Abstract
Triple-helix-forming oligonucleotides (TFOs) bind in the major groove of double-stranded DNA at oligopyrimidine⋅oligopurine sequences and therefore are candidate molecules for artificial gene regulation, in vitro and in vivo. We recently have described oligonucleotide analogues containing N3′-P5′ phosphoramidate (np) linkages that exhibited efficient inhibition of transcription elongation in vitro. In the present work we provide conclusive evidence that np-modified TFOs targeted to the HIV-1 polypurine tract (PPT) sequence can inhibit transcriptional elongation in cells, either in transient or stable expression systems. The same constructs were used in transient expression assays (target sequence on transfected plasmid) and in the generation of stable cell lines (target sequence integrated into cellular chromosomes). In both cases the only distinguishable feature between the cellular systems is the presence of an insert containing the wild-type PPT/HIV-1 sequence, a mutated version with two mismatches, or the absence of the insert altogether. The inhibitory action induced by np-TFOs was restricted to the cellular systems containing the complementary wild-type PPT/HIV-1 target, and consequently can be attributed only to a triple-helix-mediated mechanism. As a part of this study we also have applied an imaging technique to quantitatively investigate the dynamics of TFO-mediated specific gene silencing in single cells.
Triplex-forming molecules recognize the double-helical DNA by sequence-specific binding in the major groove; they are among the most specific ligands of double-stranded DNA (1), together with some recently described small molecules such as synthetic hairpin oligoamides that bind in the minor groove (2). Thus triplex-based approaches are an attractive means to achieve targeted gene regulation and gene manipulation both in vitro and in vivo. In a variety of cell-free applications, triplex formation has been used successfully (3–5). In living cells triple-helix-forming oligonucleotides (TFOs) have been used to target specific DNA modifications resulting in site-specific mutagenesis and to inhibit gene expression. Mutations can be induced by using a TFO conjugated to a DNA-damaging agent, both on an exogenous plasmid vector (6, 7) and an endogenous chromosomal locus (8): the mutations are localized around the triplex site. Intracellular inhibition of transcription initiation induced by TFOs has been reported for various clinically relevant genes. But definitive proof of a triplex-mediated mechanism of inhibition is very rarely provided (9). In one example a mutated target has been used to provide an appropriate control. The TFO target site was located in a promoter region, and the TFO was competing with transcription factor binding: inhibition of transcription initiation was observed when the triplex was preformed in vitro on a plasmid target before cell transfection. Arresting the transcription elongation complex once it is launched on the DNA turns out to be more difficult to achieve than competing with transcription factor binding. Intracellular inhibition of transcription elongation by a noncovalent triplex has not been reported previously.
In the present study we report triplex-induced inhibition of transcription elongation in cell cultures using oligonucleotide analogues containing N3′-P5′ phosphoramidate (np) linkages that we previously have characterized as the best blockers of RNA synthesis in vitro (10, 11). To unambiguously demonstrate that triplex formation is the molecular mechanism by which TFOs inhibit gene expression we have designed cellular systems that differ only by the presence or the absence of the oligopyrimidine⋅oligopurine target site, located either in a transfected plasmid or in an endogenous gene. This approach provides us with an appropriate control system because all cellular components that could have nonspecific interactions with TFOs are the same in both cases. The dynamics of triplex-induced gene inhibition also was studied and quantified by using a noninvasive imaging technique for gene expression studies in single living cells. With this kind of technique, heterogeneities in a population of cells can be identified, which is difficult or impossible with conventional techniques that generally average over many cells.
Materials and Methods
Oligonucleotides.
Oligonucleotide analogues with N3′-P5′ np linkages were synthesized as described (12–15). Acridine and psoralen-5′ modified oligonucleotides were prepared as reported (refs. 16 and 17, respectively).
Plasmids.
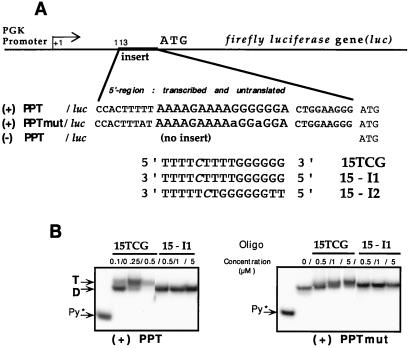
The phosphoglycerate kinase (PGK)/luc plasmids contain the firefly luciferase (Photinus pyralis) gene (luc) under the control of the murine PGK promoter. Two 34-bp inserts [5′-CCACTTTTT (PPT or PPTmut) CTGGAAGGG-3′] containing the wild-type HIV-1 polypurine tract (PPT: 5′-AAAAGAAAAGGGGGGA-3′) or a mutated sequence (PPTmut: 5′-AAAAGAAAAaGGaGGA-3′. Both were obtained after M13 mutagenesis (Sculptor kit, Amersham Pharmacia) and cloned in the 5′ transcribed but untranslated region of the luciferase gene (BamHI–HindIII fragment from the pGL2-basic vector, Promega), 113 bp downstream of the putative PGK start site (the PGK promoter is present on a retroviral vector, pTG6236, obtained from Transgene) (see Fig. 1).
Figure 1.
