Abstract
Although hydrogens comprise half of the atoms in a protein molecule and are of great importance chemically and structurally, direct visualization of them by using crystallography is difficult. Neutron crystallography is capable of directly revealing the position of hydrogens, but its use on unlabeled samples faces certain technical difficulties: the large incoherent scattering of hydrogen results in background scattering that greatly reduces the signal to noise of the experiment. Moreover, whereas the scattering lengths of C, N, and O are positive, that of hydrogen is negative and about half the magnitude. This results in density for hydrogens being half as strong and close to the threshold of detection at 2.0-Å resolution. Also, because of its opposite sign, there is a partial cancellation of the hydrogen density with that from neighboring atoms, which can lead to ambiguities in interpretation at medium resolution. These difficulties can be overcome by the use of deuterated protein, and we present here a neutron structure of fully deuterated myoglobin. The structure reveals a wealth of chemical information about the molecule, including the geometry of hydrogen bonding, states of protonation of histidines, and the location and geometry of water molecules at the surface of the protein. The structure also should be of broader interest because it will serve as a benchmark for molecular dynamics and energy minimization calculations and for comparison with NMR studies.
The direct determination of hydrogen positions in protein crystallography is difficult. Because of their low scattering power for x-rays, hydrogens generally are not seen in protein structures determined by x-ray crystallography except at very high resolution. The scattering of neutrons by hydrogen or deuterium is comparable to that of other atoms in a protein, with the result that neutron diffraction, beginning with its first application to protein crystallography (1), has been used as an experimental tool to obtain hydrogen atom positions in a protein (2–5). However, this technique is limited in several ways. The intensity of even the strongest neutron sources today is several orders of magnitude weaker than a typical laboratory x-ray source. Consequently, one needs large crystals, with a volume greater than 1 cubic mm, to produce reflections that are strong enough to be detected. Even with large crystals, exposure times are long, normally in tens of minutes per degree of rotation, leading to data collection times of weeks or months. These difficulties are being addressed by improvements in instrumentation as well as in new methods of data collection (6). Two other technical problems are intrinsic to unlabeled samples. First, hydrogen atoms in a protein produce significant amounts of background, owing to their large incoherent scattering. As hydrogens can make up half of the atoms in a protein, the incoherent background scattering from them can be dominating in a weakly diffracting system such as a protein, making the measurement of weak reflections difficult or impossible. Second, the coherent scattering length of a hydrogen atom is negative, whereas those of the other atoms are positive. This opposite sign of the scattering length will lead to cancellation, both in the summation leading to the structure factors and in the resulting density. The former makes the average magnitude of the structure factor smaller, leading to smaller intensities in the diffraction pattern; the latter makes the interpretation of the map difficult. In previous neutron studies, these problems were partially addressed by using crystals soaked in mother liquor containing D2O. This method results in replacement of the exchangeable hydrogens with deuterium, and indeed, many of the specific hydrogens directly located in these studies belong to this class (7, 8). But exchangeable hydrogens account for only about 25% of the total hydrogen atoms. The only way to replace the nonexchangeable, covalently bound hydrogens by deuterium is by the synthesis of fully deuterated protein. We report here a neutron structure of fully deuterated sperm whale myoglobin (Mb), by using a single crystal and data to 2.0-Å resolution.
Materials and Methods
Plasmid DNA Manipulations.
A detailed description of the particular plasmid constructions selected is included here because it satisfies the requirements for good yield of the deuterated product and reproduces the exact wild type needed for crystallization. Two plasmids were constructed for this study: pET13Mb and pET13cIIFxMb. The former expresses the gene for wild-type Mb with its initial Met unprocessed, whereas the latter produces a fusion protein with a cleavage site right before the first residue, Val, of Mb, for the protease factor Xa. The plasmid pET13Mb was constructed by inserting a synthetic Mb gene (9) between the NdeI and BamHI sites in a T7 expression vector pET13a (10). Restriction sites for NdeI and BamHI were engineered, respectively, at the 5′ and 3′ ends of the Mb gene, using PCR with pUC19Mb (gift of S. Sligar, University of Illinois) as the template. The gene for the fusion protein was constructed by linking the gene encoding for the first 31 N-terminal residues of phage λ_cII protein (11) and the synthetic Mb gene, in the T7 expression vector pET13a. The phage lambda cll protein was chosen for its high solubility. As shown in Fig. 1A, the NdeI–BamHI fragment that contained the first 31 N-terminal residues of phage λ cII was excised from the plasmid pLcII (gift of K. Nagai, Medical Research Council, Cambridge, U.K.) and inserted between the NdeI and BamHI of pET13a sites to produce the vector pet13cII. The coding sequence for Mb was inserted into the BamHI site of pet13cII as follows: the coding sequence for sperm whale Mb was constructed so that a blood coagulation factor Xa cleavage site was added to the 5′ end of the synthetic Mb gene by PCR, while introducing BamHI restriction sites at both ends of the insert. The correct orientation of the Mb gene was verified by restriction digests of the recombinant plasmids. The Escherichia coli strain DH5α was used for plasmid manipulations, and standard cloning protocols were followed for the T7 expression system (12). The final construct, pET13cIIFxMb, coded for a fusion protein that consisted of an N-terminal leader from the λ_cII protein fused to the Mb gene, with a factor Xa cleavage site at the fusion junction. For reasons discussed below, the fusion construct was used in this study. The construct was verified by sequencing.
