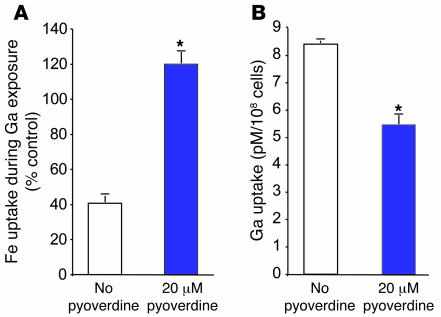
We hypothesized that the pyoverdine system was involved in Ga’s effects because it is pvdS controlled and because repression of pyoverdine could decrease Fe uptake. We tested this hypothesis in 2 ways. First, we forced pvdS expression (during Ga treatment) in a P. aeruginosa strain in which pyoverdine genes were inactivated and found that the protective effects of overriding pvdS repression were largely lost (Figure 7B). Second, we exogenously added purified pyoverdine (to the pyoverdine mutant) during Ga treatment. As shown in Figure 7C, pyoverdine addition protected P. aeruginosa from the growth-inhibitory effects of Ga (although this protection could be overcome by high Ga concentrations). Furthermore, pyoverdine addition eliminated the Ga-induced inhibition of Fe uptake, and, surprisingly, it also decreased bacterial Ga uptake (Figure 8, A and B).
Figure 8. Pyoverdine reduces the Ga-induced repression of Fe uptake and decreases Ga uptake.
(A) Pyoverdine protects P. aeruginosa from Ga-induced repression of Fe uptake. The pvdA mutant was grown in medium containing 2 μM 59Fe and 5 μM Ga, with or without 20 μM pyoverdine. Ga reduced bacterial Fe uptake in the absence of pyoverdine. Pyoverdine addition protected cells from this effect. Results are expressed as percent of Fe uptake in the absence of Ga and are the mean of 3 experiments; error bars are SEM; *P < 0.001 versus untreated control. (B) Pyoverdine protects P. aeruginosa from Ga uptake. The pvdA mutant was grown in biofilm medium containing 2 μM 67Ga with and without 20 μM pyoverdine. Pyoverdine addition decreased bacterial 67Ga uptake. Results are the mean of 3 experiments; error bars are SEM; *P < 0.001 versus untreated control.
Taken together, these data indicate that repression of pyoverdine (mediated by pvdS) contributes to Ga’s antimicrobial activity. The reduced levels of pyoverdine decrease Fe uptake and allow increased Ga uptake. These effects also likely contribute to the enhanced Ga sensitivity of Fe-starved bacteria. Ga treatment blocks protective pyoverdine secretion in Fe-limited cells as well (Figure 6C), and low Fe levels likely increase the ability of Ga to substitute for Fe at critical sites.
Ga is effective in mouse lung infection models.
Our in vitro data suggest that Ga may be an effective antipseudomonal antibiotic. The growth-inhibitory and bactericidal activity of Ga against planktonic cells could be effective in acute infections, and its antibiofilm action could be beneficial in chronic infections. To test Ga’s efficacy in a severe acute infection, we administered a lethal dose of the highly virulent P. aeruginosa strain PA103 to mice by inhalation. PA103 expresses cytotoxins by a type III secretion mechanism, and work in a number of models shows that this strain produces rapid bacterial dissemination and death (35, 36).
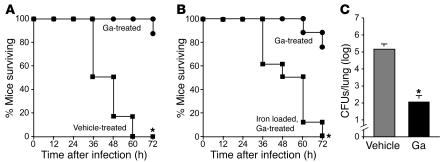
A single dose of inhaled Ga administered 3 hours after infection reduced histological evidence of lung injury (Supplemental Figure 9) and was almost completely effective in preventing death caused by PA103 sepsis (Figure 9A). Furthermore, instilling FeCl3 into the lungs immediately prior to infection eliminated Ga’s antimicrobial action (Figure 9B). These data show that Ga is effective in a severe acute infection and suggest that, as in vitro, Ga’s in vivo activity results from disruption of Fe-dependent processes.