(A) Experimental design. The PGK/luc plasmids contain the firefly luciferase gene (luc) under control of the PGK promoter. An insert of 34 bp containing either the wild-type PPT [(+)PPT/luc] or a mutated PPT [(+)PPTmut/luc] has been cloned within the 5′ transcribed but untranslated region of the luciferase gene. Alternatively, there was no insertion of a 34-bp fragment [(−)PPT/luc]. Three sequences of 15-mer np oligonucleotides were used in the described experiments. The TFO (15TCG) perfectly matches the wild-type PPT. Two control oligonucleotides were used (15-I1 and 15-I2). The cytosines were methylated at the C5 position (in italics). Two 5′ substituted conjugates containing an acridine derivative (Acr-15TCG) or a psoralen derivative (Pso-15TCG) were used in some of the experiments. (B) Specificity of triplex formation. The double-stranded target containing the wild-type PPT [(+)PPT] or mutated PPT [(+)PPTmut] (for sequences see Materials and Methods) labeled on the 5′ end of the pyrimidine strand was incubated overnight in a buffer containing 50 mM Hepes, 50 mM NaCl, 10 mM MgCl2 in the presence of increasing concentrations (as indicated on the top of the gels) of 15TCG or control 15-I1 np oligonucleotide. The samples were analyzed by electrophoresis in a 15% nondenaturing polyacrylamide gel (50 mM Hepes, 10 mM MgCl2) at 37°C. The left lane on each gel corresponds to the radiolabeled pyrimidine strand alone (Py*). D, duplex; T, triplex.
The Renilla luciferase (Renilla reniformis; RL) control vectors (Promega) were used to monitor either transfection or microinjection efficiencies. The thymidine kinase (TK)/RL and simian virus 40 (SV40)/RL vectors contained the herpes simplex virus-TK promoter region or the SV40 viral promoter, respectively.
Cell Cultures.
The P4 cells were derived from HeLa cells (18). The Rama 27 cells are rat mammary fibroblast cells (19). Both were maintained in DMEM supplemented with 10% heat-inactivated FBS.
The CEM/luc cells were derived from the human lymphoid cell line CEM-SS, and they constitutively express firefly luciferase activity. Stable expression of luciferase was obtained after electroporation of the expression vector [PGK(±)PPT/luc plasmid] and selection for G418-resistant clones. In each case (±PPT), the PGK(±) PPT/luc DNA reporter gene was integrated within the cellular genome. Each clone (±PPT) expressed comparable levels of luciferase. The CEM(±)PPT/luc cells were grown in RPMI medium 1640 with 10% serum. Cells were routinely maintained at a density of 0.5–2 × 106 cells/ml.
Gel Shift Assay.
For gel shift assays we used two DNA duplexes containing either the PPT/HIV-1 oligopyrimidine⋅oligopurine sequence (29 bp long, with the following sequence of the oligopurine-containing strand: 5′-CCACTTTTT AAAAGAAAAGGGGGGA CTGG-3′) or the mutated version (34 bp long, with the following sequence of the oligopurine-containing strand: 5′-CACATTTTAT AAAAGAAAAaGGaGGA CTGGAAGG-3′). The pyrimidine-containing strand was end-labeled by using T4 polynucleotide kinase and γ-[32P]ATP. The duplex DNA was formed by mixing the labeled strand with the complementary strand at a ratio of 1:1.2. The duplex DNA (10-8 M) was incubated with increasing concentrations of oligomers in 50 mM Hepes (pH 7.2), 10 mM MgCl2, 50 mM NaCl, 10% sucrose. The nondenaturing polyacrylamide gel (15%) was run in 50 mM Hepes and 10 mM MgCl2 buffer at 37°C.
Transient Expression Experiments with an Exogenous DNA Target.
Transfections.
P4 cells were used for transfection of oligonucleotides and reporter vectors. Transfections were performed by using Superfect (cationic activated dendrimer; Qiagen, Chatsworth, CA) or Fugene 6 (Roche) reagents. Similar results were obtained with either transfection reagent. A typical transfection experiment was performed as follows: 0.5 μg of PGK/luc and 0.5 μg of TK/RL plasmids, and various quantities of oligonucleotides were mixed with Superfect or Fugene 6 (7 μl) in a total volume of 50 μl (serum-free medium). The mixture was added to P4 cells (105 cells per well spread in 24-well plates in 350 μl of serum-containing medium). The mixture was prepared for duplicates or triplicates. Values are presented as the mean (± SD) firefly/Renilla ratio (relative luciferase activity) of duplicate or triplicate measurements from a representative experiment that was independently repeated at least three times.
Luciferase assays.
Both luciferase activities (firefly and Renilla) were measured in cell extracts by using the dual-luciferase assay kit (Promega).
Microinjection and Single-Cell Imaging.
Rama 27 cells were microinjected (Eppendorf system) with the PGK/(±)PPT/luc plasmid (0.1 μg/μl in the microcapillary) and the SV40/RL DNA (0.01 μg/μl) in the presence or absence of oligonucleotides (1,600 μM of 15TCG and 540 μM of Acr-15TCG, concentrations in the needle). A volume of 10−14 liter was routinely injected, which corresponds to 50–100 times dilution from needle to cell. The cells were cultured on the microscope (Zeiss) on a heated stage at 37°C in a humidified incubator chamber in the presence of beetle luciferin (1 mM), the substrate for the firefly luciferase. Living cells were imaged for firefly luminescence at the time intervals indicated. Images were captured by using a photon-counting intensified charge-coupled device camera (VIM Hamamatsu Photonics, Hamamatsu City, Japan), and all images were integrated for 30 min. Both slice and center of gravity images were recorded: slice images for image display and center of gravity image for quantitative analysis, as described (20).
