Abstract
The synthesis of double-stranded DNA by a rolling circle mechanism was reconstituted in vitro with a replisome consisting of the DNA polymerase-UL42 complex and the heterotrimeric helicase-primase encoded by herpes simplex virus type 1. Okazaki fragments 3 kilobases in length and leading strands that may exceed 10 kilobases are produced. Lagging strand synthesis is stimulated by ribonucleoside triphosphates. DNA replication appears to be processive because it resists competition with an excess of (dT)150/(dA)20. The single-strand DNA binding protein ICP8 is not required, and high concentrations of ICP8 can, in fact, inhibit lagging strand synthesis. The inhibition can, however, be overcome by the addition of an excess of the UL8 component of the helicase-primase. Rolling circle replication by the herpesvirus and bacteriophage T7 replisomes appears to proceed by a similar mechanism.
Herpes simplex virus type 1 (HSV-1) is a member of the α1 lineage herpesviruses (1). Herpesviruses consists of large double-stranded DNA genomes that become circular at a very early stage of the infectious cycle (2). The circular template supports a rolling circle mode of DNA replication generating DNA concatemers late in the infectious cycle. HSV-1 encodes seven proteins that are essential for DNA replication in vivo (3, 4). The origin binding protein, encoded by the UL9 gene, is required for initiation of DNA synthesis at the viral origins of replication oriS and oriL. The remaining six proteins make up a putative replisome responsible for lytic replication. It consists of a DNA polymerase with a processivity factor (the product of the UL30 and UL42 genes), a heterotrimeric helicase-primase complex (the product of the UL5, UL8, and UL52 genes), and the single-strand DNA binding protein ICP8 (the product of the UL29 gene). Several examples of physical and functional interactions between these proteins have been described (4). It is therefore likely that they act together at the replication fork in a way that resembles the macromolecular machines responsible for replication of the Escherichia coli and bacteriophages T4 and T7 genomes (5–7). The mechanism for initiating lytic DNA replication may differ between the members of the herpesvirus family, but all herpesviruses use the same enzymatic machinery for propagation of replication forks (2, 4).
Previous work has established that extracts from cells either infected with HSV-1 or with recombinant baculoviruses encoding the HSV-1 replication proteins are able to support the semiconservative replication of circular duplex templates (8, 9). DNA synthesis was independent of the origin and the origin binding protein and proceeded by a rolling circle mechanism. To understand the mechanism of rolling circle replication, we have explored the optimal conditions for leading strand synthesis by the HSV-1 DNA polymerase-UL42 complex coupled to the unwinding of duplex DNA by the HSV-1 helicase-primase (10). We found that at the appropriate ionic conditions the rate of unwinding, 60–65 bp/s, approached the rate of fork movement in vivo (10). We also noted that the rate of unwinding was not stimulated further by the HSV-1 DNA polymerase (10). The single-strand DNA binding protein ICP8 promoted the unwinding of duplex DNA by preventing the reannealing of the complementary strands generated as a consequence of helicase action (10, 11). However, high concentrations of ICP8 inhibited the UL5/52 subassembly of the helicase-primase but not of the UL5/8/52 heterotrimer, indicating a functional interaction between the UL8 protein and ICP8 (11).
Experiments that use a synthetic minicircle template with a replication fork to study coordinated synthesis of leading and lagging strands by a T7 replication complex have recently been described (12). We have used a similar strategy to examine HSV-1 DNA replication with purified proteins in vitro. Our results demonstrate efficient coordinated synthesis of leading and lagging strands by a HSV-1 replisome that gives rise to double-stranded DNA concatemers.
Materials and Methods
Materials.
Restriction endonucleases were from Roche. Oligonucleotides were from Eurogentec (Brussels). [α-32P]dGTP and [α-32P]dCTP (3,000 Ci/mmol) were obtained from Amersham Pharmacia.
Cells and Virus.
Stocks of recombinant Autographa californica nuclear polyhedrosis virus for the expression of the UL8, UL5, UL52, UL30, UL42, and UL29 gene products were produced in Spodoptera frugiperda (Sf9 and Sf21) cells grown in Sf-900 SFM medium (GIBCO/BRL) (12).