Figure 9. Ga is effective in 2 different P. aeruginosa lung infection animal models.
(A) Mice treated with Ga are protected from lethal P. aeruginosa pneumonia. P. aeruginosa strain PA103 (5 × 105 bacteria) was administered to mice by the intratracheal route. Mice were treated with a single dose of inhaled Ga (or vehicle alone) 3 hours after infection. Inhalation treatment was achieved by placing a 50-μl drop of concentrated Ga(NO3)3 (250 mm) or Ga-free vehicle on the nares of the mice. Because mice are obligate nose breathers, some of the drop was inhaled. Data are representative of 4 separate experiments (n = 6 mice in each group); *P < 0.005 versus vehicle control. Ga was also effective against acute pneumonia produced by strain PA01 (Supplemental Figure 10). (B) Preloading the lung with Fe blocks Ga’s antimicrobial effects. Ten microliters of 2 mM FeCl3 (or Fe-free vehicle) was administered to mice by the intratracheal route immediately prior to infection. Mice (n = 8 per group) were treated with Ga as described in A. Data are representative of 3 separate experiments; *P < 0.005 versus Fe-free control. (C) Inhaled Ga is effective in a mouse airway biofilm infection model. Infections were established as described in Methods. Mice were treated with vehicle alone or Ga (as described in A) 3 times a day for 3 days. Lungs were homogenized, and bacterial counts determined by plating (n = 4 mice per group). Data are representative of 3 separate experiments; *P < 0.005 versus vehicle control.
We also tested the ability of Ga to prevent biofilm formation in a chronic airway infection model (37, 38). In this model a small plastic tube is inserted in either main bronchus of mice. Planktonic P. aeruginosa is then infused, and biofilm formation occurs on the tube. Ga was administered by the inhaled route 3 times a day for 9 doses beginning 5 hours after infection. As shown in Figure 9C, Ga treatment reduced the number of bacteria in the lungs by approximately 1,000-fold. Of note, the mice appeared to tolerate 3 days of Ga treatment well. These data suggest that in addition to its activity in acute infections, Ga may have activity in infections where biofilm formation occurs.
Discussion
The competition for Fe is critical in the struggle between pathogen and host. Here we explore an antimicrobial approach that targets bacterial Fe metabolism by exploiting the chemical similarities between Fe and Ga. In in vitro studies, we found that low concentrations of Ga inhibit P. aeruginosa growth and prevent biofilm formation and that high levels kill planktonic bacteria and established biofilms. Although the available P. aeruginosa lung infection models are imperfect, we found corresponding effects in animal infections: Ga effectively cured acute lethal pneumonia, and early treatment reduced bacterial counts in an airway biofilm infection by approximately 1,000-fold. These data suggest that Ga’s ability to disrupt Fe metabolism in P. aeruginosa could be therapeutically useful in both acute and chronic infections.
Our experiments begin to shed light on Ga’s mechanism of action. We found that Ga is internalized by P. aeruginosa, interferes with Fe signaling, and decreases bacterial Fe uptake. Some of Ga’s antimicrobial activity is mediated by repression of the transcriptional regulator pvdS, as overriding pvdS repression partially protected bacteria from Ga’s growth-inhibitory action. Furthermore, our data indicate that repression of pyoverdine (by pvdS) plays an important role in Ga’s action, as the protective effect of overriding pvdS repression was lost in a pyoverdine mutant. Consistent with this, we found that the exogenous addition of pyoverdine decreased Ga uptake, prevented the Ga-induced reduction of Fe uptake, and protected bacteria from Ga’s growth inhibitory effects. Taken together, these data suggest that Ga exposure, may set bacteria on a self-destructive course. Ga reduces pyoverdine expression (via pvdS), which in turn increases Ga uptake and decreases Fe uptake. Both of these effects are likely to augment Ga’s antimicrobial activity.
