Abstract
The tetratricopeptide repeat (TPR) is a 34-aa sequence motif, typically found in tandem clusters, that occurs in proteins of bacteria, archea, and eukaryotes. TPRs interact with other proteins, although few details on TPR–protein interactions are known. In this paper we show that a portion of a loop in the homeodomain of the DNA-binding protein α2 is required for its recognition by the TPRs of the corepressor Ssn6. The amino acid sequence of this loop is similar to the sequences recognized by the TPRs of an entirely different protein, Pex5, which directs peroxisomal import. We further show that α2 can be made to bind specifically in vitro to the TPRs of Pex5 and that a point mutation that disrupts the α2-Ssn6 interaction also disrupts the α2-Pex5 interaction. These results demonstrate that two different TPR proteins recognize their target by a similar mechanism, raising the possibility that other TPR-target interactions could occur through the same means.
With the completion of increasing numbers of genome sequences, families of related proteins are expanding at a rapid rate. In some cases, it is possible to recognize the cellular process in which a protein participates from its sequence. For example, the presence of a homeodomain indicates that the protein is likely to be a regulator of transcription. However, for other protein families, the existence of a shared motif does not identify the cellular process in which the family members participate. One such protein family is defined by the presence of tetratricopeptide repeats (TPRs). The TPR is a 34-aa degenerate sequence motif that was first recognized 9 years ago (1, 2). Although these tandem repeats originally were found in only five proteins, this family has grown to contain more than 100 members, in organisms ranging from bacteria to humans. Saccharomyces cerevisiae alone has 26 TPR proteins, representing nearly one half a percent of the yeast's total gene products. TPRs usually are found in tandem arrays of between three and 16 copies, and they function in a wide variety of cellular processes, including transcriptional regulation, protein import into organelles, cell cycle regulation, and splicing (for review see ref. 3). The TPR motif originally was proposed to mediate protein–protein interactions (1) and over the past few years this has been shown to be the case in several systems (4–8). Despite this progress, we still do not know the common features through which TPRs recognize their target proteins.
Two distinct biological systems in which TPRs mediate protein–protein interactions have been relatively well studied, and these systems can be used to address whether common elements exist among different TPR-protein interactions. Ssn6 is a transcriptional repressor found in the nucleus of S. cerevisiae (9, 10). It contains 10 TPR repeats (2, 10), and together with its corepressor, Tup1, is responsible for the repression of more than 150 genes in yeast (9, 11). The Ssn6-Tup1 complex is not capable of binding DNA on its own, but is brought to the promoters of the genes it represses by interactions with sequence-specific DNA binding proteins (9, 12, 13). In all, the Ssn6 TPRs must interact with at least nine proteins (sequence-specific DNA binding proteins, Tup1, and perhaps histones and downstream components of the repression machinery), which share no gross sequence similarity to one another. The interaction between Ssn6 and the sequence-specific DNA binding protein α2 has been studied in some detail. It was found that the homeodomain of α2 could interact directly with single TPR repeats or with an array of TPR repeats (4). The other eight proteins mentioned above also must interact with some or all of the TPR repeats, and these results, as well as the fact that Tup1 can interact with at least some of the same TPRs bound by α2 (4, 6), suggest that a particular repeat is not dedicated to binding only one target protein. However, a given repeat may show a higher affinity for one target protein over another (6).
The second well-studied system involving a TPR repeat protein is the import of proteins into peroxisomes. Pex5, which contains eight TPRs, functions as a peroxisomal import receptor, and the repeats in Pex5 have been shown to bind directly to a degenerate tripeptide sequence found at the C terminus of proteins to be imported (for a review of peroxisomal import see ref. 14). This tripeptide, first identified as SKL (15), is called the peroxisomal targeting sequence 1 (PTS1), and is required for Pex5-mediated import into peroxisomes (16). Although the proteins that interact directly with Pex5 contain no gross overall sequence similarity to one another, they all do contain this short consensus sequence, which directs the interaction with the TPRs of Pex5, and thus, import into the peroxisomes.
In this paper, we demonstrate, both in vivo and in vitro, that a sequence similar to the PTS1 peroxisomal import sequence is required for the interaction of the DNA-binding protein α2 with TPRs of the Ssn6 corepressor. Moreover, we show that this sequence also can mediate a nonphysiological, in vitro interaction between α2 and the TPRs of Pex5. Identification of a general TPR-targeting sequence within α2 helps explain how it can bind to each of the TPRs in Ssn6 and suggests a common feature of at least some TPR–protein interactions.
Materials and Methods
Plasmid Construction.
Plasmids used in this study are outlined in Table 1 and are presented in the order they are used in the paper. All plasmids were confirmed by sequencing.
Table 1.