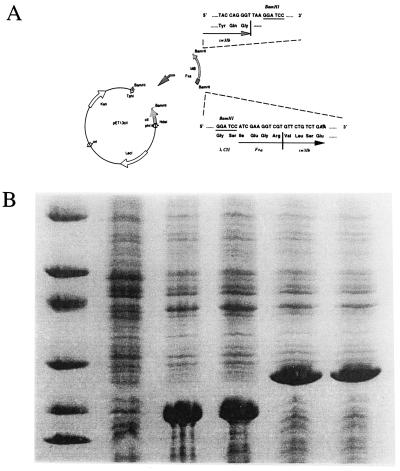
Figure 1.
Cloning and expression of Mb. (A) Construction of plasmid containing the Mb gene. pET13cIIFxMb was constructed by fusing the first 32 codons for the λ cII gene with the coding sequence for Mb. A cleavage site for the protease factor Xa was engineered at the junction between the cII and Mb coding sequences to allow cleavage right before the first residue of Mb. The coding sequence for the fusion protein was inserted between the NdeI and BamHI sites in vector pET13a. (B) Levels of overexpression of recombinant Mb in different expression vectors as visualized on a 5–25% PAGE gel, stained with Coomassie blue. Lane 1 shows molecular weight markers. Lane 2 contains a cell lysate of the culture containing the plasmid pUC19Mb in late saturation phase (kindly donated by S. Sligar). Lanes 3 and 4 contain lysates of cells containing the plasmid pET13Mb 3 h after induction at 37°C, in the strains BL21(DE3) and BL26(DE3), respectively. Lanes 5 and 6 contain lysates of cells containing the plasmid pET13cIIFxMb 3 h after induction at 37°C, in the strains BL21(DE3) and BL26(DE3), respectively. The level of expression is substantially higher in the T7 expression system (lanes 3–6). The plasmid pET13cIIFxMb in BL26(DE3) was used in this work.
Expression and Deuteration of Fusion Mb.
Fully deuterated Mb was expressed as a fusion apo-protein in the E. coli strain BL26(DE3), in fully deuterated minimal media [2 g/liter ND4Cl, 6 g/liter KD2PO4, 12 g/liter Na2DPO4, 0.4% (wt/vol) D6-succinate, 2 mM MgSO4, 25 mg/liter FeSO4·7D2O, 25 mg/liter kanamycin in 99.6% D2O]. First, E. coli cells were adapted, stepwise, to increasing concentrations of D2O in the medium, with unlabeled succinate as the carbon source. When the cells were adapted to medium containing 99.6% D2O, they were given deuterated succinate as a carbon source and grown in a closed system in which the air supply first was filtered through drierite, and then saturated with 99.6% D2O. When the OD of the culture at 600 nm reached 1.0, the cells were induced by the addition, from a 50 mM stock solution in D2O, of isopropyl thiogalactoside (IPTG), to a final concentration of 0.1 mM. The temperature of the incubator was 37°C before induction and was reduced to 25°C immediately after the addition of IPTG. The cells were harvested 12–15 h after induction and stored at −76°C until further use.
Purification and Digestion of Fusion Mb.
All procedures were done at 0–4°C unless otherwise noted. Between 6 and 10 g of frozen BL26(DE3) cells were thawed at room temperature for about 10 min and resuspended in a glass-Teflon homogenizer in 50 ml of lysis buffer [50 mM Tris⋅HCl, pH 8.0/25% (wt/vol) sucrose/5 mM Na2EDTA/1 mM DTT/0.05 mM PMSF]. The cells were lysed by addition of 50 mg lysozyme and 0.08% sodium deoxycholate, followed by incubation on ice for 45 min. The lysed cells then were treated with 1 mg DNase I in the presence of 10 mM MgCl2 and 1 mM MnCl2 for 15 min. The DNA was precipitated by the dropwise addition of polyethylenimine to a final concentration of 0.3%. The cell debris, precipitated DNA, and other insoluble material were removed by centrifugation at 12,000 g force for 30 min. The pellet was extracted by resuspension in lysis buffer and recentrifugation, and the supernatant was combined with the previous one. Most of the overexpressed Mb was present in the supernatant in the apolipoprotein form. For its reconstitution with heme, the pH of the supernatant was increased to 9.0 by adding concentrated CH3NH2. Solid heme from Sigma was dissolved in a minimal volume of 0.2 M NaOH and added to the protein solution until the ratio of absorption of the protein solution at 410 nm and 280 nm stopped increasing. The pH of the supernatant containing the reconstituted Mb was adjusted to 6.0 by using concentrated acetic acid, and then the sample was loaded onto a 2.5-cm × 30.0-cm S-Sepharose Fast-Flow (Amersham Pharmacia) column equilibrated with 50 mM Na/K PO4, pH 6.0/0.05 mM PMSF/1 mM DTT. The fusion Mb was eluted by a linear gradient of 0–0.4 M NaCl. Fractions containing Mb were collected, concentrated to a volume of about 10 ml, and dialyzed into digestion buffer (50 mM Tris⋅HCl, pH 9.0/0.1 M NaCl/1 mM CaCl2). Before digestion, the protein sample was further concentrated to more than 8 mg/ml. Trypsin (Sigma) instead of factor Xa, was used to liberate genuine Mb (13); 90 mg of trypsin was used for 1 g of Mb. The reaction mixture was incubated at room temperature for 12–16 h; then, the trypsin was inhibited by the addition of 1 mM PMSF. Mb was separated from the partially digested products on a 2.5-cm × 35-cm S-Sepharose Fast-Flow (Amersham Pharmacia) column equilibrated with 50 mM Na/K PO4, pH 6/0.05 mM PMSF. Mb eluted at about 0.24 M NaCl, and the partially digested proteins came off at about 0.16 M NaCl when a 0–0.4 M NaCl linear gradient was applied. The appropriate Mb fractions then were applied to a 2.5-cm × 120-cm S-100 filtration column equilibrated with 50 mM Na/K PO4, pH 6/0.05 mM PMSF/0.5 M NaCl for final purification.