Enzyme Purification.
The UL5/UL52, UL8, and ICP8 proteins were isolated from Sf9 cells as described (12). The UL30/UL42 complex was purified from Sf21 cells following a previously described protocol (13). The purity of the protein samples was at least 95% as determined by SDS/PAGE followed by Coomassie blue staining. The proteins were frozen in liquid nitrogen and were stored at −80°C.
Preparation of Minicircle Template.
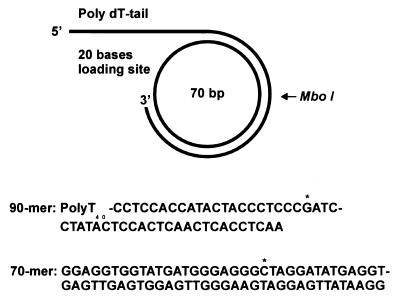
The 70-mer oligonucleotide 5′-GGAATATTGAGGATGAAGGGTTGAGGTGAGTTGAGTGGAGTATAGGATCGGGAGGGTAGTATGGTGGAGG-3′ was converted to a single-stranded circle in the following way. The 70-mer was phosphorylated and hybridized to the 20-mer 5′-TCAATATTCCCCTCCACCAT-3′. The 20-mer is complementary to 10 bases at both ends of the 70-mer and thus promotes the covalent circularization of the 70-mer by T4 DNA ligase. The hybridization and ligation was performed in 120 ml of 50 mM Tris⋅HCl (pH 7.6), 10 mM MgCl2, 10 mM DTT, 1 mM ATP, and 25 mg/ml BSA containing 8 mmol of the 70-mer and 8 mmol of the 20-mer. The reaction mixture was incubated for 30 min at 20°C. Three hundred and sixty units of T4 DNA ligase were then added, and the incubation was allowed to continue at 20°C overnight. The reaction was stopped with 25 mM EDTA. The reaction mixture was extracted with phenol/chloroform, and the oligonucleotides were recovered by ethanol precipitation. About 50% of the reaction products were monomeric circles. The 70-mer circle was purified by electrophoresis through a 10% polyacrylamide gel containing 6 M urea in TBE (14). The circular product (300 pmol) was eluted from the gel and annealed to 400 pmol of a partially complementary 90-mer 5′-T40CCTCCACCATACTACCCTCCCGATCCTATACTCCACTCAACTCACCTCAA-3′ in 1 ml of 100 mM NaCl, 20 mM Tris⋅HCl (pH 7.6). The resulting minicircle with a replication fork was purified by electrophoresis through a 12% nondenaturing polyacrylamide gel.
DNA Synthesis.
The minicircle containing a replication fork (35 fmol) was added to a 25-μl standard reaction mixture containing 25 mM Tris⋅HCl (pH 7.6), 8 mM magnesium acetate, 5 mM DTT, 100 μg/ml BSA, 40 mM creatine phosphate, 5 mg creatine kinase, 4 mM ATP, 250 μM GTP, 250 μM CTP, 250 μM UTP, 10% glycerol, 100 μM dATP, and 100 μM dTTP. To measure leading strand synthesis, 10 μM dCTP, 2 μCi [α-32P]dCTP, and 100 μM dGTP were added. Lagging strand synthesis was measured after the addition of 10 μM dGTP, 2 μCi [α-32P]dGTP, and 100 μM dCTP.
HSV-1 helicase-primase, either UL5/52 or UL5/52/8, (350 fmol) was preincubated for 5 min at 4°C in reaction buffer before addition of 50 fmol DNA polymerase/UL42 and 500 fmol ICP8. Incubation was continued at 37°C for 45 min. The reaction mixtures were analyzed either by denaturing agarose gel electrophoresis or by restriction enzyme cleavage followed by polyacrylamide gel electrophoresis.
Denaturing agarose gel electrophoresis was performed by adding 6 μl of alkaline loading buffer containing 18% (wt/vol) ficoll, 300 mM NaOH, 60 mM EDTA (pH 8.0), 0.15% (wt/vol) bromocresol green, and 0.25% (wt/vol) xylene cyanol FF to the reaction mixture. The samples were subjected to electrophoresis through an 0.8% alkaline agarose gel in 50 mM NaOH and 1 mM EDTA at 1.5 V/cm for 20 h.