Plasmids
| pAJ# | Other name | Description | Source |
|---|---|---|---|
| pRS315 | LEU2, Ars/CEN | (37) | |
| pAJ786 | pAV116 | MATα cassette in pRS315 | Andrew Vershon |
| pAJ264 | pAV115 (R173A) | MATα cassette w/α2 R173A | (17) |
| pAJ233b | pAV100 | α2 E. coli expression vector | Andrew Vershon |
| pAJ1011 | — | Above with R173A | This study |
| pAJ584 | Ssn6 1–9 | GST:Ssn6 TPR 1–9 | (4) |
| pAJ558 | Ssn6 TPR6 | GST:Ssn6 TPR6 | (4) |
| pGAD.RI | Pex5 PCR template | (38) | |
| pAJ552 | — | GST:Pex5 TPRs 2–8 | This study |
| pAJ705 | pGEX1 | GST fusion vector | Amrad |
| pAJ21 | pCK1 | α2/Mcm1 operator sites | (33) |
| pAJ331 | pCG71 | Asy operator sites | Caroline Goutte |
pAJ1011, the α2 R173A bacterial expression vector, was made by cutting a 580-bp BglII–BamHI fragment from pAJ264 and ligating this fragment to similarly cut pAJ233b to replace the wild-type sequence.
pAJ552 is an in-frame fusion of glutathione S-transferase (GST) to TPRs 2–8 of Pex5 from S. cerevisiae. TPRs 2–8 of Pex5 were made by PCR using pGAD.R1 as a template and the following primers: 5′-GAATATTTTAAGGATCCTAATGCTTAT-3′ and 3′ end-5′-TTTATTATTAGAATTCACTTCATGCAT-3′; the restriction sites are underlined. The resulting PCR product was cut with EcoRI and BamHI and ligated into similarly cut pGEX1.
Strain Construction, Yeast Media, and Transformations.
Other strains were generated by lithium acetate transformation using standard procedures. Yeast media used in these experiments are described in ref. 32.
Interaction Assays.
Extracts from bacteria containing 1.5 μg partially purified α2 protein (12) were diluted in binding buffer II (BBII) + the salt concentration indicated in the figure legends to a final volume of 250 μl/assay. The diluted protein was dialyzed on a microdialyzer (GIBCO/BRL), 15 ml/h, 1 h against BBII + indicated salt concentration, spun for 45′, 4°C, 14K rpm. Glutathione agarose containing immobilized fusion proteins (see ref. 4) was diluted with unconjugated beads, for a final fusion protein concentration of 2.5 μM. The resin was equilibrated by washing 3 × 750 μl elution buffer, followed by 4 × 1 ml BBII + indicated salt concentration. A total of 250 μl of the appropriate extract containing α2 was added to the resin, and the tubes were incubated at 25°C for 20 min on a Nutator platform. The resin was pelleted at 2K rpm, 2 min, 4°C, and the supernatant was removed and saved. The resin was washed once in 250 μl BBII + indicated salt concentration, with the salt concentration being identical to that used in the loading step, and the wash was saved. The resin was washed twice more in 250 μl BBII, and supernatant from these washes was discarded. The resin was resuspended in 250 μl elution buffer and incubated for 10 min, 25°C on the Nutator platform. The resin was pelleted, and the supernatant was collected and saved as the elution fraction. The resin was washed once in 250 μl elution buffer, and the elution supernatants were combined. All fractions were spun at 2 K for 5 min, and the supernatant was transferred to a new tube to remove any remaining resin. The samples were trichloroacetic acid-precipitated and resuspended in 25 μl Laemmli buffer. Samples were neutralized with NH4OH vapors, boiled for 5 min, and spun for 5 min. Two microliters of each sample was loaded on a 15% SDS-polyacrylamide gel and transferred to membrane for Western blotting.
The experiments depicted in Fig. 3, α2 vs. Ssn6 TPR6 1–9 and α2 vs. GST, were conducted at 175 mM final salt concentration. α2 vs. Pas 10 TPRs 2–8 and α2 vs. Ssn6 TPR6 were conducted at 150 mM final salt. The BBII is 20 mM Hepes, pH 7.6/1 mM EDTA/2 mM DTT/1 mM PMSF/2 mM benzamidine. BBII + 175 mM final salt also contains 164.5 mM potassium acetate, 7.5 mM sodium acetate, and 3 mM magnesium acetate. BBII + 150 mM final salt also contains 141 mM potassium acetate, 6.4 mM sodium acetate, and 2.6 mM magnesium acetate. Elution buffer is 50 mM Hepes, pH 7.6/1 mM EDTA/1 mM EGTA/1 M NaCl/0.05% NP-40/2 mM DTT/2 mM PMSF.
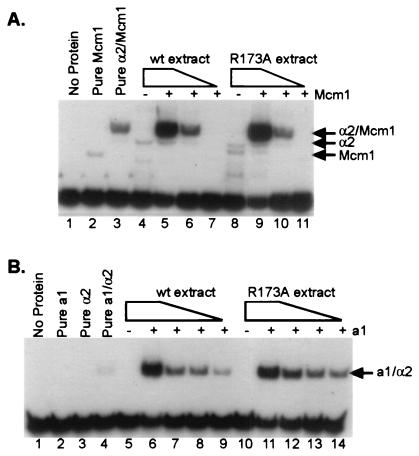
Figure 3.