At the end of the experiment the luciferin-containing medium was washed off the cells, and an image was taken to ensure that no residual firefly luciferase luminescence remained. Then coelenterazine (5 μM), the substrate for the RL was added to the cells. Renilla luminescence was measured 15 min after coelenterazine addition (corresponding to the peak of luminescence) with a 5-min integration period. This Renilla image was used as a control to identify which cells had been successfully microinjected with the DNA mixture.
Stable Expression Experiments on an Endogenous Target.
Reversible cell permeabilization.
Streptolysin-O (SLO; Sigma) was used to reversibly permeabilize cells, as described (21). Briefly, CEM/luc cells were washed and resuspended in serum-free medium (5 × 106 cells in 200 μl); an optimized amount of SLO and 100 μM of various oligonucleotides were added. After a 10-min incubation at 37°C that allowed SLO-induced cell permeabilization, 1 ml of serum-containing medium was added for 20 min to reseal the cells. The cells then were diluted (10 ml at 0.5 × 106/ml), and samples were taken for flow cytometry, Northern blotting, and protein analysis 24 h after initiation of permeabilization. Results are presented as the mean (± SD) of duplicate measurements from a representative experiment that was independently repeated at least three times. Using the method described previously (22, 23), the amount of oligonucleotide per living cell was estimated at 10-17 mole.
RNA analysis.
Total cellular RNA was prepared by the guanidinium thiocyanate acid phenol method. After denaturing agarose gel electrophoresis, RNAs were transferred to nylon membranes (Amersham Pharmacia) by using standard procedures. Radiolabeled antisense RNA probes were used. They were obtained by using in vitro transcription (Ambion kit) of linearized DNA templates: pTri-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) plasmid (Ambion) for a 383-nt GAPDH probe and pGEM-luc plasmid (Promega) for a 1,660-nt luciferase probe. Quantification of Northern blots was obtained by PhosphorImager analysis. The GAPDH signal was used as a control to normalize; results are presented as an RNA ratio luc/GAPDH.
Protein analysis.
Samples were removed from cell suspensions; 0.5 ml from a total of 10 ml was used for lysate (passive lysis buffer, Promega) and 1 ml was stained with propidium iodide for flow cytometry analysis, which permitted the determination of the fraction of viable cells in the original culture. From every lysate both the firefly luciferase activity [expressed in relative light units (RLU)] and protein concentration [Bradford reagent from Bio-Rad, expressed in optical density (OD) were determined. The luciferase activity shown in Fig. 4 (in arbitrary units) corresponds to the luciferase activity for an equal quantity of living cells [RLU/(% viable cells ×) OD].
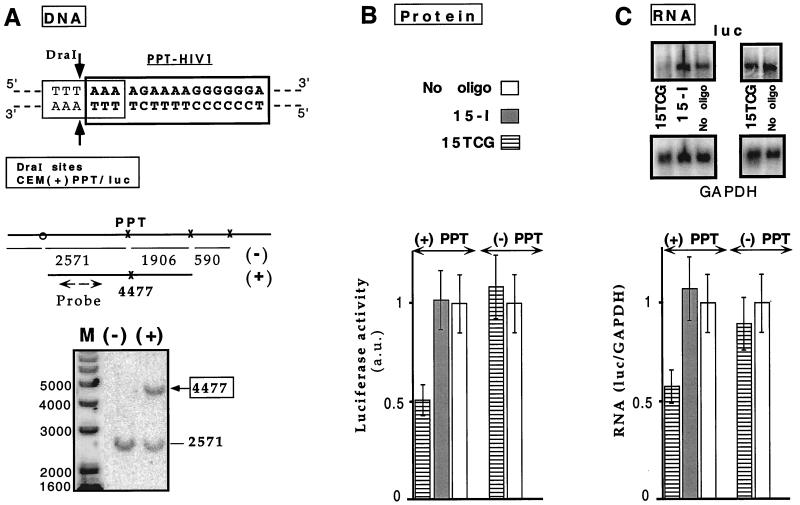
Figure 4.
Triplex inhibition of transcription elongation of an endogenous gene. (A) Target accessibility: DraI protection assay. (Upper) The PPT oligopyri-midine⋅oligopurine sequence (in bold type) overlaps DraI restriction site (thin line). The two arrows indicate the cleavage sites by the enzyme. DraI and AscI sites around the triplex formation site are indicated with x or o, respectively. AscI cleavage is used because no DraI site is present between the 5′ end of the PPT site and the integration site on the retroviral vector. The lengths of the fragments obtained after complete digestion are displayed. Specific triplex formation on the PPT site is revealed by the appearance of a longer fragment of 4,477 bp because of inhibition of DraI cleavage at the PPT site. The probe hybridization site is shown by a broken line. (Lower) An oligonucleotide psoralen conjugate (Pso-15TCG) was used as a probe for accessibility of the PPT target in the CEM(+)PPT/luc cell line. Cell nuclei were incubated in the presence [lane (+)] or in the absence [lane (−)] of Pso-15TCG and irradiated. DNA was extracted, digested by DraI and AscI, and analyzed by Southern blotting. Fragment lengths are indicated besides the gel. Lane M: DNA marker (1-kb marker, GIBCO/BRL). (B and C) Two cell lines expressing firefly luciferase reporter gene and that differ from each other only by the presence or the absence of a 34-bp insert containing the wild-type PPT sequence [CEM(+)PPT/luc and CEM(−)PPT/luc, respectively] were reversibly permeabilized with SLO in the absence (−) (empty bars) or presence of 15TCG or 15-I (referring to either 15-I1 or 15-I2 olignucleotides that exhibited equivalent effect). (B) Protein analysis. Luciferase activity per living cell was measured 24 h after oligonucleotide delivery (see Materials and Methods). Normalized values with respect to the SLO-treated cells in the absence of oligonucleotides are reported [luciferase activity in arbitrary units (a.u.)]. (C) RNA analysis. (Upper) Northern blots of total RNA extracted 24 h after oligonucleotide introduction were hybridized with luc and GAPDH probes (Left: CEM(+)PPT/luc cells; Right: CEM(−)PPT/luc cells). (Lower) Bar graph shows relative abundance of luc transcript compared with GAPDH mRNA (luc/GAPDH), normalized with the value obtained in the absence of oligonucleotide.