Samples to be analyzed by nondenaturing PAGE were treated with proteinase K, 100 μg/ml, and 0.5% SDS for 20 min at 37°C. The samples were diluted to 175 μl and were extracted first with phenol/chloroform and then with chloroform. The aqueous phase (150 μl) was precipitated by addition of 2.5 M ammonium acetate and 2.5 volumes of ethanol. The pellet was dissolved in 20 μl of a buffer containing 100 mM NaCl, 50 mM Tris⋅HCl (pH 7.9), 10 mM MgCl2, and 1 mM DTT. MboI (5 units) was added to each reaction. The samples were then incubated for 4–8 h at 37°C and were analyzed by 12% PAGE in TBE (14).
Southern Blotting.
DNA synthesis was carried out in a total volume of 75 μl under the same reaction conditions described above with 100 μM each of dATP, dGTP, dCTP, and dTTP. The reaction was stopped after 45 min with 18 μl of the alkaline loading buffer (see above). The sample was divided in two parts and was loaded onto a 0.8% alkaline agarose gel. After electrophoresis, the DNA was transferred to a nylonfilter (N-Hybond+, Amersham Pharmacia). The membrane was cut in two pieces and was hybridized with either the 70-mer 5′-GGAATATTGAGGATGAAGGGTTGAGGTGAGTTGAGTGGAGTATAGGATCGGGAGGGTAGTATGGTGGAGG-3′ to detect leading strand products or the 50-mer 5′-CCTCCACCATACTACCCTCCCGATCCTATACTCCACTCAACTCACCTCAA-3′ to detect lagging strand products.
DNA Competition.
The reaction mixture (150 μl) was prepared as described above. Competitor DNA, consisting of (dA)20 annealed to (dT)150, was added to the reactions as indicated. The ratio of competitor DNA to template was 200:1. Samples (25 μl) were removed at the indicated times and were stopped by the addition of 100 μg/ml proteinase K and 0.5% SDS. The samples were then analyzed by restriction enzyme cleavage and PAGE as described above.
Results
Synthesis of Leading and Lagging Strands.
A synthetic minicircle template with a replication fork has recently been described that can be used to examine the synthesis of leading and lagging strands during rolling circle replication (ref. 12; Fig. 1). The template offers several experimental advantages: The products of leading and lagging strand synthesis can be specifically radiolabeled; oligonucleotide probes can be designed that unambiguously detect leading and lagging strands, and restriction enzyme cleavage can be used to distinguish between single- and double-stranded DNA products.
Figure 1.
Minicircle template for rolling circle DNA replication. The minicircle template was prepared as described in Materials and Methods. [α-32P]dCTP was used to preferentially label the leading strand, and [α-32P]dGTP was used to label the lagging strand fragments. The single G and C in the templates for lagging and leading strands are indicated with an asterisk.
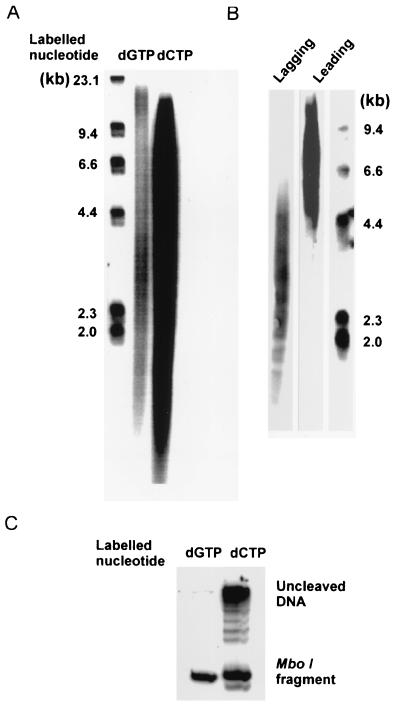
We have used such a minicircle template to look for coupled leading and lagging strand synthesis using the purified HSV-1 encoded replication proteins DNA polymerase-UL42 complex, helicase-primase, and ICP8. We found that a moderate excess of the heterotrimeric and heterodimeric helicase-primase complexes, UL5/8/52 or UL5/52, and ICP8, resulted in maximum stimulation of DNA synthesis by the DNA polymerase-UL42 complex (results not shown). High concentrations of ICP8, however, inhibited synthesis of the lagging strand (see below). When products were analyzed by alkaline agarose gel electrophoresis, the products labeled with [α-32P]dGTP, corresponding to the lagging strand, showed a bimodal distribution (Fig. 2A). Presumably, the shorter products were Okazaki fragments, and the long products were leading strand DNA. A hybridization experiment using oligonucleotide probes showed that the short 3-kb fragments were indeed produced by lagging strand synthesis and the long (>10 kb) products by leading strand synthesis (Fig. 2B). Restriction enzyme cleavage showed that ≈50% of the products labeled with [α-32P]dCTP were double-stranded and could be cleaved by MboI (Fig. 2C).This finding suggests that approximately half of the active templates were used for coordinated synthesis of leading and lagging strands. Incomplete cleavage of the replication products by MboI gave rise to a ladder of fragments, demonstrating that double-stranded DNA was produced by rolling circle replication (Fig. 2C, second lane).