The mechanism through which pyoverdine protects cells from Ga uptake is unknown. One possibility is suggested by the finding that Fe uptake by pyoverdine involves Fe reduction (31). Since Ga cannot be reduced, pyoverdine-bound Ga may not enter cells. If this is true, secreted pyoverdine may act like flypaper or a sponge, binding up Ga and keeping it away from Fe-requiring cellular machinery. Another possibility relates to pyoverdine’s action as a signaling molecule that can modulate bacterial gene expression (34). In this case, pyoverdine could induce some function that decreases Ga uptake. While additional work will be required to understand the mechanism, our data raise the possibility that pyoverdine may serve the previously unrecognized function of limiting the uptake of Ga and perhaps other toxic trivalent metals. This possibility is evolutionarily plausible, as the natural soil and water environments of P. aeruginosa contain low (nanomolar) concentrations of Ga (39).
While our work implicates pvdS and pyoverdine in Ga’s action, it is likely that Ga has other, more direct antimicrobial effects. In support of this idea, we found that higher Ga levels could overcome the protective effect of pyoverdine. Given Ga’s general ability to substitute for Fe, it could interfere with many Fe-requiring enzymes, including ribonucleotide reductase, which catalyzes the first step in DNA synthesis (40); superoxide dismutase and catalase, which protect against oxidant stress (41); enzymes involved in oxidative phosphorylation such as cytochromes; and others. It is also possible that Ga could act on several targets simultaneously. If this is true, fully defining Ga’s mechanism of action may be difficult. However, this would also suggest that mutation of a single intracellular target is unlikely to produce high-level Ga resistance.
Our work involves several assumptions and limitations. In the Ga and Fe uptake studies, we used a more concentrated tryptic soy broth (TSB) medium (with higher Fe levels) than was used in the biofilm and growth experiments. This was necessary because we found that Ga and Fe uptake were markedly reduced if growth was inhibited by Ga, probably due to secondary effects related to low metabolic activity. In interpreting the uptake data, we made the assumption that the trends in Ga and Fe uptake observed in conditions involving minimal growth inhibition reflect the primary actions of Ga that mediate its effects. We think this is a reasonable assumption, but it is possible that different Ga and Fe uptake dynamics occur in the course of Ga’s antimicrobial activity.
The in vivo experiments also have limitations. The nasal aspiration treatment method we used does not replicate aerosol treatment modalities used in humans. It is possible that inhalation of aerosol droplets (used in human treatment) could have different effects. Furthermore, we have not ruled out the possibility that some of Ga’s therapeutic action is due to antiinflammatory effects; Ga has been shown to have antiinflammatory activity in vitro and in vivo (42). Another limitation is that in our biofilm infection studies, we initiated treatment soon (5 hours) after bacterial inoculation. While these experiments suggest that Ga may be effective during the early stages of biofilm formation in vivo, we have not determined whether Ga has efficacy against biofilm infections that have been established for prolonged periods. Moreover, it is unclear whether any P. aeruginosa animal model predicts efficacy in human infections.
Further work and, ultimately, human studies will be needed to determine whether Ga is an effective antimicrobial treatment. Several of our findings suggest that it holds promise and could offer some advantages over conventional therapies. First, antibiotic-resistant organisms (even those with multidrug resistance) are generally sensitive to Ga. This is likely due to the fact that Ga works by a completely different mechanism than conventional drugs. Second, Ga’s ability to prevent biofilm formation and to kill established biofilms (if high levels can be tolerated) raise the possibility that it may be effective against chronic infections in which biofilm growth has been implicated.
Our finding that Ga is particularly effective against cells located in the center of biofilm structures is also encouraging. Centrally located bacteria are relatively resistant to conventional antibiotics such as tobramycin, and Ga appears to exploit the Fe limitation that occurs in these regions. Third, our experiments indicate that Ga enhances the antimicrobial actions of lactoferrin, and it also may augment other Fe-withholding defense mechanism. Because lactoferrin is known to act synergistically with other host defenses (43), enhancing its activity could have amplified effects. Finally, the fact that Fe levels are so low in human tissues, and that Ga’s activity is increased when Fe is limited, raises the possibility that Ga may be more effective in vivo than our in vitro data indicate.
Methods
Chemicals, media, and bacteria.