DNA analysis for triplex detection.
Covalent triplex was obtained by using a psoralen-modified oligonucleotide and UV light irradiation. Cell nuclei were prepared according to a digitonin-based protocol to bypass any limitations caused by oligonucleotide delivery, and then treated (5 × 107 cells/ml) with the psoralen-TFO (10 μM) and irradiated at 365 nm as described (17). Adducts were analyzed by DraI protection assay and standard Southern blotting.
Results and Discussion
Demonstration of Triplex Involvement in Inhibition of Transcription Elongation: Design of Cellular Systems.
TFOs previously have been used in cell cultures to inhibit transcription initiation through competition with transcription binding to DNA. To our knowledge no report has described the arrest of transcription elongation by TFOs, most probably because unmodified oligonucleotides do not form stable enough complexes with their DNA target. We previously have reported that oligomers with N3′-P5′ np linkages formed more stable triplexes and that transcription elongation could be inhibited in an in vitro assay using cell extracts (10, 11). The demonstration that TFOs could inhibit transcription elongation would considerably extend the range of applications of the antigene strategy in as much as both intronic and exonic sequences could be targeted in genes of interest.
The unambiguous demonstration of triplex involvement in the observed inhibition can be achieved only by direct detection of TFO binding to its double-stranded target and/or by the use of appropriate cellular control systems. The oligopyrimidine⋅oligopurine target that we have chosen is a 16-bp sequence abbreviated PPT that is present in the coding regions of two HIV-1 viral genes (integrase and nef). An oligonucleotide containing thymines, cytosine, and guanines (15TCG; see sequence in Fig. 1A) is the best ligand of the PPT sequence under conditions close to physiological ones (24). The temperature of half-dissociation of the 15TCG (np) from the PPT sequence (determined by spectroscopic measurement) was 62°C compared with 32°C for the phosphodiester isosequential oligonucleotide (at 1 μM triplex in a 10 mM sodium cacodylate buffer, pH 7.0 containing 100 mM NaCl and 10 mM MgCl2; ref. 10).
To rigorously test the mechanism of action of the 15TCG (np) in cell cultures, three different constructs were designed. They all contained the reporter firefly luciferase gene under the control of the PGK promoter. They only differed by the absence [in PGK(−)PPT/luc construct] or the presence of a 34-bp insert containing the wild-type sequence [PPT: 5′-AAAAGAAAAGGGGGGA-3′; in PGK(+)PPT/luc construct] or a mutated version [PPTmut: 5′-AAAAGAAAAaGGaGGA-3′, two mismatches indicated in lowercase; in PGK(+)PPTmut/luc construct]. The oligopyrimidine⋅oligopurine sequence was placed in the 5′ transcribed part of the luc gene, a region that is not translated (see Fig. 1A). These luciferase-expressing vectors were used both for transient expression assays and the establishment of two clonal cell lines (CEM/luc) constitutively expressing the firefly luciferase reporter gene after integration within the cellular genome of either the PGK(−)PPT/luc DNA [CEM(−)PPT/luc] or the PGK(+)PPT/luc construct [CEM(+)PPT/luc].
Specificity of triplex formation was studied in vitro on the wild-type and mutated target sequences by using a gel shift assay (Fig. 1B). Triplex DNA with 0.5 μM of 15TCG oligonucleotide was completely formed on the PPT/HIV-1 target whereas no triplex DNA was detected on the mutated PPT DNA target under the same conditions (Fig. 1B, lanes 15TCG/0.5 μM). In the case of mutPPT target, when increasing amounts of 15TCG are added the duplex is progressively retarded. This behavior probably is caused by unstable associations that tend to dissociate during gel migration and are unable to produce transcription inhibition (see Fig. 2). The control oligonucleotide with an inverted sequence (15-I1 = 3′-T4CT4G6–5′) was unable to bind either the wild-type or the mutated target even at 5 μM concentration. It can be noted that nine contiguous triplets theoretically could be formed with the T4CT4 symmetric sequence on the two target DNAs [(+)PPT and (+)PPTmut] with both oligonucleotides (15TCG and 15-I1). Another sequence (15-I2 = 3′-T5CTG6T2-5′) was used to abolish this partial complementarity of the T4CT4 sequence but maintain the G6 portion of the oligonucleotide.