Figure 2.
Products of leading and lagging strand synthesis. Products formed by rolling circle DNA replication in vitro were analyzed by alkaline agarose gel electrophoresis and restriction enzyme cleavage as described in Materials and Methods. The reaction mixtures contained 50 fmol of DNA polymerase-UL42, 350 fmol of the UL5/UL52/UL8 heterotrimer, 500 fmol of ICP8, and 35 fmol of the minicircle template. (A) Autoradiograph of an alkaline agarose gel. First lane, DNA size markers; second lane, products labeled by [α-32P]dGTP; third lane, products labeled by [α-32P]dCTP. (B) Autoradiograph of Southern blot analyses of replication products. First lane, the oligonucleotide probe is complementary to the lagging strand; second lane, the oligonucleotide probe is complementary to the leading strand; third lane, DNA size markers. (C) Products labeled with [α-32P]dCTP and [α-32P]dGTP were cleaved with MboI and were subjected to polyacrylamide gel electrophoresis as described in Materials and Methods.
Ribonucleoside Triphosphates Stimulate Lagging Strand Synthesis.
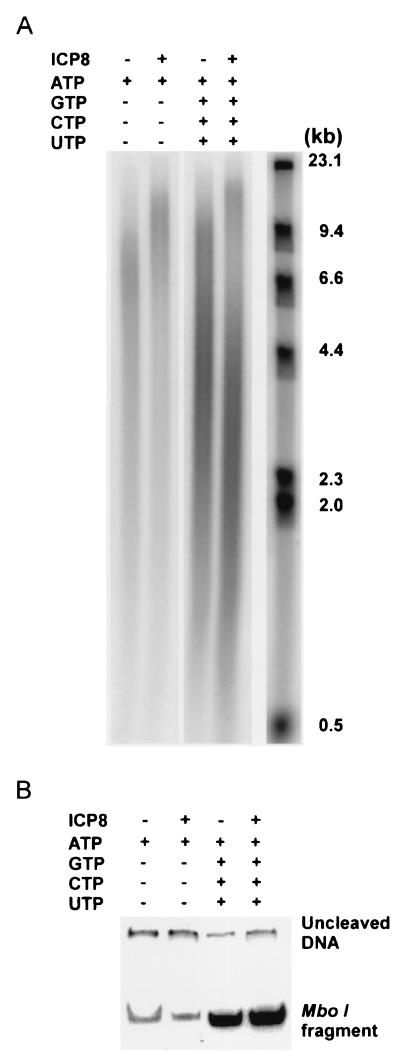
Initiation of Okazaki fragment formation requires the synthesis of an RNA primer catalyzed by primase action. We observed that the synthesis of Okazaki fragments labeled by [α-32P]dGTP and detected by alkaline agarose gel electrophoresis was stimulated by the four ribonucleoside triphosphates (Fig. 3A). The synthesis of double-stranded DNA was also greatly enhanced by all four ribonucleoside triphosphates as judged by restriction enzyme analysis (Fig. 3B). We also found that, although the synthesis of double-stranded DNA was strictly dependent on the presence of helicase-primase (see below), ICP8 was not required (Fig. 3B).