Crystallization and Diffraction Data Collection.
Crystals for diffraction studies were grown by using the sitting-drop vapor diffusion method in 9-well depression plates sealed in plastic boxes. Each well contained 100–200 μl of 30 mg/ml protein in 45% saturated (NH4)2SO4/50 mM Na/K PO4, pH 6.2. The plate was placed in a plastic box with an equilibration reservoir of 25 ml of 70% (NH4)2SO4. The protein solution was equilibrated overnight against the precipitant, then was seeded with a 10-4 dilution of a small crystal that was finely crushed in 100 μl of 65% saturated (NH4)2SO4/50 mM Na/K PO4, pH 6.2. A large crystal of about 2.5 mm3 in volume was transferred to D2O-based mother liquor 8 months before it was mounted in a 5-mm quartz capillary for data collection. Neutron data were collected on the H3A station at the High Flux Beam Reactor at Brookhaven National Laboratory (14). The data were 52% complete to 1.9 Å and 95% complete to 2.0 Å. The data were integrated and merged by using madnes (15) to give a merging R factor of 10.8% on intensities. The x-ray coordinates of fully deuterated Mb, obtained by refinement against 1.5-Å x-ray data, were used as a starting point for neutron refinement. The initial x-ray analysis was carried out on a perdeuterated met-Mb crystal at 1.5-Å resolution and is described in the Ph.D. thesis of F.S. (16). The structure refined to an R factor of 17.1% by using a combination of x-plor and shell solvent refinement. The initial model for neutron refinement was calculated by applying the appropriate neutron-scattering lengths (17) to these refined x-ray coordinates, with all deuteriums except the ones bound to histidines based on stereo chemistry. Special care was taken when deuterium atoms were built in the model. Nonexchangeable deuterium atoms were first included in the model, then were checked for existence by running simulated annealing omit maps (18) omitting one residue at a time. Strong positive density at deuterium positions in these omit maps of ordered residues confirmed the high-level deuteration of Mb by the method. Deuterium atoms bonded to the 12 histidines were omitted in the initial model, then were added based on examination of difference Fourier maps. Adjustment was made later on in the refinement by running simulated annealing omit maps on each specific histidine residue. A refined MbCO structure was used as a reference to check the protonation of histidines (19). The 8,360 unique reflections between 6.0 and 2.0 Å were used to determine and refine the structure by using x-plor (18), which was modified to include all hydrogen atoms. There are a total of 2,544 atoms in the holo Mb, 30 hydrogens (all in heme), 1,254 deuteriums, and 1,260 others. Both stereo chemical and energy constraints, as defined in x-plor, were used, resulting in 7,623° of freedom compared with 8,360 independent observations. During positional refinement using x-plor, water molecules were added and restrained to their locations based on features in difference maps with peaks 3.0 sigma (20). All measured reflections were used. During refinement, 10% of the data were left out for cross-validation using the free R factor (21). After four cycles of positional and grouped B factor refinement, with manual adjustment on the graphics display between cycles, the R factor dropped from 30.4% to 15.6% and Rfree from 30.4% to 24.6%. The refined model has very good geometry as judged by procheck (22), with a deviation from ideal geometry of 0.011 Å in bond lengths and 1.205° in bond angles.
Results
High-Level Overexpression of Mb.
This project required a number of preparative obstacles to be overcome. The synthetic gene for Mb (9) when expressed results in a protein with an N-terminal methionine, which does not produce the large P21 crystals characteristic of naturally occurring sperm-whale Mb. Thus a fusion product was constructed that would liberate the authentic protein upon proteolytic cleavage, following the general strategy used previously for the beta globin chain of hemoglobin (11) or porcine Mb (23). Second, the T7 expression system was used to obtain very high yields of protein. Fusion Mb was overexpressed in E. coli to a very high level, in both protonated and deuterated media, comprising 40–50% of the total cellular protein as assessed by scanning densitometry of Coomassie-stained SDS polyacrylamide gels. As a comparison, Fig. 1B shows the level of expression of Mb, in both its intact and fusion forms, in its original plasmid pUC19 (9) and the T7 expression plasmid pET13cIIFxMb that we constructed in this study. The T7 system clearly expressed the gene at levels at least 10 times higher than the pUC19 system, which allowed us to obtain the quantities required to grow large crystals for neutron diffraction.