α2 R173A binds to DNA cooperatively and with wild-type (wt) affinity with its partners, Mcm1 and a1. (A) Gel mobility-shift assay using a Ste6 (a-specific gene) operator as probe. The gel mobility shifts shown here were produced by using decreasing amounts of either wild-type α2 or α2 R173A in combination with constant levels of purified Mcm11–96. Lane 1 contains free probe, and lane 2 contains added Mcm11–96 (2.3 × 10-9 M). All other lanes with added Mcm11–96 contain 4.6 × 10-10 M Mcm1 (lanes 3, 5–7, and 9–11). In addition to Mcm1, lane 3 contains purified α2 (1 × 10-8M). Lanes 5–7 and 9–11 contain 10-fold serial dilutions of either wild-type α2 or α2 R173A, respectively. The dilution series begins at 5 × 10-8 M and ends at 5 × 10-10 M, as indicated by the gradient above the lanes. Lanes 4 and 8 each contain 5 × 10-8 M α2 or α2 R173A, respectively, in the absence of any added Mcm11–96. (B) Gel mobility-shift assay using a haploid-specific gene operator as probe. The gel mobility shifts shown here were produced by using decreasing amounts of either wild-type α2 or α2 R173A in combination with constant levels of purified a1. Lane 1 contains free probe, lane 2 contains added purified a1 (8.4 × 10-8 M) in the absence of α2, and lane 3 contains purified α2 (1 × 10-9 M) in the absence of added a1. Lane 4 contains both purified a1 (8.4 × 10-8 M) and purified α2 (2 × 10-9 M). All other lanes with added a1 contain 8.4 × 10-8 M a1. Lanes 6–9 and 11–14 contain 2-fold serial dilutions of either wild-type α2 or α2 R173A, respectively. The dilution series begins at 1.6 × 10-9 M and concludes at 2.1 × 10-10 M, as indicated by the gradient above the lanes. Lanes 5 and 10 each contain 1.6 × 10-9 M wild-type α2 or α2 R173A, respectively, in the absence of added a1. Extract dilutions for each of the above gel mobility shifts were compared by Western blot to ensure that roughly equivalent levels of protein were used for each set of matched lanes (data not shown).
Liquid β-Galactosidase assays.
Liquid β-galactosidase assays were performed as described (33) except that cell lysis was performed according to CLONTECH MATCHMAKER kit directions. Comparisons of liquid β-galactosidase assays conducted by each of the above methods did not reveal any significant differences in the numbers achieved. The remainder of the assay was performed as described previously.
Whole-Cell Extracts for Western Blotting.
Extracts for the blots used in Fig. 3C were prepared by using the trichloroacetic acid-Western protocol (34) and transferred according to ref. 35. The α2 blot was probed with anti-α2 antibody at a 1:1,000 dilution. Anti-α2 was used at 1:1,000, and anti-rabbit-horseradish peroxidase (HRP) was used at 1:5,000. HRP was detected by using NEN Renaissance reagents.
Gel Mobility Shift Assays.
The α2/Mcm1 gel mobility shifts used a labeled wild-type Ste6 operator fragment as probe. The probes and shift conditions are as in ref. 17, except the following buffer was used 25 mM Tris, pH 8.0/0.5 mM EDTA/5% glycerol/5 mM MgCl2/0.1% NP-40/50 μg/ml polydIdC (ICN)/10 mg/ml BSA. Purified Mcm11–96 was generously provided by S. Soisson and C. Wolberger, Johns Hopkins University School of Medicine, Baltimore.
Gel mobility shifts on the assay site were performed in 60 mM Tris, pH 8.0/0.55 mM EDTA/5 mM MgCl2/0.1% NP-40/50 μg/ml polydIdC/10 mg/ml BSA/10% glycerol. Remaining conditions were described in ref. 36.
Results
α2 Contains a PTS1-Like Sequence in the Region that Interacts with Ssn6.
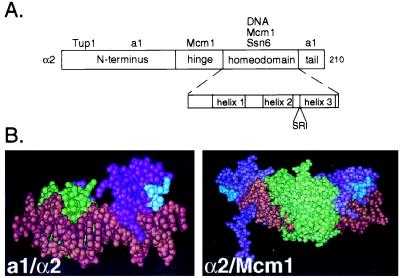
Previous work had demonstrated that the homeodomain of α2 is responsible for the interaction of α2 with the TPRs of Ssn6 (4). Point mutations within the loop between helices 2 and 3 of the homeodomain were known to affect α2's ability to direct repression in vivo (17). On closer inspection, we realized that the sequence of this loop was closely related to the PTS1 sequence that directs interactions with the TPRs of Pex5 (Fig. 1A). The PTS1 sequence is degenerate; and among the sequences that have been demonstrated to promote peroxisomal import are S/A/C in the first position, K/H/R in the second position, and L or M in the third position (15, 18). The PTS1-like sequence in the loop of the α2 homeodomain is SRI. Because the structures of the α2 homeodomain bound to DNA alone (19) and with each of its two binding partners, a1 and Mcm1 (20, 21) are known, we could easily determine whether this sequence was surface exposed and therefore available in principle to interact with the TPRs of Ssn6 when α2 was bound to DNA. In each of these protein–DNA structures, the SRI is indeed surface exposed (Fig. 1B).