Figure 3.
Lagging strand synthesis is stimulated by ribonucleoside triphosphates. The reaction mixtures were prepared as described in Materials and Methods. ICP8 and the ribonucleoside triphosphates GTP, CTP, and UTP were omitted as indicated. (A) Products were analyzed by alkaline agarose gel electrophoresis. (B) The products were cleaved with MboI and were analyzed by PAGE.
Functional Interaction Between UL8 and ICP8.
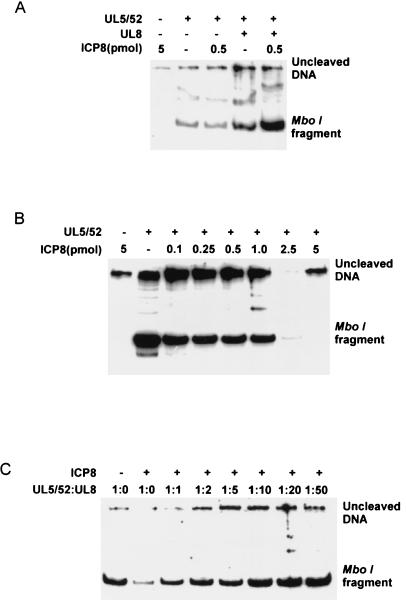
Leading strand synthesis promoted by the DNA polymerase-UL42 complex was stimulated by high concentrations of ICP8 (Fig. 4A). However, double-stranded DNA was produced only in the presence of the helicase-primase (Fig. 4A). Double-stranded DNA was synthesized in the absence of both ICP8 and UL8. However, the addition of these proteins stimulated synthesis of double-stranded DNA (Fig. 4A).
Figure 4.
Functional interaction between UL8 protein and ICP8. (A) The reaction mixture contained [α-32P]dGTP 350 fmol of the UL5/52 subassembly and 50 fmol of DNA polymerase-UL42. 700 fmol UL8 and 0.5 or 5 pmol ICP8 were added as indicated. MboI cleavage and PAGE was performed as described in Materials and Methods. (B) The reaction mixture contained [α-32P]dGTP, 350 fmol of the UL5/52 subassembly, and 50 fmol of DNA polymerase-UL42. Increasing amounts of ICP8 were added as indicated. The samples were processed as above. (C) The reaction mixture contained [α-32P]dGTP, 350 fmol of the UL5/52 subassembly, 50 fmol of DNA polymerase-UL42, and 1 pmol of ICP8. Increasing amounts of UL8 protein were added as indicated. The molar ratio of UL5/52 to UL8 is shown.
Previous experiments have provided evidence that ICP8 and the UL8 protein interact both physically and functionally (11, 15–17). We therefore wished to investigate the functional interaction between these proteins during rolling-circle replication (Fig. 4 B and C). We found that the synthesis of double-stranded DNA supported by the helicase-primase subassembly UL5/52 was inhibited by high concentrations of ICP8 (Fig. 4B). We then investigated the ability of the UL8 protein to stimulate the synthesis of double-stranded DNA in a reaction containing the DNA polymerase-UL42 complex, the helicase-primase subassembly UL5/52, and ICP8 (Fig. 4C). We found that the addition of the UL8 protein in amounts that would reconstitute the helicase-primase heterotrimer suppressed the inhibition produced by ICP8. The addition of excess UL8 protein led to a further stimulation of double-stranded DNA synthesis. These findings provide functional evidence for a direct interaction between the UL8 protein and ICP8. It has been shown that the UL8 protein may also interact with the origin binding protein and the HSV-1 DNA polymerase (18). Possibly, the UL8 protein facilitates the assembly of the HSV-1 replisome and coordinates the synthesis leading and lagging strands.
Leading and Lagging Strand Synthesis Are Coupled and Processive.