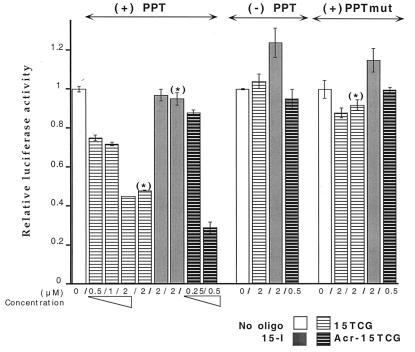
Figure 2.
P4 cell extracts were assayed for firefly and Renilla luciferase activities 24 h after transfection by cationic lipids. PGK/luc reporter constructs [(+)PPT, (+)PPTmut, (−)PPT] were introduced with a RL-expressing vector (TK/RL) in the presence or absence of various oligonucleotides, as indicated. Results are presented as normalized firefly/Renilla luminescence intensity ratios (relative luciferase activity). The use of either control oligonucleotide (15-I1 or 15-I2, indicated as 15-I) provided equivalent results. If two isolated transfections were performed to introduce plasmids first and oligonucleotides 3–7 h later, the same levels of modulation were observed [conditions indicated by (*)].
Triplex-Induced Inhibition of Transcription Elongation in Cells on an Exogenous Plasmid Target: Transient Expression Assay.
We evaluated the effect of TFOs on firefly luciferase expression from the different DNA templates that either contained or lacked the PPT/HIV-1 target sequence. P4 cells were transiently transfected with the PGK/luc reporter constructs [(+)PPT, (+)PPTmut, or (−)PPT], together with a RL-expressing vector (TK/RL), and various oligonucleotides. Both luciferase activities were measured on the same cell extracts. The RL expression provided an internal control for transfection efficiency and was used to normalize the firefly gene expression level. Consequently, the ratio of luminescence (firefly/Renilla) from the two luciferase activities was used to quantitate the modulation of the firefly luciferase expression induced by oligonucleotide treatment (Fig. 2; relative luciferase activity).
The 15TCG oligonucleotide induced an inhibition of firefly luciferase expression from the PGK(+)PPT/luc whereas the control oligonucleotides (15-I1 and 15-I2, globally referred as 15-I) did not have any effect. Deletion or mutation of the triplex target sequence abolished the inhibitory effect of the 15TCG oligonucleotide because PGK(−) PPT/luc and PGK(+)PPTmut/luc expression were not affected by the presence of the 15TCG oligonucleotide. These data demonstrated that inhibition was caused by interaction of the 15TCG oligonucleotide with the triplex target sequence. An effect of the oligonucleotide on the PPT sequence present on the mRNA can be excluded. In fact, the 15TCG can form a 9-bp duplex with the symmetric sequence present on the 5′ part of the PPT RNA (A4GA4), but exactly the same complex could be formed with the mRNA transcribed from the DNA containing the mutated PPT [(+)PPTmut], the expression of which is unaffected.
When the plasmid and oligonucleotide are mixed together with the transfection reagent, it cannot be excluded that the triplex is formed before cell uptake, during DNA (plasmid and oligonucleotide) association with the transfection reagent. To allow the triplex to unambiguously form inside cells, we performed two successive transfections. Using the same transfection reagents, the plasmids (PGK/luc and TK/RL) and the oligonucleotide were successively transfected within 3- to 7-h intervals. In this context the 15TCG was still able to inhibit firefly luciferase synthesis from the PGK(+)PPT/luc plasmid to the same extent as seen in the single transfection experiments (Fig. 2; conditions indicated by *). The control oligonucleotides (15-I) had no effect on the expression of either the PGK(+)PPT/luc or PGK(+)PPTmut/luc plasmid. These data demonstrated both the possibility of forming a triplex inside cells and the ability of a noncovalent complex formed with the 15TCG np to arrest transcription elongation.
The efficacy of triplex-induced inhibition was studied by using various np oligonucleotides in dose-response experiments, and comparison was made with a 15TCG oligonucleotide covalently linked to an acridine derivative (Acr-15TCG). Acridine derivatization enhanced the inhibitory effect of 15TCG on the expression of the wild-type reporter construct because 50% inhibition was obtained with 0.5 μM of Acr-15TCG as compared with 2 μM of unmodified 15TCG (Fig. 2). These results are consistent with the increase in triplex thermal stability that has been described previously for the isosequential phosphodiester oligonucleotides and is caused by the intercalation of the acridine moiety at the duplex-triplex junction (25).
Dynamics of Oligonucleotide-Induced Gene Expression Inhibition: Real-Time Imaging in a Single Living Cell.
Evaluation of triplex-induced effect on cell extracts reflects an average value concerning a cell population at a given time point. The use of luminescent reporter genes such as luciferases allows the imaging of gene expression noninvasively in real time and at the level of single living cells (26, 27). We therefore have applied this approach to investigate the dynamics of gene expression and its response to TFOs. The short half-life of firefly luciferase protein in the presence of luciferin (<1 h) makes it a suitable reporter gene for kinetic studies of gene expression. The nuclei of Rama 27 cells were microinjected with both the PGK(+)PPT/luc construct and a Renilla expression vector (SV40/RL) in the presence of various oligonucleotides. The plasmid expressing RL was coinjected as an internal control and as a marker for injected cells. Firefly luciferase expression can be directly measured. The cells were cultured under the microscope in the presence of luciferin that was stable in the culture medium for several days, and the intracellular concentrations of the other cofactors O2 and ATP were not limiting. Thus the intracellular luciferase luminescence was directly proportional to the level of gene expression and could be monitored in single living cells by using sensitive charge-coupled device cameras. Adjacent fields of cells were injected with the plasmids in the presence or absence of oligonucleotide before simultaneous imaging (Fig. 3A).