Ga(NO3)3, lactoferrin, FeCl3, sodium citrate, l-arabinose, and antibiotics were purchased from Sigma-Aldrich. For the in vitro studies, Ga(NO3)3 was dissolved in water and adjusted to pH 7. For the in vivo studies, Ga(NO3)3 (250 mM) in 100 mM sodium citrate adjusted to pH 7 was used. TSB diluted in water was used as the medium for most experiments; 1% TSB (biofilm medium) was used for growth, biofilm, and microarray experiments; 10% TSB was used in the metal uptake experiments. Cultures were incubated at 37°C unless otherwise indicated. BM2 medium consisted of 40 mM K2HPO4, 22 mM KH2PO4, 7 mM (NH4)2SO4, 34 mM succinic acid, and 1 mM MgSO4 adjusted to pH 7. Pyoverdine was provided by C. Cox (University of Iowa, Iowa City, Iowa, USA). Bacterial strains (and their construction) are described in Supplemental Table 1.
Growth, killing, and biofilm experiments.
Growth experiments used 104 mid-log-phase cells grown in 1% TSB, and bactericidal assays used 108 stationary-phase PA01. Studies of biofilm formation were performed and imaged (at 25°C) as described previously (9). Some biofilms were stained with propidium iodide: 4 μM to counterstain; 30 μM to detect nonviable cells (44). Ga susceptibility of clinical isolates was determined by measuring the OD590 in BM2 medium with 0.1% casamino acids (BD) after 24 hours of growth. Pyoverdine measurements in culture supernatants were performed as described in ref. 45.
Transcriptome analysis.
Duplicate cultures of PA01 were grown in 1% TSB with 1 μM Ga(NO3)3 or 3 μM NaNO3 (as a control) at 37°C with shaking. RNA was isolated from 2 × 109 mid-log cells. RNA purification, cDNA synthesis, labeling, processing of microarrays (Affymetrix), and data analysis were performed as previously described (46, 47). Gene expression levels were averaged from duplicate chips, and expression was characterized as being increased or decreased according to a P threshold of 0.01, a posterior probability of differential expression of greater than 0.90, and a fold change of at least 3.0 was used. 4,523 genes were expressed above the detection limit, and 1,047 genes were not significantly expressed under either condition.
Metal uptake studies.
5 × 106 organisms/ml were mixed with 59Fe citrate, 67Ga nitrate, and in some cases unlabeled metal and incubated in 10% TSB medium with or without 0.05% arabinose at 37°C for 6 hours. A 1-ml aliquot was withdrawn, OD600 measured, and centrifuged at 10,000 g at 4°C for 10 minutes. The cell pellet was washed 3 times to remove noninternalized metal using 5 mM ascorbate, pH 4.5 (for Fe), or 1 mM EDTA (for Ga), and bacteria-associated radioactivity measured using a gamma counter. To calculate the picomoles of metal acquired per 108 organisms, 2–4 nmol of 59Fe citrate and 67Ga nitrate was counted.
Infection models.
All experiments were approved in advance by the Animal Care and Use Committee of the University of Iowa. For the acute infection experiments, 8- to 12-week-old C57BL/6 pathogen-free mice (The Jackson Laboratory) were anesthetized, and 50 μl of PBS containing 1 × 106 cfu/ml of PA103 were administered intratracheally. Fifty microliters of 250-mM Ga(NO3)3 or vehicle was given 3 hours later by intranasal administration. Some animals were pretreated with 10 μl of 2 mM FeCl3 in PBS with 2 mM sodium citrate, or sodium citrate alone. Animals were euthanized if they became moribund, distressed, or unable to eat or drink. The biofilm infection model was performed as described in refs. 37 and 38 using 2 × 107 CFU/ml mid-log-phase PA01 in 50 μl of PBS. Intranasal treatment with 50 μl of 250 mM Ga(NO3)3 or vehicle was begun 5 hours later and continued 3 times daily for 3 days. Animals were then sacrificed, the lungs were removed and homogenized, and bacteria enumerated by plate counting.
Statistics.