Figure 1.
α2 contains a surface exposed PTS1-like sequence in the homeodomain. (A) A schematic diagram of α2 illustrating its domain structure. Interacting proteins are placed above the domain where they are known to make contacts. The homeodomain is shown in more detail in the Inset. The PTS1-like sequence is placed below its position in the linear sequence of the homeodomain. (B) The PTS1-like sequence in the homeodomain of α2 is surface exposed. (Left) The structure of the α2 homeodomain (purple) is shown bound as a trimeric complex with the homeodomain of a1 (green) and DNA (brown) (21). (Right) The α2 homeodomain (purple) is shown bound to DNA (brown) and a dimer of Mcm1 (green) (20). In each of the structures the PTS1 sequence in the homeodomain of α2 is shown in blue.
The −SRI in the Homeodomain of α2 Is Required for the Interaction with the TPRs of Ssn6.
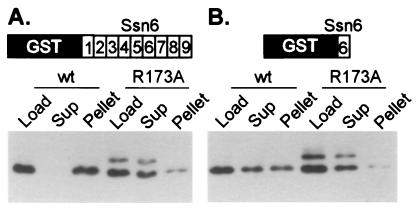
We addressed the relevance of the SRI sequence in the homeodomain of α2 by mutating its sequence and testing the ability of the resulting proteins to interact with the TPRs of Ssn6. Wild-type α2 and α2 containing a mutation in the SRI sequence (R173A, in the second position) were overexpressed in bacteria. Partially purified extracts containing either wild-type or mutant α2 were incubated with two GST-TPR fusion proteins, each of which contained a different set of TPRs from Ssn6, immobilized on glutathione agarose. The beads were washed and the bound protein was eluted. Fig. 2A demonstrates that wild-type α2 binds efficiently to the fusion protein containing TPRs 1–9 of Ssn6: the wild-type α2 was almost completely depleted from the supernatant fraction, and it reappeared in the bound or pellet fraction. Many other proteins present in the extract were not bound by the beads, showing the specificity of the α2-TPR interaction (not shown, see ref. 4). In contrast, α2 containing the R173A mutation was only minimally depleted from the supernatant fraction, and only minimally detectable amounts reappeared in the eluate fraction. These results indicate that the PTS1-like sequence in the homeodomain of α2 (SRI) is required for the efficient interaction of α2 with the TPRs of Ssn6. We repeated this experiment by using a single TPR (TPR6) immobilized on the beads, instead of the tandem array. As seen in Fig. 2B, wild-type protein was retained by the beads, whereas the α2 R173A was not. This result indicates that a single TPR repeat recognizes the same determinant on α2 as does the tandem array.
Figure 2.
An intact PTS1 sequence in the homeodomain of α2 is required for TPR binding. Wild-type or mutant (R173A) α2 extracts (Load) were incubated with resin containing immobilized GST-TPRs. After the incubation, the supernatant (Sup) contains unbound protein, and bound protein is pelleted with the resin and later eluted (Pellet). Equivalent volumes of sample were loaded in all lanes of a SDS/PAGE gel. Shown are Western blots of these gels probed with anti-α2 antibody. (A) This blot shows the result of incubating bacterial extracts containing either partially purified wild-type α2 or α2 R173A with resin containing immobilized GST:Ssn6 TPRs 1–9. (B) The resin contained immobilized GST:Ssn6 TPR6. The experiment of A was conducted at a final salt concentration of 175 mM, whereas the experiment in B was conducted at 150 mM final salt. The band of larger molecular weight present in the lanes containing α2 R173A is a covalent dimer of α2. The fraction of covalent dimer present in wild-type preparations is known to vary (39) and because the R173A protein behaves like the wild-type with respect to DNA binding and protein–protein interactions with a1 and Mcm1 (see below) we do not believe the presence of this dimer affects the interpretation of these experiments.