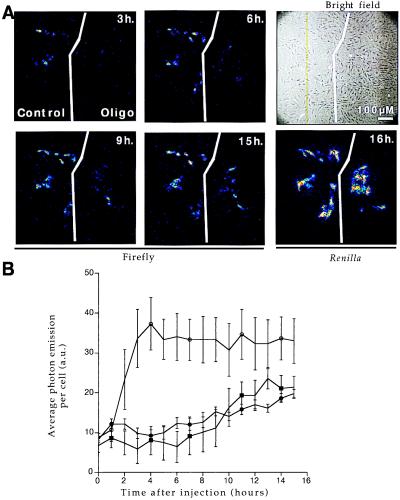
Figure 3.
RAMA 27 nuclei were comicroinjected with PGK(+)PPT/luc construct and a Renilla-expressing vector (SV40/RL) in the presence or absence of various oligonucleotides (15TCG at 1,600 μM, Acr-15TCG at 540 μM concentrations in the microcapillary). Cells were cultivated in the presence of 1 mM luciferin on the microscope stage so that firefly activity could be directly measured by photon output at regular intervals as indicated. Renilla expression was measured at the end of the experiment to determine the number of successfully microinjected cells. (A) Single-cell imaging. In the absence (control field) or presence (oligo field) of 540 μM Acr-15TCG oligonucleotide, firefly expression is shown at different times after injection: 3, 6, 9, and 15 h. Renilla expression was measured at 16 h. Bright-field image shows all of the cells in the microinjected area. (B) Quantification of firefly luciferase activity. Time course of firefly luciferase activity for different treatments: ○, control; ●, 15TCG at 1,600 μM; and ■, Acr-15TCG at 540 μM. Average photon emission per cell with integration periods of 30 min was determined by using Renilla activity to count the number of positive cells (n = 32, 19, and 18 for ○, ●, and ■, respectively). Control injections in the absence of oligonucleotide or presence of a control oligonucleotide (15-I1 at 1,600 μM) produced undistinguishable levels of firefly luciferase gene expression. Five arbitrary units (a.u.) corresponds to background photon emission level.
In the absence or in the presence of the control oligonucleotides, average photon output per cell increased to reach steady levels after 4–6 h and remained constant up to 16 h (Fig. 3B). In the presence of TFO light emission was significantly lower, reflecting inhibition of firefly luciferase expression. Several concentrations were used (data not shown), and a 3- to 4-fold decrease in the inhibitory concentrations was obtained by using the acridine-modified oligonucleotide as compared to the unmodified 15TCG.
It should be noted that the experimental conditions in the microinjection procedure are completely different from the ones used in the transfection assays described above. In the microinjection technique, the TFO was delivered and colocalized with the plasmid target at the initial time, in the cell nucleus where transcription takes place. In transfection mediated by cationic agents the two partners were delivered to the cytoplasm (most likely via endocytic vesicles) and they could migrate independently of each other to the nucleus.
In addition, the use of low-light imaging of single cells in real time allowed us to observe that there is significant variability in the level of gene expression in single cells over time and markedly different temporal responses to oligonucleotide treatment among individual and unsynchronized cells (Fig. 3A). A number of reasons may be considered for this observed heterogeneity, including intracellular variations in the expression of transcription factors because of cell cycle (independently of oligonucleotide treatment) and the modification of the intranuclear oligonucleotide concentration caused by cell division, for example.
Despite the variations mentioned above, our results clearly show that triplex-mediated inhibition of gene expression occurs at the single-cell level.
Triplex-Induced Inhibition of Transcription Elongation on an Endogenous Target: Proof of Concept.
For chromosomally located targets there has been no direct evidence yet that the inhibitory effect induced by a TFO really is caused by triplex formation on the oligopyrimidine⋅oligopurine target sequence. This is the reason stable cell lines were established containing the PGK(±)PPT/luc DNA fragment integrated in their genome and constitutively expressing the firefly luciferase [CEM(±)PPT/luc]. Two clonal cell lines were selected; they nearly expressed the same level of luciferase and differed only by the presence or absence of the 34-bp insert containing the PPT triplex site downstream of the PGK promoter region. This system allowed us to assess the potential of TFOs to block transcription elongation on a chromosomally located gene.
First, accessibility of the PPT sequence was directly analyzed by using a photochemical tool, the psoralen-modified oligonucleotide (Pso-15TCG) as described on another cellular system (17). Briefly, after photoactivation the triplex formed with the Pso-15TCG can be converted into an irreversible lesion (a psoralen crosslink) on cellular DNA. Cellular DNA then can be purified and analyzed by a DraI protection assay and Southern blotting. A DraI site overlaps the PPT triplex site, and triplex formation inhibits DraI cleavage at the PPT site (Fig. 4A Upper). Thus triplex formation on cellular DNA can be directly detected and quantitated by the modified DraI cleavage pattern (disappearance of the 2,571-bp fragment and appearance of a longer 4,477-bp fragment). In addition, the lack of protection of the neighboring sites reflects triplex specificity. To overcome any limitation in uptake and to deliver a nonlimiting concentration of TFO, a digitonin-based procedure was used (see Materials and Methods). This method allowed delivery of large amounts of TFO to the nucleus. Digitonin-treated cells were incubated with Pso-15TCG and irradiated; genomic DNA was purified and analyzed with the DraI assay. In the absence of oligonucleotide [Fig. 4A, lane (−)] the 2,571-bp fragment was observed; in the presence of Pso-15TCG [Fig. 4A, lane (+)] the 4,477-bp fragment appeared. Up to 50% of the PPT site could be protected from DraI cleavage. These data reflect the accessibility of the PPT sequence within the chromatin structure of the cellular DNA of CEM(+)PPT/luc cells and lend support to the possibility of inhibiting gene expression by a triplex-directed mechanism.