Survival was followed for 72 hours and the data were plotted on Kaplan-Meier curve. The Student’s t test (2-tailed) was performed for statistic analysis. A P value less than 0.005 was used to indicate a significant difference.
Supplementary Material
Acknowledgments
We thank S. Moskowitz, J. Burns, L. Saiman, and M. Olson for supplying clinical isolates and M. Vasil, C. Manoil, E. Banin, and E. Greenberg for supplying P. aeruginosa mutants. We thank M. Welsh, D. Hornick, M. Parsek, C. Manoil, R. Siehnel, E. Banin, and E. Greenberg for helpful discussions. This work was supported by the Cystic Fibrosis Foundation and the NIH.
Footnotes
Nonstandard abbreviations used: CF, cystic fibrosis; TSB, tryptic soy broth.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:877–888 (2007). doi:10.1172/JCI30783
Matthew Thoendel and Oyebode Olakanmi contributed equally to this work.
References
- 1.Norrby S.R., Nord C.E., Finch R. Lack of development of new antimicrobial drugs: a potential serious threat to public health. Lancet Infect. Dis. 2005;5:115–119. doi: 10.1016/S1473-3099(05)01283-1. [DOI] [PubMed] [Google Scholar]
- 2.Flamm R.K., et al. Factors associated with relative rates of antibiotic resistance in Pseudomonas aeruginosa isolates tested in clinical laboratories in the United States from 1999 to 2002. Antimicrob. Agents Chemother. 2004;48:2431–2436. doi: 10.1128/AAC.48.7.2431-2436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prince A.S. Biofilms, antimicrobial resistance, and airway infection. N. Engl. J. Med. 2002;347:1110–1111. doi: 10.1056/NEJMcibr021776. [DOI] [PubMed] [Google Scholar]
- 4.Costerton J.W., Stewart P.S., Greenberg E.P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 5.Stewart P.S. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 2002;292:107–113. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- 6.Bullen J.J., Rogers H.J., Spalding P.B., Ward C.G. Iron and infection: the heart of the matter. FEMS Immunol. Med. Microbiol. 2005;43:325–330. doi: 10.1016/j.femsim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 7.Griffiths, E. 1999. Iron in biological systems. In Iron and infection: molecular, physiological and clinical aspects. D.J. Bullen and E. Griffiths, editors. 2nd edition. John Wiley & Sons Ltd. Chichester, United Kingdom. 1–26. [Google Scholar]
- 8.Forsberg C.M., Bullen J.J. The effect of passage and iron on the virulence of Pseudomonas aeruginosa. J. Clin. Pathol. 1972;25:65–68. doi: 10.1136/jcp.25.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh P.K., Parsek M.R., Greenberg E.P., Welsh M.J. A component of innate immunity prevents bacterial biofilm development. Nature. 2002;417:552–555. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- 10.Rogan M.P., et al. Loss of microbicidal activity and increased formation of biofilm due to decreased lactoferrin activity in patients with cystic fibrosis. J. Infect. Dis. 2004;190:1245–1253. doi: 10.1086/423821. [DOI] [PubMed] [Google Scholar]
- 11.Banin E., Vasil M.L., Greenberg E.P. Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11076–11081. doi: 10.1073/pnas.0504266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berlutti F., et al. Iron availability influences aggregation, biofilm, adhesion and invasion of Pseudomonas aeruginosa and Burkholderia cenocepacia. Int. J. Immunopathol. Pharmacol. 2005;18:661–670. doi: 10.1177/039463200501800407. [DOI] [PubMed] [Google Scholar]
- 13.Xu Q., Dziejman M., Mekalanos J.J. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. U. S. A. 2003;100:1286–1291. doi: 10.1073/pnas.0337479100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown M.R.W., Anwar H., Lambert P.A. Evidence that mucoid Pseudomonas aeruginosa in the cystic fibrosis lung grows under iron-restricted conditions. FEMS Microbiol. Lett. 1984;21:113–117. [Google Scholar]