A trivial explanation for the effect of the R173A mutation on the interaction of α2 with the TPRs of Ssn6 is that the substitution affects the structure of the α2 homeodomain and thereby nonspecifically disrupts the interaction. To rule out this possibility, we tested whether the mutation affected the ability of the mutant α2 to bind DNA and to interact with two of its partner proteins, a1 and Mcm1. Fig. 3A shows that α2 R173A is able to bind an a-specific gene operator cooperatively with Mcm1, and Fig. 3B shows that α2 R173A binds cooperatively with a1 to a haploid specific gene operator. In both cases the affinities were indistinguishable from wild type. These results indicate that the mutation does not affect the local structure of the homeodomain, which is required to bind DNA, and also does not affect the structure of the remaining portions of the protein required to interact with Mcm1 and a1 (17, 22–24). In addition to the R173A mutation, a triple alanine mutation (S172A; R173A; I174A) which changed the entire PTS1-like sequence in the loop was constructed. As expected, this mutant α2 also failed to bind the TPRs of Ssn6; however it showed a decreased binding affinity to DNA in combination with Mcm1, suggesting that the mutant α2 had a structural defect. For this reason we limited our interpretation to experiments performed with the R173A mutant.
The −SRI in the Homeodomain of α2 Is Required for Repression in Vivo.
We next tested whether the SRI in the homeodomain of α2 disrupted the interaction with Ssn6 in vivo. A library of mutations in the homeodomain of α2 previously had been constructed and analyzed (17). A number of these mutations altered the DNA-binding surface of α2 and, predictably, disrupted DNA binding and therefore biological function. In contrast, most mutations on the non-DNA binding surface of the homeodomain seemed to have little or no effect on the biological activity of α2. One of the few mutations located outside the DNA-binding surface (see ref. 19, and above) that did disrupt biological function was the R173A substitution, and, as described below, we carefully reexamined its effect in vivo.
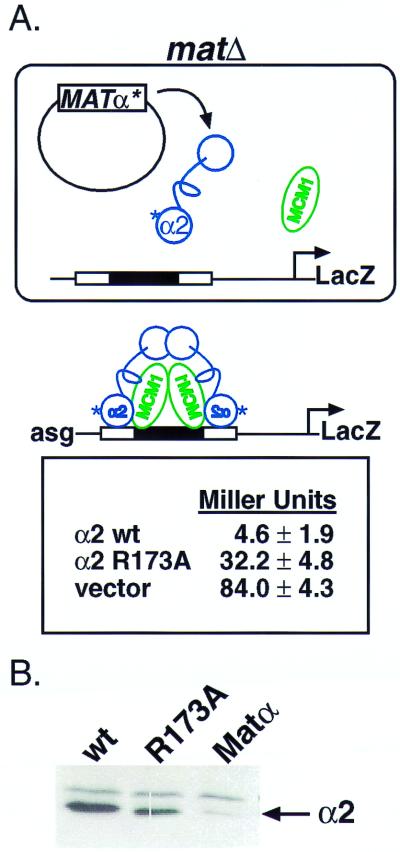
In α cells, Ssn6 and Tup1 are brought to the promoters of a-specific genes by association with DNA-bound α2 and Mcm1 (9, 25). To test the ability of the mutant α2 protein to bring about repression of the a-specific gene set, we transformed a plasmid that expresses either wild-type or mutant α2 into a yeast strain whose MAT locus had been deleted. This strain also carries an a-specific gene reporter, and β-galactosidase assays thereby provide a quantitative readout of the ability of α2 to direct repression (17). As seen in Fig. 4A, matΔ cells that express wild-type α2 show strong repression of the reporter, whereas matΔ cells expressing α2 R173A are defective, but not completely inactive, in repression. We know that Tup1 interacts with the N terminus of α2 (12) and we believe that the residual repression observed for the mutant α2 activity results from the recruitment of Ssn6-Tup1 through the Tup1 interaction.
Figure 4.
The PTS1 in the homeodomain of α2 is required for repression of a-specific genes. (A) (Upper) A schematic diagram illustrating the reporter gene system used in this experiment to quantitate repression. A matΔ strain contains a reporter gene construct integrated into the chromosome. In the reporter, upstream regulatory sequences control the activity of the LacZ gene. Here, these regulatory sequences contain an α2/Mcm1 operator. For repression to occur, α2 must bind its operator with Mcm1. In the absence of repression, the strain expresses high levels of β-galactosidase (vector). (Lower) Data from liquid β-galactosidase assays, effectively quantitating the amount of repression. In these experiments, we have conducted liquid β-galactosidase assays, in triplicate, on three unique transformants. Our data agrees with that previously published in ref. 17. (B) α2 and α2 R173A are expressed at comparable levels within the cells. Western blots of whole-cell extracts from the strains used in the liquid β-galactosidase assays above were probed with anti-α2 antibody. wt, wild type.
A trivial explanation for this result is that the mutant protein is present at lower intracellular levels than the wild-type protein. However, the result of Fig. 4B demonstrates that α2 containing the R173A mutation is present at levels approximately the same as those of the wild-type protein. Thus, these results demonstrate that the R173A is defective in repression of a reporter construct and is consistent with the biochemical observation that the R173A mutation disrupts the interaction of α2 with Ssn6.