This possibility was tested on the same cell line with oligonucleotides being delivered by SLO reversible permeabilization. Typically this procedure allowed us to deliver oligonucleotide to more than 90% of the cell nuclei with less than 10% cell death (as determined by flow cytometry and confocal microscopy analysis, data not shown). The half-lives of the luciferase mRNA and protein in CEM(±)PPT/luc were determined to be 2 and 4 h, respectively, using actinomycin and cycloheximide to inhibit transcription and translation, respectively. The RNA and protein levels were analyzed 24 h after initiation of permeabilization, as indicated in Fig. 4. A specific decrease (50%) in luciferase protein level was observed in the presence of 15TCG in CEM(+)PPT/luc cells whereas no effect was detected in CEM(−)PPT/luc cells that do not contain the triplex site in the luc gene. The control oligonucleotide (15-I) did not affect luciferase expression in either CEM(+)PPT/luc or CEM(−)PPT/luc (Fig. 4B). Total RNA was analyzed for luciferase mRNA levels, and the GAPDH mRNA was used as an internal control (Fig. 4C). The 15TCG-induced reduction of luciferase protein expression was associated with a significant decrease of luciferase RNA expression (40%). The demonstration that transcription can be inhibited by oligophosphoramidates targeted to the transcribed region of a gene raises possibilities for the development of triplex-based strategies.
Conclusions
In the present work we have shown that transcription elongation can be sequence-specifically inhibited in cell cultures by using a TFO analogue with N3′-P5′ np linkages. Covalent attachment of an intercalating acridine derivative at the 5′ end of the oligomer enhanced the efficacy of the TFO as expected on the basis of the stabilization caused by intercalation at the triplex-duplex junction. The mechanism of transcription inhibition was demonstrated: (i) by using control oligonucleotides with the same base composition but different sequences (while maintaining the G6 tract that could have led to the formation of multistranded structures; ref. 28); and (ii) by mutating the target sequence so as to destabilize the triple-helical complexes or by suppressing the target site from the reporter gene. In addition, the dynamics of triplex-induced modulation of gene expression was followed by using an imaging technique, and the gene-specific inhibition by TFO also occurs at the single-cell level. The association of this kind of approach with fluorescence microscopy is likely to provide very powerful tools to correlate intracellular oligonucleotide distribution and gene expression modulation. This kind of information certainly will help the rational design of more potent oligonucleotides.
Even though triplex formation presently is restricted to oligopyrimidine⋅oligopurine sequences, the fact that triplex sites can be located downstream of the transcription start site, including exons and introns, considerably increases the number of potential target genes for triplex-based strategies. In addition, the present work as well as previous reports (8, 17, 29) have shown that target accessibility within the chromatin structure might not be as difficult to achieve as sometimes believed. TFO analogues represent an appropriate tool to investigate DNA accessibility as a function of e.g., cell cycle progression or induction of gene expression.
We can anticipate that a substantial increase in intracellular efficacy of triplex-induced effects will be achieved by modifications that could improve nuclease resistance, cellular uptake, absence of toxicity, and, especially, triplex stability of TFOs. At the present stage np-modified oligonucleotides have some of the greatest potential as antigene reagents and could be used either for the development of therapeutic drugs or as tools for further understanding of DNA-associated functions in chromatin context.
Acknowledgments
We thank D.G. Spiller and R.V. Giles for helpful technical assistance and discussions and D.M. Perrin for critical reading of the manuscript. We thank Hamamatsu Photonics for provision of equipment. We thank S. Gryaznov, D. Lloyd, and J.K. Chen for providing the first samples of oligophosphoramidates using the oxidative phosphorylation chemistry, and L. DeDionisio and A. Raible for many of the np syntheses via the np amine-exchange methods. This work was supported by Agence Nationale de Recherche sur le Sida and Centre International des Etudiants et Stagiaires. M.F. is supported by Comissao de Aperfeiçoamento de Pessoal de Nival Superior.