A third problem was that at 37°C, although we had high levels of expression, only about 30% of the product was soluble. Extraction of the protein in urea followed by renaturation resulted in unacceptably low yields of the final product. This problem was circumvented by induction of the cells at 25°C instead of 37°C, with less isopropyl thiogalactoside, 0.1 mM instead of 0.4 mM. At this lower induction temperature, about 85% of the overexpressed protein was soluble. Using the modified procedures, we obtained about 40–50 mg of pure fully deuterated Mb per liter of culture.
The deuteration level of the sample was determined by NMR on the protein solution obtained by dissolving small crystals soaked in the same mother liquor as the crystal used for data collection. The NMR result showed that the apo-protein was over 98% deuterated, whereas the heme was hydrogenated as expected (G. Gupta, personal communication).
Crystal Space Group.
The recombinant Mb, in both protonated and fully deuterated forms, gave the genuine wild-type Mb sequence after trypsin cleavage. The postcleavage Mb in both forms has identical absorption spectrum as the wild type. They both crystallized in the same P21 space group as native Mb, with cell parameters of a = 64.5 Å, b = 30.9 Å, c = 34.8 Å, and β = 105.8°. It has been reported that synthetic Mb with the initiator Met crystallizes in the P6 space group and is too small for neutron studies (24).
Enhancement of Density at H Atoms.
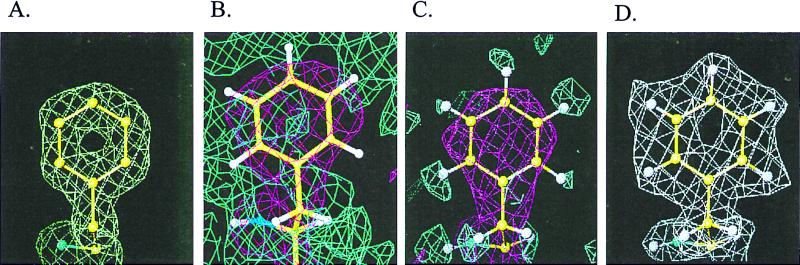
Fourier density maps from the data on fully deuterated Mb clearly are superior to those obtained previously by neutron diffraction. At least part of the improvement in the data are caused by a 3-fold improvement in the signal-to-noise ratio over unlabeled crystals in D2O mother liquor (unpublished results). The advantage of fully deuterating the protein to locate hydrogen positions in a neutron map can be seen in Fig. 2. Fig. 2A shows that a 2Fo−Fc x-ray map even at 1.5 Å cannot reveal the positions of hydrogens. A neutron 2Fo−Fc at 2-Å resolution (19) on unlabeled Mb shows that the level of noise makes it difficult to visualize hydrogen atoms directly (Fig. 2B). Even the best-case scenario for an unlabeled protein, an Fc neutron map calculated from the current model with all deuteriums replaced by hydrogens and using equivalent experimental data at 2.0 Å (Fig. 2C), shows that the dispersion of the neutron density from hydrogens and the neighboring carbon atoms gives rise to significant overlap, which results in cancellation between the negative contribution from the H atoms and positive contribution from the C atoms. The cancellation results in apparent cavities near the C atoms in the positive contour map and pushes the negative hydrogen density peak away from the center of the hydrogen atoms. Because the negative density peaks of hydrogens do not reside on top of the hydrogen centers makes unambiguous determination of their positions difficult. Further, the location of the negative peaks are strongly affected by properties of neighboring atoms, such as their temperature factor and the accuracy of their positions (5). The negative densities for hydrogens are also closer to the threshold of noise, again leading to difficulties in interpretation. These limitations all are overcome when a fully deuterated sample is used. As a consequence, deuterium positions can easily be determined because they have the same positive density as other types of atoms (Fig. 2D).
Figure 2.
Deuterium (hydrogen) atoms can be located directly as positive peaks in 2Fo−Fc maps as illustrated by residue Phe-43. (A) The 2Fo−Fc x-ray map of fully deuterated Mb using 6.0- to 1.5-Å data, contoured at +1.0 σ (0.84 e-/Å3). (B) A 2Fo−Fc neutron map on unlabeled Mb (19) calculated to 2.0-Å resolution, with the pink map contoured at 1.0 σ and the blue map contoured at −1.0 σ. (C) An Fc neutron map generated by using equivalent experimental reflections in 6.0 to 2.0 Å calculated from the current protein model except that D was replaced with H. The pink map is contoured at +1.0 σ; the blue map is contoured at −2.0 σ. (D) The 2Fo−Fc neutron map of fully deuterated Mb using 6.0- to 2.0-Å data, contoured at +1.0 σ (1.03 fermi/Å3).
Histidine Protonation States.