The library of α2 mutations mentioned above also included a mutation in the isoleucine in the third position of the PTS1-like sequence, I174A. This mutation, not retested here, showed a defect in repression significantly smaller than that of the R173A mutation. These results suggest that although both mutations affect the interaction the arginine (R173) has the more pronounced effect.
Peptides Containing the PTS1-like Sequence Are Not Sufficient to Direct the TPR-Target Interaction.
The above results demonstrate that a PTS1-like sequence is necessary for the interaction between α2 and the TPRs of Ssn6. To address whether this sequence is sufficient for the interaction, we constructed synthetic peptides. The peptides were 7 aa long and contained either the SRI sequence or this PTS1-like sequence reversed (IRS), as a control (see ref. 5), and we performed two types of experiments. In the first, we attempted to block by competition α2 binding to GST:TPRs with the peptides, and in the second we directly measured the binding of radiolabeled peptides to GST:Ssn6. In neither case could it be shown conclusively that the peptide bound specifically to the TPRs of Ssn6 or Pex5. Finally, we crosslinked the peptides to a solid affinity resin and determined whether a purified TPR construct selectively interacted with the immobilized peptides. Again, we could not demonstrate a conclusive interaction between the peptides and the TPRs. Several scenarios could explain these negative results, including the possibilities that the peptides failed to fold into the proper conformation and that additional Ssn6-contact points on α2 lie outside this sequence.
Mutation of the PTS1-Like Sequence in α2 Disrupts an Interaction with the TPRs of Pex5.
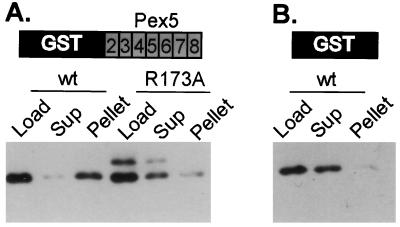
TPRs constitute a highly degenerate repeated sequence motif. Because α2 can interact with a number of different TPRs from Ssn6, we were interested to determine whether the PTS1-like sequence in α2 could bind to the TPRs from a completely different protein in vitro. In the experiment of Fig. 5, extracts containing wild-type or mutant α2 were passed over an affinity column containing immobilized TPRs 2–8 of Pex5 (the contiguous block of Pex5 TPRs). The results show that α2 is indeed capable of binding an affinity column containing the array of Pex5 TPRs. Furthermore, the R173A mutation in α2 disrupts this interaction. These experiments indicate that the PTS1-like sequence on α2 is recognized by the TPRs from both Ssn6 (a natural interaction) and Pex5 (a presumed nonphysiological interaction) and support the idea that different TPR-target protein interactions can occur by a common mechanism.
Figure 5.
The PTS1-like sequence in α2 is required for the interaction with the TPRs of Pex5. The experiment was conducted as those in Fig. 2 with the exception that in A the resin contained immobilized GST:Pex5 TPRs 2–8, schematized by shading the TPRs. (B) This experiment serves as a binding control. In this experiment the resin contained only immobilized GST. The experiment of B was conducted at a final salt concentration of 175 mM, whereas the experiment in A was conducted at 150 mM final salt.
Discussion
In this paper we have demonstrated both in vivo and in vitro that the TPRs of Ssn6 recognize a loop in the homeodomain of α2. The amino acid sequence of this loop is very similar to the tripeptide signal (known as PTS1) that directs the interaction of peroxisomal proteins with the TPRs of the peroxisomal import receptor, Pex5. The similarity between the way that α2 interacts with the TPRs of Ssn6 and the way the peroxisomal proteins interact with the TPRs of Pex5 is underscored by the demonstration that wild-type α2 can be made to interact in vitro with the TPRs of Pex5. Moreover, a mutation in α2 (R173A) that disrupts the α2-Ssn6 interaction also disrupts the α2-Pex5 interaction. These experiments support the idea that different TPRs, either from the same protein or from different proteins, can recognize the same short amino acid sequences.
Where Does the Specificity Come From?
If the TPRs of Pex5 can recognize α2 why doesn't α2 end up in the peroxisome? The likely answer is that the PTS1-like sequence is only one of a number of factors that provide specificity to the α2-Ssn6 interaction. At the most fundamental level, cellular localization could be an important determinant of specificity. For example, α2 is a nuclear protein; except for the brief moments after its translation and before its import into the nucleus, it is not in the appropriate part of the cell to be recognized by Pex5 and imported into the peroxisome. Additionally, it is possible that the nuclear localization signals on α2 simply could override those potentially directing peroxisomal import. An example of this type of signal prioritization is seen for the protein carnitine-acetyltransferase, the Cat2 gene product in S. cerevisiae, which contains signals for both mitochondrial and peroxisomal import. Only when Cat2 is missing the mitochondrial-targeting signal, generated through the use of an alternative initiation codon, is it imported into the peroxisome (26).