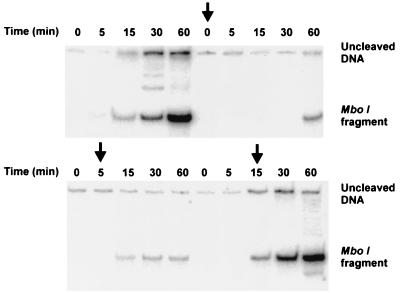
The experiments described above suggest that double-stranded DNA may be produced by the coupled synthesis of leading and lagging strands promoted by the HSV-1 encoded replisome. It is, however, still possible that continuous and discontinuous DNA synthesis are uncoupled events. We therefore performed an experiment in which the putative replisome was challenged at various times during the reaction by a 200-fold excess of (dT)150/(dA)20 (Fig. 5). The competitor DNA efficiently prevented the synthesis of double-stranded DNA labeled with [α-32P]dGTP when added early in the reaction. In contrast, when the competitor was added late, it failed to inhibit synthesis of double-stranded DNA. These findings suggest that leading and lagging strand synthesis are coupled and rolling circle replication is a processive process. They also indicate that the assembly of the HSV-1 replisome at a preformed replication fork is a relatively slow process requiring at least 5 min at 37°C. Once assembled, however, it appears to be stable and to be able to carry out processive DNA synthesis for an extended period.
Figure 5.
Rolling circle replication by the HSV-1 replisome is resistant to challenge by (dT)150/(dA)20. Reaction conditions were as described in Materials and Methods. (dT)150/(dA)20was added to a final concentration of 0.32 μM at 0, 5, and 15 min as indicated by the arrows. Samples were processed by restriction enzyme cleavage and PAGE as described in Materials and Methods. No competitor DNA was added to the experiment shown in the first six lanes.
Discussion
The synthesis of double-stranded DNA requires the coordination of several enzymatic activities to achieve the speed and fidelity consistent with DNA synthesis in vivo (5–7). A considerable body of evidence suggests that a macromolecular complex, a replisome, is formed at the replication fork. The replisome, however, is only stable at the fork, and it is held together by transient protein-protein and protein-DNA interactions.
The HSV-1 replisome consists of the DNA polymerase-UL42 heterodimer and the helicase-primase heterotrimer (4). The single-strand DNA binding protein ICP8 may stimulate the DNA polymerase-UL42 complex (19) and assist the helicase-primase by preventing the reannealing of complementary single strands produced by its helicase action (10). The HSV-1 specific initiator protein OBP is not required for the propagation of the replication fork. In fact, OBP cannot replace the helicase-primase in leading strand synthesis (M.F., unpublished observations). The minimal herpesvirus replisome may contain two DNA polymerase-UL42 heterodimers and one helicase-primase heterotrimer. The helicase-primase complex would be expected to translocate along the lagging strand template in the 5′ to 3′-direction. However, the stoichiometry of the components of the functional replisome has not been determined.
Several protein-protein interactions have been described that may serve to promote the assembly of a herpesvirus replisome and to coordinate its enzyme activities. The C terminus of the DNA polymerase binds specifically to the UL42 protein (20–23). Furthermore, the DNA polymerase interacts physically with the C terminus of the UL8 protein (24). The UL8 protein is easily incorporated into the UL5/52 subassembly of the helicase-primase and may be regarded as a subunit of the UL5/8/52 helicase-primase (4, 11, 25). The UL8 protein also interacts physically and functionally with ICP8 (11, 15–17). Thus, the UL8 protein, because of its many protein-protein interactions, may serve to keep the replisome intact.
The HSV-1 origin binding protein also interacts with components of the replisome. The C-terminal DNA binding domain of OBP forms a stable complex with ICP8 (26). OBP may also interact with the UL8 and UL42 proteins (18, 27). Because OBP is not needed at the fork, these interactions may also help to promote the assembly of the herpesvirus replisome.
It is surprising that UL8 and ICP8 are not strictly required for synthesis of duplex DNA in vitro. They are, however, essential for DNA replication in vivo (3). ICP8 may be a multifunctional protein. It may promote initiation of DNA replication through interaction with the C terminus of the HSV-1 origin binding protein. In fact, an OBP mutant that no longer binds ICP8 but has retained helicase and origin binding activity has a reduced ability to support origin-dependent DNA synthesis in vivo (28). It has also been shown that temperature-sensitive mutants that map in the N-terminal and C-terminal parts of ICP8 display functional complementation in trans (29). These mutations do not, however, map in the DNA binding domain. Further studies are clearly needed to define the interactions between ICP8 and the replication machinery. It is interesting to note that the bacteriophage T7 specific single-strand DNA binding protein is not required for DNA synthesis in vitro, but it is needed for coupled synthesis of leading and lagging strands in vivo (12). It has therefore been argued that an important function would be to protect exposed stretches of single-stranded DNA from proteins and enzymes involved in recombination and repair (30). Perhaps ICP8 has a similar function in the eukaryotic nucleus.