- 15.Snyder J., et al. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 2004;72:6373–6381. doi: 10.1128/IAI.72.11.6373-6381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Windus D.W., Stokes T.J., Julian B.A., Fenves A.Z. Fatal Rhizopus infections in hemodialysis patients receiving deferoxamine. Ann. Intern. Med. 1987;107:678–680. doi: 10.7326/0003-4819-107-5-678. [DOI] [PubMed] [Google Scholar]
- 17.Cornelis P., Matthijs S. Diversity of siderophore-mediated iron uptake systems in fluorescent pseudomonads: not only pyoverdines. Environ. Microbiol. 2002;4:787–798. doi: 10.1046/j.1462-2920.2002.00369.x. [DOI] [PubMed] [Google Scholar]
- 18.Chitambar C.R., Narasimhan J. Targeting iron-dependent DNA synthesis with gallium and transferrin-gallium. Pathobiology. 1991;59:3–10. doi: 10.1159/000163609. [DOI] [PubMed] [Google Scholar]
- 19.Bernstein L.R. Mechanisms of therapeutic activity for gallium. Pharmacol. Rev. 1998;50:665–682. [PubMed] [Google Scholar]
- 20.Smith E.E., et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chitambar C.R., Matthaeus W.G., Antholine W.E., Graff K., O’Brien W.J. Inhibition of leukemic HL60 cell growth by transferrin-gallium: effects on ribonucleotide reductase and demonstration of drug synergy with hydroxyurea. Blood. 1988;72:1930–1936. [PubMed] [Google Scholar]
- 22.Pina S.E., Mattingly S.J. The role of fluoroquinolones in the promotion of alginate synthesis and antibiotic resistance in Pseudomonas aeruginosa. Curr. Microbiol. 1997;35:103–108. doi: 10.1007/s002849900220. [DOI] [PubMed] [Google Scholar]
- 23.Rachid S., Ohlsen K., Witte W., Hacker J., Ziebuhr W. Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob. Agents Chemother. 2000;44:3357–3363. doi: 10.1128/aac.44.12.3357-3363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi A., Yomoda S., Ushijima Y., Kobayashi I., Inoue M. Ofloxacin, norfloxacin and ceftazidime increase the production of alginate and promote the formation of biofilm of Pseudomonas aeruginosa in vitro. J. Antimicrob. Chemother. 1995;36:743–745. doi: 10.1093/jac/36.4.743. [DOI] [PubMed] [Google Scholar]
- 25.Hoffman L.R., et al. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005;436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 26.Vorland L.H. Lactoferrin: a multifunctional glycoprotein. APMIS. 1999;107:971–981. doi: 10.1111/j.1699-0463.1999.tb01499.x. [DOI] [PubMed] [Google Scholar]
- 27.Weiner R., Hoffer P.B., Thakur M.L. Lactoferrin: its role as a Ga-67-binding protein in polymorphonuclear leukocytes. J. Nucl. Med. 1981;22:32–37. [PubMed] [Google Scholar]
- 28.Bjarnsholt T., et al. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology. 2005;151:373–383. doi: 10.1099/mic.0.27463-0. [DOI] [PubMed] [Google Scholar]
- 29.Walters M.C., 3rd, Roe F., Bugnicourt A., Franklin M.J., Stewart P.S. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 2003;47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown M.R., Collier P.J., Gilbert P. Influence of growth rate on susceptibility to antimicrobial agents: modification of the cell envelope and batch and continuous culture studies. Antimicrob. Agents Chemother. 1990;34:1623–1628. doi: 10.1128/aac.34.9.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wandersman C., Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 32.Kim E.J., Sabra W., Zeng A.P. Iron deficiency leads to inhibition of oxygen transfer and enhanced formation of virulence factors in cultures of Pseudomonas aeruginosa PAO1. Microbiology. 2003;149:2627–2634. doi: 10.1099/mic.0.26276-0. [DOI] [PubMed] [Google Scholar]