Separation of function by cellular compartmentalization cannot readily explain all instances of TPR-mediated specificity, as there are many TPR proteins that reside in the same compartment. It seems likely that features of the target proteins in addition to the PTS1-like sequence are involved. Thus interactions with the TPR through additional sequence determinants, with other parts of the TPR protein or with other proteins complexed with the TPR protein may be required for a productive interaction to occur. For example, Ssn6 is always found complexed with Tup1 (27) and we know that α2 interacts with both Ssn6 and Tup1 (4, 12). As shown in this paper, a loop in the homeodomain of α2 interacts with Ssn6; previous work has shown that a determinant at the other end of the protein (the amino terminus) interacts with Tup1 (12). The successful interaction of α2 with Ssn6-Tup1 requires both sets of interactions, because mutations in either binding determinant are defective in repression (ref. 12, this work). Finally, it has been shown that the attachment of a PTS1 to the end of a heterologous protein is not always sufficient to direct import into the peroxisome; additional sequences present on the target protein sometimes are required (28, 29). These additional sequences might either contribute to the interaction with Pex5 or mediate interactions with other components of the import machinery. Just as multiple recognition events might contribute to specificity in the systems described above, this second recognition step might provide a specificity check on proteins delivered to the peroxisome by Pex5; the resident peroxisomal membrane proteins could confirm that the peroxisome is the intended destination of the protein by checking for other determinants on its surface.
Can Other Signals Direct TPR Target Interactions?
The structure of a TPR protein, PP5, recently has been solved, and it reveals that each TPR has two distinct faces (30). The tandem array of TPRs in the structure resembles a cupped hand with an inner and an outer surface, and the authors suggest that the inside of the hand could provide an interaction surface for an α-helix. Recent work on Hsp90 has demonstrated that it binds this inner TPR groove, by using a non-PTS1 based signal (31). Given the size of this groove, it seems unlikely that multiple proteins fit into it simultaneously. It is plausible that the PTS1-mediated interactions might occur along the outer face of the TPR structure. Because of the curvature of the TPR array, it should be possible for several PTS1-containing targets to interact simultaneously with different TPRs along the outside of the array.
The experiments reported here, in combination with work using other TPR systems, suggests that there are at least two mechanisms by which TPRs recognize target proteins. One mechanism, that exemplified by Hsp90, requires interactions across a whole array of TPRs and so is fairly rigid in its assembly. The other, exemplified by α2 and the PTS1 sequence, is directed, at least in part by a consensus sequence on the target protein that allow it to occupy any one repeat in the array. This latter mode of binding would allow for considerable flexibility in the assembly to TPR-target complexes a feature that has been proposed to contribute to tight transcriptional repression by Ssn6 (4).
Acknowledgments
We thank Martha Stark for sharing purified α2 and a1 homeodomain proteins, Stephen M. Soisson and Cynthia Wolberger for sharing purified Mcm11–96, Andreas Hartig for providing the plasmid pGAD.RI that was used as a PCR template to generate the GST:Pex5 TPRs 2–8 construct, and Stephen Rader for help with Fig. 1. We are indebted to Wendell Lim and Cynthia Wolberger for valuable discussions and comments on the manuscript. We also thank Corrie Detweiler for stimulating discussions. Finally, we thank the members of the Johnson Laboratory for their insight, assistance, patience, and critical comments on the manuscript. This work was supported by a grant from the National Institutes of Health to A.D.J.
Abbreviations
- TPR
tetratricopeptide repeat
- PTS1
peroxisomal targeting sequence 1
- GST
glutathione S-transferase
- BBII
binding buffer II
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.070506797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.070506797
References
- 1.Hirano T, Kinoshita N, Morikawa K, Yanagida M. Cell. 1990;60:319–328. doi: 10.1016/0092-8674(90)90746-2. [DOI] [PubMed] [Google Scholar]
- 2.Sikorski R S, Boguski M S, Goebl M, Hieter P. Cell. 1990;60:307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- 3.Lamb J R, Tugendreich S, Hieter P. Trends Biochem Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- 4.Smith R L, Redd M J, Johnson A D. Genes Dev. 1995;9:2903–2910. doi: 10.1101/gad.9.23.2903. [DOI] [PubMed] [Google Scholar]