Abbreviations
- TFO
triple-helix-forming oligonucleotide
- PPT
polypurine tract
- np
phosphoramidate
- PGK
phosphoglycerate kinase
- RL
Renilla luciferase
- SV40
simian virus 40
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- TK
thymidine kinase
- SLO
streptolysin-O
References
- 1.Thuong N T, Hélène C. Angew Chem Int Engl. 1993;32:666–690. [Google Scholar]
- 2.White S, Szewczyk J W, Turner J M, Baird E E, Dervan P D. Nature (London) 1998;29:468–471. doi: 10.1038/35106. [DOI] [PubMed] [Google Scholar]
- 3.Wils P, Escriou V, Warnery A, Lacroix F, Lagneaux D, Ollivier M, Crouzet J, Mayaux J F, Sherman D. Gene Ther. 1997;4:323–330. doi: 10.1038/sj.gt.3300388. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z, Rana T M. Proc Natl Acad Sci USA. 1997;94:6688–6693. doi: 10.1073/pnas.94.13.6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giovannangeli C, Hélène C. Antisense Nucleic Acid Drug Dev. 1997;7:413–421. doi: 10.1089/oli.1.1997.7.413. [DOI] [PubMed] [Google Scholar]
- 6.Wang G, Levy D D, Seidman M M, Glazer P M. Mol Cell Biol. 1995;15:1759–1768. doi: 10.1128/mcb.15.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faruqi A F, Krawczyk S H, Matteucci M D, Glazer P M. Nucleic Acids Res. 1997;25:633–640. doi: 10.1093/nar/25.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Majumdar A, Khorlin A, Dyatkina N, Lin F-L M, Powel J, Liu J, Fei Z, Khripine Y, Watanabe K A, George J, et al. Nat Genet. 1998;20:212–214. doi: 10.1038/2530. [DOI] [PubMed] [Google Scholar]
- 9.Grigoriev M, Praseuth D, Guieysse A L, Robin P, Thuong N T, Hélène C. C R Acad Sci. 1993;316:492–495. [PubMed] [Google Scholar]
- 10.Escudé C, Giovannangeli C, Sun J S, Lloyd D H, Chen J K, Gryaznov S, Garestier T, Hélène C. Proc Natl Acad Sci USA. 1996;93:4365–4369. doi: 10.1073/pnas.93.9.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giovannangeli C, Perrouault L, Escudé C, Gryaznov S, Hélène C. J Mol Biol. 1996;261:386–398. doi: 10.1006/jmbi.1996.0471. [DOI] [PubMed] [Google Scholar]
- 12.Gryaznov S M, Chen J K. J Am Chem Soc. 1994;116:3143–3144. [Google Scholar]
- 13.Nelson J S, Fearon K L, Nguyen M Q, McCurdy S N, Frediani J E, Foy M F, Hirschbein B L. J Org Chem. 1997;62:7278–7287. doi: 10.1021/jo970801t. [DOI] [PubMed] [Google Scholar]
- 14.McCurdy S N, Nelson J S, Hirschbein B L, Fearon K L. Tetetrahedron Lett. 1997;38:207–210. [Google Scholar]
- 15.Fearon K L, Hirschbein B L, Nelson J S, Foy M F, Nguyen M Q, Okruszek A, McCurdy S N, Frediani J E, DeDionisio L A, Raible A M, et al. Nucleic Acids Res. 1998;26:3813–3824. doi: 10.1093/nar/26.16.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reed M W, Adams A D, Nelson J S, Meyer R B. Bioconjugate Chem. 1991;2:217–225. doi: 10.1021/bc00010a005. [DOI] [PubMed] [Google Scholar]
- 17.Giovannangeli C, Diviacco S, Labrousse V, Gryaznov S, Charneau P, Hélène C. Proc Natl Acad Sci USA. 1997;94:79–84. doi: 10.1073/pnas.94.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charneau P, Alizon M, Clavel F. J Virol. 1992;66:2814–2820. doi: 10.1128/jvi.66.5.2814-2820.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rudland P S, Twiston Davies A C, Tsao S W. J Cell Physiol. 1984;120:364–376. doi: 10.1002/jcp.1041200315. [DOI] [PubMed] [Google Scholar]
- 20.White M R G, Masuko M, Amet L, Elliott G, Braddock M, Kingsman A J, Kingsman S M. J Cell Sci. 1995;108:441–455. doi: 10.1242/jcs.108.2.441. [DOI] [PubMed] [Google Scholar]
- 21.Giles R V, Grzybowski J, Spiller D G, Tidd D M. Nucleosides Nucleotides. 1997;16:1155–1163. doi: 10.1080/07328319908044855. [DOI] [PubMed] [Google Scholar]
- 22.Spiller D G, Tidd D M. Anticancer Drug Des. 1992;7:115–129. [PubMed] [Google Scholar]
- 23.Spiller D G, Giles R V, Grzybowski J, Tidd D M, Clark R E. Blood. 1998;91:4738–4746. [PubMed] [Google Scholar]
- 24.Giovannangeli C, Montenay-Garestier T, Thuong N T, Hélène C. Proc Natl Acad Sci USA. 1992;89:8631–8635. doi: 10.1073/pnas.89.18.8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun J S, Giovannangeli C, François J C, Kurfurst R, Montenay-Garestier T, Asseline U, Saison-Behmoaras T, Thuong N T, Hélène C. Proc Natl Acad Sci USA. 1991;88:6023–6027. doi: 10.1073/pnas.88.14.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rutter G A, Kennedy H J, Wood C D, White M R H, Tavaré J M. Chem Biol. 1998;11:285–290. doi: 10.1016/s1074-5521(98)90287-3. [DOI] [PubMed] [Google Scholar]
- 27.Takasuka N, White M R H, Wood C D, Robertson W R, Davis J R E. Endocrinology. 1998;139:1361–1368. doi: 10.1210/endo.139.3.5826. [DOI] [PubMed] [Google Scholar]
- 28.Hawley P, Nelson J S, Fearon K L, Zon G, Gibson I. Antisense Nucleic Acid Drug Dev. 1999;9:61–69. doi: 10.1089/oli.1.1999.9.61. [DOI] [PubMed] [Google Scholar]
- 29.Belousov E S, Afonina I A, Kutyavin I V, Gall A A, Reed M W, Gamper H B, Wydro R M, Meyer R B. Nucleic Acids Res. 1998;26:1324–1328. doi: 10.1093/nar/26.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]