The synthesis of duplex DNA by the HSV-1 replisome is not inhibited by the addition of competing DNA, indicating that the synthesis of leading and lagging strands are coupled and processive. It is unclear how this is achieved. The HSV-1 DNA polymerase does not form a stable dimer in solution, and there is no evidence for an interaction between the leading strand DNA polymerase and the helicase-primase as it translocates along the lagging strand template. Possibly, the lagging strand polymerase is tethered to the helicase-primase and cycles processively during the synthesis of consecutive Okazaki fragments. The rate of fork movement is then set by the events occurring on the discontinuous strand. The processive polymerase synthesizing the continuous strand may only have to adjust to the rate of fork movement. More work is needed to identify a connection between the leading strand DNA polymerase and the enzymes involved in lagging strand synthesis.
The replisomes that have been carefully characterized thus far fall in to two categories (7, 12). The E. coli and T4 replisomes have a dimeric DNA polymerase and a helicase that moves along the lagging strand template in the 5′ to 3′ direction. Direct interactions between the leading strand polymerase and the helicase greatly enhance the rate of unwinding. The polymerase on the lagging strand recycles in an ATP-dependent manner and requires a sliding clamp and a clamp loader. The T7 replisome, on the other hand, uses a helicase-primase that translocates along the lagging strand template in the 5′ to 3′ direction (12), and recycling of the processive polymerase-thioredoxin complex is independent of ATP. There is no evidence for the existence of a dimeric DNA polymerase. Possibly, the physical interaction between the helicase and the leading strand polymerase serves to coordinate leading and lagging strand synthesis (31). The HSV-1 and T7 replisomes may therefore belong to the same category. If this is true, it might indicate an evolutionary relationship that is reflected in the structure of the replisome components.
The identification of conditions that allow herpesvirus-specific rolling circle replication in vitro permits us to look for the factors that participate in the repair of the lagging strand. It will also be of interest to determine how the viral replisome responds to damaged DNA templates and how it interacts with repair systems in the eukaryotic cell.
Acknowledgments
This work was supported by the Swedish Cancer Society Grant 2552-B99–13XCC and a grant from the Swedish Foundation for Strategic Research to P.E and National Institutes of Health Grant AI 20538 to I.R.L.
Abbreviation
- HSV-1
herpes simplex virus type 1
References
- 1.McGeoch D J, Davison A J. In: Origin and Evolution of Viruses. Domingo E, Webster R, Holland J, editors. New York: Academic; 1999. pp. 441–465. [Google Scholar]
- 2.Roizman B, Sears A E. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott; 1996. pp. 2297–2342. [Google Scholar]
- 3.Challberg M D. Proc Natl Acad Sci USA. 1986;83:9094–9098. doi: 10.1073/pnas.83.23.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehman I R, Boehmer P E. J Biol Chem. 1999;274:28059–28062. doi: 10.1074/jbc.274.40.28059. [DOI] [PubMed] [Google Scholar]
- 5.Alberts B M, Morris C, Mace D, Sinha N, Bittner M, Moran L. In: DNA Synthesis and Its Regulation. Goulian M, Hanawalt P, Fox C F, editors. Menlo Park, CA: Benjamin; 1975. pp. 241–269. [Google Scholar]
- 6.Kornberg A, Baker T A. DNA Replication. New York: Freeman; 1992. [Google Scholar]