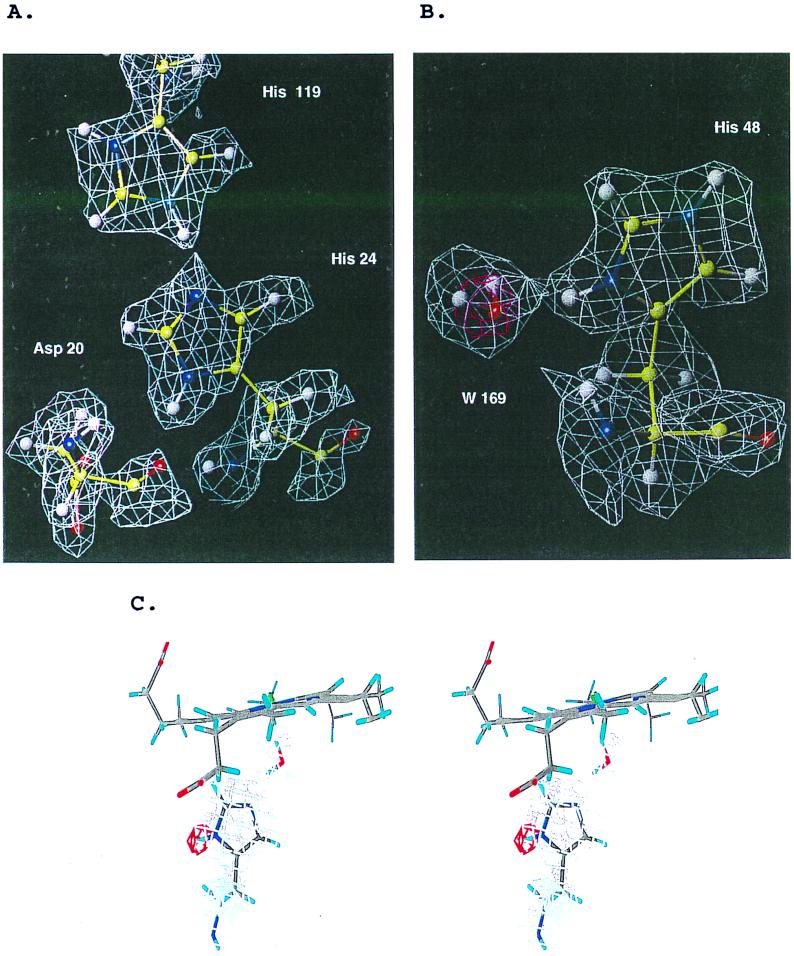
In this study, the protonation states of all 12 histidines in Mb at pH 6.2 can be determined (Table 1). There are four histidine residues (His-12, His-36, His-113, and His-116) that show slow exchange (19). An additional five histidines are buried (His-24, His-64, His-82, His-93, and His-97). The shared hydrogen between the histidine pair His-24 and His-119 is shown in Fig. 3A. Fig. 3B shows His-64 and His-93 at the heme pocket: Both of them are deprotonated at Nɛ position at pH 6.2. The protonation of His-64 is of special interest. A previous neutron study of met Mb at pH 5.6 showed that His-64 was protonated at the Nɛ position, deprotonated at Nδ (25), just as in oxy-Mb at pH 8.2 (4). But our data show that at pH 6.2, His-64 is deprotonated at the Nɛ position, just as in MbCO at pH 5.6 (19). The map in Fig. 3B indicates that the ligand water is hydrogen-bonded to the distal histidine His-64 Nɛ whereas the oxygen forms the sixth coordination of the ion.
Table 1.
Protonation of histidines in sperm whale Mb at pH 6.2
| Residue | Tautomer | Q | pKa | pKb | pKc |
|---|---|---|---|---|---|
| 12 | ɛ | 0.70 | — | 6.38 | 6.29 |
| δ | 0.49 | 6.28 | 6.39 | 6.26 | |
| 24 | ɛ | 0.00 | — | — | — |
| δ | 1.00 | — | — | — | |
| 36 | ɛ | 0.76 | 7.97 | 8.18 | 8.06 |
| δ | 1.00 | — | — | 7.91 | |
| 48 | ɛ | 0.73 | 6.73 | 5.48 | 5.25 |
| δ | 0.96 | — | 5.52 | 5.30 | |
| 64 | ɛ | 0.00 | — | — | <5 |
| δ | 0.94 | — | — | <5 | |
| 81 | ɛ | 0.88 | 6.17 | 6.73 | 6.68 |
| δ | 0.45 | — | 6.69 | 6.53 | |
| 82 | ɛ | 0.95 | — | <4.8 | <5 |
| δ | 0.25 | — | <4.8 | <5 | |
| 93 | ɛ | 0.43 | — | — | <5 |
| δ | 0.86 | — | — | <5 | |
| 97 | ɛ | 1.00 | — | — | 5.63 |
| δ | 0.29 | — | — | — | |
| 113 | ɛ | 0.26 | 5.49 | ≈5 | 5.44 |
| δ | 1.00 | — | ≈5 | 5.36 | |
| 116 | ɛ | 0.06 | 6.55 | 6.60 | 6.49 |
| δ | 1.00 | — | 6.59 | 6.50 | |
| 119 | ɛ | 0.64 | — | 6.09 | 6.13 |
| δ | 0.84 | 5.46 | 6.20 | 6.13 |
Figure 3.