- 33.Ochsner U.A., Wilderman P.J., Vasil A.I., Vasil M.L. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 2002;45:1277–1287. doi: 10.1046/j.1365-2958.2002.03084.x. [DOI] [PubMed] [Google Scholar]
- 34.Lamont I.L., Beare P.A., Ochsner U., Vasil A.I., Vasil M.L. Siderophore-mediated signaling regulates virulence factor production in Pseudomonasaeruginosa. Proc. Natl. Acad. Sci. U. S. A. 2002;99:7072–7077. doi: 10.1073/pnas.092016999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pankhaniya R.R., et al. Pseudomonas aeruginosa causes acute lung injury via the catalytic activity of the patatin-like phospholipase domain of ExoU. Crit. Care Med. 2004;32:2293–2299. doi: 10.1097/01.ccm.0000145588.79063.07. [DOI] [PubMed] [Google Scholar]
- 36.Miyazaki S., Matsumoto T., Tateda K., Ohno A., Yamaguchi K. Role of exotoxin A in inducing severe Pseudomonas aeruginosa infections in mice. J. Med. Microbiol. 1995;43:169–175. doi: 10.1099/00222615-43-3-169. [DOI] [PubMed] [Google Scholar]
- 37.Kaneko Y., et al. Effects of parenterally administered ciprofloxacin in a murine model of pulmonary Pseudomonas aeruginosa infection mimicking ventilator-associated pneumonia. Chemotherapy. 2001;47:421–429. doi: 10.1159/000048553. [DOI] [PubMed] [Google Scholar]
- 38.Kaneko Y., et al. Clarithromycin inhibits overproduction of muc5ac core protein in murine model of diffuse panbronchiolitis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2003;285:L847–L853. doi: 10.1152/ajplung.00216.2002. [DOI] [PubMed] [Google Scholar]
- 39.Langodegard M., Wibetoe G. Determination of gallium in soil by slurry-sampling graphite-furnace atomic-absorption spectrometry. Anal. Bioanal. Chem. 2002;373:820–826. doi: 10.1007/s00216-002-1365-0. [DOI] [PubMed] [Google Scholar]
- 40.Narasimhan J., Antholine W.E., Chitambar C.R. Effect of gallium on the tyrosyl radical of the iron-dependent M2 subunit of ribonucleotide reductase. Biochem. Pharmacol. 1992;44:2403–2408. doi: 10.1016/0006-2952(92)90686-d. [DOI] [PubMed] [Google Scholar]
- 41.Hassett D.J., et al. Cloning and characterization of the Pseudomonas aeruginosa sodA and sodB genes encoding manganese- and iron-cofactored superoxide dismutase: demonstration of increased manganese superoxide dismutase activity in alginate-producing bacteria. J. Bacteriol. 1993;175:7658–7665. doi: 10.1128/jb.175.23.7658-7665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drobyski W.R., Ul-Haq R., Majewski D., Chitambar C.R. Modulation of in vitro and in vivo T-cell responses by transferrin-gallium and gallium nitrate. Blood. 1996;88:3056–3064. [PubMed] [Google Scholar]
- 43.Singh P.K., Tack B.F., McCray P.B., Jr., Welsh M.J. Synergistic and additive killing by antimicrobial factors found in human airway surface liquid. Am. J. Physiol. Lung Cell Mol. Physiol. 2000;279:L799–L805. doi: 10.1152/ajplung.2000.279.5.L799. [DOI] [PubMed] [Google Scholar]
- 44.Banin E., Brady K.M., Greenberg E.P. Chelator-induced dispersal and killing of Pseudomonas aeruginosa cells in a biofilm. Appl. Environ. Microbiol. 2006;72:2064–2069. doi: 10.1128/AEM.72.3.2064-2069.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyer J.M., Abdallah M.A. The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physicochemical properties. J. Gen. Microbiol. 1978;107:319–328. [Google Scholar]
- 46.Schuster M., Lostroh C.P., Ogi T., Greenberg E.P. Identification, timing and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lequette Y., Lee J.H., Ledgham F., Lazdunski A., Greenberg E.P. A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit. J. Bacteriol. 2006;188:3365–3370. doi: 10.1128/JB.188.9.3365-3370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.