- 7.Baker T A, Bell S P. Cell. 1998;92:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- 8.Skaliter R, Lehman I R. Proc Natl Acad Sci USA. 1994;91:10665–10669. doi: 10.1073/pnas.91.22.10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skaliter R, Makhov A M, Griffith J D, Lehman I R. J Virol. 1996;70:1132–1136. doi: 10.1128/jvi.70.2.1132-1136.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falkenberg M, Elias P, Lehman I R. J Biol Chem. 1998;273:32154–32157. doi: 10.1074/jbc.273.48.32154. [DOI] [PubMed] [Google Scholar]
- 11.Falkenberg, Bushnell D A, Elias P, Lehman I R. J Biol Chem. 1997;272:22766–22770. doi: 10.1074/jbc.272.36.22766. [DOI] [PubMed] [Google Scholar]
- 12.Lee J, Chastain P D, Kusakabe T, Griffith J D, Richardson C C. Mol Cell. 1998;1:1001–1010. doi: 10.1016/s1097-2765(00)80100-8. [DOI] [PubMed] [Google Scholar]
- 13.Crute J J, Lehman I R. J Biol Chem. 1989;264:19266–19279. [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 15.Sherman G, Gottlieb J, Challberg M D. J Virol. 1992;66:4884–4892. doi: 10.1128/jvi.66.8.4884-4892.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Gac N T, Villani G, Hoffman J S, Boehmer P E. J Biol Chem. 1996;271:21645–21651. doi: 10.1074/jbc.271.35.21645. [DOI] [PubMed] [Google Scholar]
- 17.Hamatake R K, Bifano M, Hurlburt W W, Tenney D J. J Gen Virol. 1997;78:857–865. doi: 10.1099/0022-1317-78-4-857. [DOI] [PubMed] [Google Scholar]
- 18.McClean G W, Abbotts A P, Parr M E, Marsden H S, Stow N D. J Gen Virol. 1994;75:2699–2706. doi: 10.1099/0022-1317-75-10-2699. [DOI] [PubMed] [Google Scholar]
- 19.Hernandez T R, Lehman I R. J Biol Chem. 1990;265:11227–11232. [PubMed] [Google Scholar]
- 20.Stow N D. Nucleic Acids Res. 1993;21:87–92. doi: 10.1093/nar/21.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Digard P, Bebrin W R, Weisshart K, Coen D M. J Virol. 1993;67:398–406. doi: 10.1128/jvi.67.1.398-406.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsden H S, Murphy M, McVey G L, MacEachran K A, Owsianka A M, Stow N D. J Gen Virol. 1994;75:3127–3135. doi: 10.1099/0022-1317-75-11-3127. [DOI] [PubMed] [Google Scholar]
- 23.Digard P, William K P, Hensley P, Brooks I S, Dahl C E, Coen D M. Proc Natl Acad Sci USA. 1995;92:1456–1460. doi: 10.1073/pnas.92.5.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marsden H S, McLean G W, Barnard E C, Francis G J, MacEachran K, Murphy M, McVey G, Cross A, Abbotts A P, Stow N D. J Virol. 1997;71:6390–6397. doi: 10.1128/jvi.71.9.6390-6397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crute J J, Olivo P D, Challberg M D, Mocarski E S, Lehman I R. Proc Natl Acad Sci USA. 1989;86:2186–2189. doi: 10.1073/pnas.86.7.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boehmer P E, Lehman I R. Proc Natl Acad Sci USA. 1993;90:8444–8448. doi: 10.1073/pnas.90.18.8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monahan S J, Grinstead L A, Olivieri W, Parris D S. Virology. 1998;241:122–130. doi: 10.1006/viro.1997.8953. [DOI] [PubMed] [Google Scholar]
- 28.Boehmer P E, Craigie M C, Stow N D, Lehman I R. J Biol Chem. 1994;269:29329–29334. [PubMed] [Google Scholar]
- 29.Gao M, Knipe D M. J Virol. 1993;67:786–885. doi: 10.1128/jvi.67.2.876-885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park K, Debyser Z, Tabor S, Richardson C C, Griffith J D. J Biol Chem. 1998;273:5260–5270. doi: 10.1074/jbc.273.9.5260. [DOI] [PubMed] [Google Scholar]
- 31.Notarnicola S M, Mulcahy H L, Lee J, Richardson C C. J Biol Chem. 1997;272:18425–18433. doi: 10.1074/jbc.272.29.18425. [DOI] [PubMed] [Google Scholar]