Protonation states of histidines were determined from omit maps using the simulated annealing procedure in x-plor (A). Shared hydrogen between His-119 and His-24. (B) The pink density around His-48 is the x-ray 2Fo−Fc map contoured at +2.0 σ, which shows the water oxygen position. The protonation at both tautomers are obvious. A water molecule is hydrogen-bonded to the Nɛ of His-48. (C) Detail of the protonation determination showing a stereo view of His-64 with the ɛ and δ hydrogens omitted from the refinement. The 2Fo−Fc Fourier detail in gray is contoured at the 1-σ level while the red contours shows the Fo−Fc map contoured at 2.5 σ. This map shows clearly that Nɛ is protonated and that Nδ is deprotonated.
The pK value for His-48 has been measured and predicted by several groups (26–28). Our result, along with results from these groups, is listed in Table 1. The map in Fig. 3C shows clearly that Nδ is protonated with a water molecule hydrogen-bonded to it. It agrees with the pK value 6.7 as observed by Botelho and Gurd (26), but is in disagreement with recent NMR studies, which showed pK values of 5.5 (27) and 5.25 (28) for the His-48.
Water Orientation.
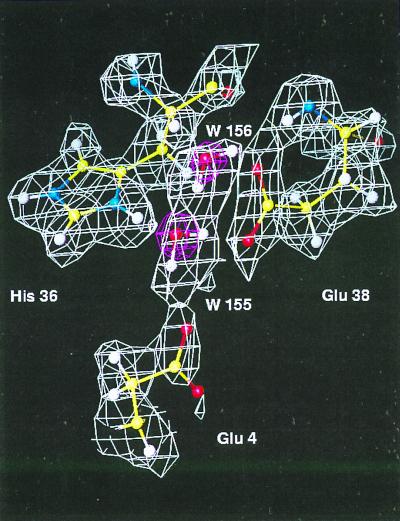
Because the scattering factor of deuterium is nearly twice that of hydrogen and of the same sign as that of oxygen, the visibility of water molecules in Fourier maps is enhanced. Water molecules W-155 and W-156 are shown in Fig. 4. The x-ray data reveal the positions of the water oxygens so that the deuterium (hydrogen) atoms therefore can be placed into the density with less uncertainty. As shown in the map, water W-155 forms four hydrogen bonds: one of its D(H) is hydrogen-bonded to Glu-4 Oδ1 (D-O distance = 1.73 Å); the other D(H) hydrogen-bonds to Glu-38 Oδ1 (D-O distance = 1.78 Å); its oxygen hydrogen-bonds to His-36 on one side, and to another water molecule W-156 on the other side. The distances between its oxygen and hydrogen donor Hs are 2.10 Å and 1.46 Å, respectively. A well-ordered histidine His-36 is shown in the back and is protonated at both N positions. In the current refined model, there are 69 water molecules; the majority of them are within 0.3 Å of their positions in the refined x-ray structure (there are 89 water molecules in the x-ray model) (16). The average B factors are 9.3 Å2, 14.7 Å2, and 33.04 Å2 for main-chain, side-chain, and water atoms, respectively. The B factors of the two water molecules, W-155 and W-156, shown here are 22 Å2 and 15 Å2, respectively.
Figure 4.
Orientation of water molecules as determined in neutron density maps. The white map is a 2Fo−Fc map calculated from 6.0- to 2.0-Å neutron data from fully deuterated Mb. The red one is the 2Fo−Fc x-ray map generated using a 1.5-Å data set collected on a fully deuterated Mb crystal, which was refined to an R factor of 17.5%.
Discussion
This study reveals the power of using fully deuterated samples in neutron crystallography to obtain information regarding hydrogen atoms in a protein. As can be seen from Fig. 2A, hydrogens are not apparent even at a resolution of 1.5 Å in x-ray diffraction map, because the x-ray scattering of atoms is proportional to the atomic number, so that the scattering of H (or D) by x-rays is 6–8 times smaller than other types of atoms. In contrast, neutron diffraction can see H (or D), because the scattering length of H (−0.374 × 10-12 cm) or D (0.667 × 10-12 cm) is comparable to those for other atoms (about 0.6 × 10-12 cm). However, the negative sign of the scattering length of H causes difficulties in interpretation. At finite resolution, the neutron scattering density from a Fourier synthesis of the diffraction data will have spatial broadening. When there is overlap of densities with opposite sign, such as those from H and C, cancellation will occur. Apart from this cancellation, the broadening also results in a weakening of the density. The combination of cancellation and weakening caused by peak broadening at finite resolution can reach a point at which the visualization of negative density at the hydrogen atom position becomes impossible because of noise in the data. A resolution that is high enough to completely overcome the cancellation problem is not normally achieved by protein crystals. In Fig. 2C, the negative map was contoured at −2.0 σ, as otherwise negative peaks caused by noise dominate the contours in a manner similar to Fig. 2B. It appears that a resolution of 2.0 Å is roughly the minimum resolution to unambiguously define hydrogens. In practice, the visualization of the negative density is further complicated by its sensitivity to the correct assignment of temperature factor to its connecting atoms and to the accuracy of their atomic positions. As can be seen in Fig. 2D, these problems are eliminated by deuteration of the protein: at 2.0-Å resolution, the neutron map from fully deuterated Mb clearly shows the deuterium atom positions.