- 5.Terlecky S R, Nuttley W M, McCollum D, Sock E, Subramani S. EMBO J. 1995;14:3627–3634. doi: 10.1002/j.1460-2075.1995.tb00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tzamarias D, Struhl K. Genes Dev. 1995;9:821–831. doi: 10.1101/gad.9.7.821. [DOI] [PubMed] [Google Scholar]
- 7.Shpungin S, Liberzon A, Bangio H, Yona E, Katcoff D J. Proc Natl Acad Sci USA. 1996;93:8274–8277. doi: 10.1073/pnas.93.16.8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radanyi C, Chambraud B, Baulieu E E. Proc Natl Acad Sci USA. 1994;91:11197–11201. doi: 10.1073/pnas.91.23.11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keleher C A, Redd M J, Schultz J, Carlson M, Johnson A D. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 10.Schultz J, Marshall-Carlson L, Carlson M. Mol Cell Biol. 1990;10:4744–4756. doi: 10.1128/mcb.10.9.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 12.Komachi K, Redd M J, Johnson A D. Genes Dev. 1994;8:2857–2867. doi: 10.1101/gad.8.23.2857. [DOI] [PubMed] [Google Scholar]
- 13.Tzamarias D, Struhl K. Nature (London) 1994;369:758–761. doi: 10.1038/369758a0. [DOI] [PubMed] [Google Scholar]
- 14.Waterham H R, Cregg J M. BioEssays. 1997;19:57–66. doi: 10.1002/bies.950190110. [DOI] [PubMed] [Google Scholar]
- 15.Gould S J, Keller G A, Hosken N, Wilkinson J, Subramani S. J Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCollum D, Monosov E, Subramani S. J Cell Biol. 1993;121:761–774. doi: 10.1083/jcb.121.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vershon A K, Jin Y, Johnson A D. Genes Dev. 1995;9:182–192. doi: 10.1101/gad.9.2.182. [DOI] [PubMed] [Google Scholar]
- 18.Swinkels B W, Gould S J, Subramani S. FEBS Lett. 1992;305:133–136. doi: 10.1016/0014-5793(92)80880-p. [DOI] [PubMed] [Google Scholar]
- 19.Wolberger C, Vershon A K, Liu B, Johnson A D, Pabo C O. Cell. 1991;67:517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- 20.Tan S, Richmond T J. Nature (London) 1998;391:660–666. doi: 10.1038/35563. [DOI] [PubMed] [Google Scholar]
- 21.Li T, Stark M R, Johnson A D, Wolberger C. Science. 1995;270:262–269. doi: 10.1126/science.270.5234.262. [DOI] [PubMed] [Google Scholar]
- 22.Ho C Y, Adamson J G, Hodges R S, Smith M. EMBO J. 1994;13:1403–1413. doi: 10.1002/j.1460-2075.1994.tb06394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mak A, Johnson A D. Genes Dev. 1993;7:1862–1870. doi: 10.1101/gad.7.10.1862. [DOI] [PubMed] [Google Scholar]
- 24.Mead J, Zhong H, Acton T B, Vershon A K. Mol Cell Biol. 1996;16:2135–2143. doi: 10.1128/mcb.16.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keleher C A, Passmore S, Johnson A D. Mol Cell Biol. 1989;9:5228–5230. doi: 10.1128/mcb.9.11.5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elgersma Y, van Roermund C W, Wanders R J, Tabak H F. EMBO J. 1995;14:3472–3479. doi: 10.1002/j.1460-2075.1995.tb07353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams F E, Varanasi U, Trumbly R J. Mol Cell Biol. 1991;11:3307–3316. doi: 10.1128/mcb.11.6.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Distel B, Gould S J, Voorn-Brouwer T, van der Berg M, Tabak H F, Subramani S. New Biol. 1992;4:157–165. [PubMed] [Google Scholar]
- 29.Blattner J, Swinkels B, Dorsam H, Prospero T, Subramani S, Clayton C. J Cell Biol. 1992;119:1129–1136. doi: 10.1083/jcb.119.5.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das A K, Cohen P T W, Barford D. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russel L C, Whitt S R, Chen M S, Chinkers M. J Biol Chem. 1999;274:20060–20063. doi: 10.1074/jbc.274.29.20060. [DOI] [PubMed] [Google Scholar]
- 32.Sherman F. In: Guide to Yeast Genetics and Molecular Biology. Guthrie C, Fink G R, editors. Vol. 194. San Diego: Academic; 1991. pp. 3–20. [Google Scholar]
- 33.Keleher C A, Goutte C, Johnson A D. Cell. 1988;53:927–936. doi: 10.1016/s0092-8674(88)90449-7. [DOI] [PubMed] [Google Scholar]
- 34.Peter M, Gartner A, Horecka J, Ammerer G, Herskowitz I. Cell. 1993;73:747–760. doi: 10.1016/0092-8674(93)90254-n. [DOI] [PubMed] [Google Scholar]
- 35.Moazed D, Johnson A D. Cell. 1996;86:667–677. doi: 10.1016/s0092-8674(00)80139-7. [DOI] [PubMed] [Google Scholar]
- 36.Stark M R, Johnson A D. Nature (London) 1994;371:429–432. doi: 10.1038/371429a0. [DOI] [PubMed] [Google Scholar]
- 37.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brocard C, Kragler F, Simon M M, Schuster T, Hartig A. Biochem Biophys Res Commun. 1994;204:1016–1022. doi: 10.1006/bbrc.1994.2564. [DOI] [PubMed] [Google Scholar]
- 39.Sauer R T, Smith D L, Johnson A. Genes Dev. 1988;2:807–816. doi: 10.1101/gad.2.7.807. [DOI] [PubMed] [Google Scholar]