The protonation states of specific histidines in Mb have long been a topic of interest. They not only are related to the function of the protein, but also serve as a basis to model electrostatic interactions in proteins (26–30). Neutron crystallography can directly determine the protonation states of specific histidines (5, 19). However, although neutron diffraction on unlabeled Mb soaked in D2O mother liquor was able to determine the protonation states of exposed histidines, its ability to determine the state of buried histidines, for which exchange is restricted, is limited by the uncertainty in interpreting the negative density of the hydrogen atoms.
Unlike the case in previous neutron studies, every hydrogen is replaced by deuterium in the protein. This allowed the determination of protonation as positive peaks near the Nɛ or Nδ atoms of histidines. As mentioned in Materials and Methods, the accurate determination of the protonation of these histidines requires extra care to overcome problems caused by the negative density from hydrogens. In this study, the protonation of the various histidines was determined by the positive peaks in the difference Fourier maps when the corresponding hydrogens had not been included in the starting model, and their occupancies were refined in the final step of refinement. Moreover all residues were checked by simulated annealing omit maps during refinement. Fig. 3A shows a shared hydrogen between His-119 and His-24. His-24 is deprotonated whereas His-119 is protonated. This finding is consistent with the previous neutron study on MbCO by Cheng and Schoenborn (19) and also agrees with the NMR titration study by Botelho and Gurd (26) (see Table 1). His-93 is deprotonated at the Nɛ position as observed in the crystal structures of all forms of Mb and in NMR titration studies. However, the protonation of His-64 depends on the Mb oxidation status. In the neutron structure of MbCO at pH 5.6, it is deprotonated at the Nɛ position (19). In met-Mb at pH 5.6, it is protonated (25). In oxy-Mb at pH 8.4, it is protonated (4). In this work on met-Mb at pH 6.2, it is deprotonated. Although Mb has been studied extensively, an explanation for its ability to favor certain ligands remains unknown (31, 32). Our data on the His-64 protonation suggest that the O2 and H2O ligands are mediated by H-bonds to the Nɛ of His-64. The H2O ligand, with the stronger H-bond to this histidine (Fig. 3B), is the more stable. On the other hand, CO does not form an H-bond to this histidine; were it able to do so, it would be even more stable and increase the CO affinity even more.
Because D2O molecules diffract neutrons strongly (0.635 fermi/Å3), ordered water molecules can be detected as easily in a neutron map as visualizing a sulfate ion in an x-ray map. With the help of an initial model obtained from x-ray diffraction, the procedure can be made more reliable, because the oxygen atom positions in the water density can be located by reference to the x-ray electron density map. The deuterium atoms of the waters then can be put into the density, from which the water orientation relative to the protein residues can be obtained.
In the current refined model, there are 69 water molecules with their corresponding deuterium positions, from which the required H-bonding geometry parameters can be obtained. This information eventually will help to define accurate parameters for H-bond geometry, which is badly needed for calculating hydrogen bond energy (33), used widely in applications such as molecular dynamics.
After initial enthusiasm in the 1970s and 1980s, the use of neutron crystallography has waned considerably. This is in part because of the development of techniques such as NMR that can answer questions that could previously only be answered by using neutrons. However, even for those problems for which neutron crystallography could provide the most definitive answers, its use has declined because of the unreliability of neutron sources over the last decade and the limitations of neutron instrumentation. This situation is changing. Both the reliability of sources as well as advances in neutron detectors and data collection strategies (6) suggest that the technique will undergo a resurgence.
This study demonstrates that a crystal of modest size, when it is fully deuterated, can produce good-quality neutron data to 2.0-Å resolution that give easily interpretable maps for the location of hydrogen (deuterium) atoms. Thus, deuteration of proteins should greatly expand the applicability of neutron crystallography and make possible work on proteins for which it is impossible to produce extremely large crystals. Apart from the additional information this reveals pertaining to the chemistry of Mb, this work provides a complete structural description of the molecule that can be used as a reference in other studies such as NMR, energy calculations, and molecular dynamics.
Acknowledgments
We acknowledge the late Z. Richard Korszun for his advice and help with this study. We also thank S. E. Gerchman, V. Graziano, and H. Kycia for advice and help with cloning, expressing, and deuterating Mb; G. Gupta for estimating deuteration levels by NMR; S. G. Sligar and K. Nagai for providing plasmids; and J. J. Dunn, R. M. Sweet, and D. K. Schneider for valuable discussions. This work was supported by the Office of Health and Environmental Research, U.S. Department of Energy.
Abbreviation
- Mb
myoglobin
Footnotes
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1CQ2).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.060024697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.060024697
References
- 1.Schoenborn B P. Nature (London) 1969;224:143–146. doi: 10.1038/224143a0. [DOI] [PubMed] [Google Scholar]
- 2.Schoenborn B P. Cold Spring Harbor Symp Quant Biol. 1971;36:569–575. doi: 10.1101/sqb.1972.036.01.071. [DOI] [PubMed] [Google Scholar]
- 3.Norvell J C, Nunes A C, Schoenborn B P. Science. 1975;190:568–570. doi: 10.1126/science.1188354. [DOI] [PubMed] [Google Scholar]
- 4.Phillips S E, Schoenborn B P. Nature (London) 1981;292:81–82. doi: 10.1038/292081a0. [DOI] [PubMed] [Google Scholar]
- 5.Kossiakoff A A, Spencer S A. Biochemistry. 1981;20:6462–6474. doi: 10.1021/bi00525a027. [DOI] [PubMed] [Google Scholar]
- 6.Niimura N, Minezaki Y, Nonaka T, Castagna J C, Cipriani F, Hoghoj P, Lehmann M S, Wilkinson C. Nat Struct Biol. 1997;4:909–914. doi: 10.1038/nsb1197-909. [DOI] [PubMed] [Google Scholar]
- 7.Kossiakoff A A. Annu Rev Biophys Bioeng. 1983;12:159–182. doi: 10.1146/annurev.bb.12.060183.001111. [DOI] [PubMed] [Google Scholar]
- 8.Kossiakoff A A. Annu Rev Biochem. 1985;54:1195–1227. doi: 10.1146/annurev.bi.54.070185.005555. [DOI] [PubMed] [Google Scholar]
- 9.Springer B A, Sligar S G. Proc Natl Acad Sci USA. 1987;84:8961–8965. doi: 10.1073/pnas.84.24.8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerchman S E, Graziano V, Ramakrishnan V. Protein Expression Purif. 1994;5:242–251. doi: 10.1006/prep.1994.1037. [DOI] [PubMed] [Google Scholar]
- 11.Nagai K, Thogersen H C. Nature (London) 1984;309:810–812. doi: 10.1038/309810a0. [DOI] [PubMed] [Google Scholar]
- 12.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:61–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 13.Varadarajan R, Szabo A, Boxer S G. Proc Natl Acad Sci USA. 1985;82:5681–5684. doi: 10.1073/pnas.82.17.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoenborn B P. In: Neutron in Biology. Schoenborn B P, editor. Vol. 27. New York: Plenum; 1984. pp. 261–279. [Google Scholar]
- 15.Messerschmidt A, Pflugrath J W. J Appl Crystallogr. 1987;20:306–315. [Google Scholar]
- 16.Shu F. Ph.D. thesis. Stony Brook: State University of New York; 1994. [Google Scholar]
- 17.Bacon G E. Neutron Diffraction. Oxford: Clarendon; 1975. [Google Scholar]
- 18.Brunger A T. x-plor. New Haven, CT: Yale Univ.; 1993. , Version 3.1. [Google Scholar]
- 19.Cheng X D, Schoenborn B P. J Mol Biol. 1991;220:381–399. doi: 10.1016/0022-2836(91)90020-7. [DOI] [PubMed] [Google Scholar]
- 20.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 21.Brunger A T. Nature (London) 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 22.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 23.Dodson G, Hubbard R E, Oldfield T J, Smerdon S J, Wilkinson A J. Protein Eng. 1988;2:233–237. doi: 10.1093/protein/2.3.233. [DOI] [PubMed] [Google Scholar]
- 24.Phillips G, Jr, Arduini R M, Springer B A, Sligar S G. Proteins. 1990;7:358–365. doi: 10.1002/prot.340070407. [DOI] [PubMed] [Google Scholar]
- 25.Raghavan N V, Schoenborn B P. Basic Life Sci. 1984;27:247–259. doi: 10.1007/978-1-4899-0375-4_15. [DOI] [PubMed] [Google Scholar]
- 26.Botelho L H, Gurd F R N. Biochemistry. 1978;17:5188–5196. doi: 10.1021/bi00617a019. [DOI] [PubMed] [Google Scholar]
- 27.Cocco M J, Kao Y-H, Phillips A T, Lecomte T J. Biochemistry. 1992;31:6481–6491. doi: 10.1021/bi00143a018. [DOI] [PubMed] [Google Scholar]
- 28.Bashford D, Case D A, Dalvit C, Tennant L, Wright P E. Biochemistry. 1993;32:8045–8056. doi: 10.1021/bi00082a027. [DOI] [PubMed] [Google Scholar]
- 29.Botelho L H, Friend S H, Matthew J B, Lehman L D, Hanania G I H, Gurd F R N. Biochemistry. 1978;17:5197–5205. doi: 10.1021/bi00617a020. [DOI] [PubMed] [Google Scholar]
- 30.Dalvit C, Wright P E. J Mol Biol. 1987;194:313–327. doi: 10.1016/0022-2836(87)90378-0. [DOI] [PubMed] [Google Scholar]
- 31.Service R F. Science. 1995;269:921–922. doi: 10.1126/science.7638612. [DOI] [PubMed] [Google Scholar]
- 32.Lim M, Jackson T, Anfinrud P A. Science. 1995;269:962–966. doi: 10.1126/science.7638619. [DOI] [PubMed] [Google Scholar]
- 33.Boobbyer D N, Goodford P J, McWhinnie P M, Wade R C. J Med Chem. 1989;32:1083–1094. doi: 10.1021/jm00125a025. [DOI] [PubMed] [Google Scholar]