Abstract
The crystal structure of native chicken fibrinogen has been determined at a resolution of 5.5 Å. The full-length molecule is 460 Å in length and sigmoidally shaped. The structure includes the full sweep of the coiled coils that connect the central and terminal domains; the chain paths of the central domain confirm a predicted scheme of planar disulfide rings in apposition with each other. Electron density maps have revealed the outlines of disordered αC domains nestled within the confines of the sinuous coiled coils. The amino-terminal segments of the α- and β-chains, including the fibrinopeptides A and B, are also disordered.
Keywords: central domain, coiled coils, x-ray structure
Fibrinogen is a large (molecular mass = 340 kDa) hexameric (α2β2γ2) glycoprotein found in the blood plasma of all vertebrate animals; it is converted into fibrin clots by the thrombin-catalyzed removal of peptides from the amino-terminal regions of the α- and β-chains. Tens of thousands of studies have been conducted on this system in the past, and its general properties are well known. Shadow-cast electron microscope images long ago revealed a triglobular structure approximately 470 Å in length (1), the subdomainal structure of which has been partially visualized by negative staining electron microscopy and low resolution x-ray crystallography (2, 3). The native protein has proved very difficult to crystallize, however, and no diffraction-grade crystals have been reported. There have been some recent successes in determining x-ray structures of certain core fragments. The first of these to be reported was a 30-kDa recombinant protein corresponding to the carboxyl-terminal domain of the γ-chain of human fibrinogen (4); it was followed shortly thereafter by a structure for the 86-kDa fragment D, also from human fibrinogen, and its crosslinked equivalent from fibrin (5–7). The D fragments account for about half the mass of the fibrinogen molecule. Most recently, the partial structure of a 285-kDa moiety prepared from bovine fibrinogen was reported (8).
One of the problems in obtaining good crystals of native fibrinogen has been attributed to the presumed mobility of the carboxyl domains of the α-chains. These domains (αC) are the most variable parts of the molecule on a species-to-species basis (9). They are easily trimmed away by proteolysis and are often referred to as “free-swimming appendages.” Moreover, most contain a series of repeated sequences (10, 11) that are thought to add to the flexible nature of the region. As it happens, chicken fibrinogen α-chains lack these repeats (12), an attribute that led us to attempt crystallization of the native protein. Indeed, we were able to obtain large, easily managed crystals in short order. Like crystals of modified bovine fibrinogen (3, 8), these crystals have a high solvent content and diffract anisotropically.
We now report a structure for native chicken fibrinogen at 5.5 Å, determined by the method of molecular replacement by using high-resolution structures of fragment D (5–7) as search models. The full extent of the interdomainal coiled coils and the chain paths and connections within the central domain has been identified. Electron density maps have also revealed the outlines of the disordered carboxyl regions of the α-chains. The amino termini of the α- and β-chains, including the fibrinopeptides A and B, are also disordered, a feature attributable to the extreme flexibility of these regions.
Experimental Procedures
Sequence Numbering.
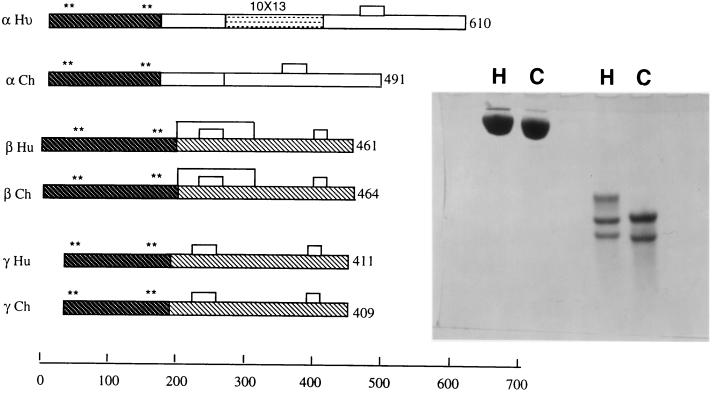
Overall, the amino acid sequence of chicken fibrinogen is about 60% identical with that of the human protein. Different regions are conserved to different extents, however. Although the β- and γ-chains are both about 67% identical with their human counterparts, the α-chains share only about 37% identity, mostly because the carboxyl-terminal region changes so rapidly during evolution, even allowing for the absence of the aforementioned repeats in chicken fibrinogen (Fig. 1). For purposes of comparison, all residue identification is given with the human numbering (10) unless otherwise specified.
Figure 1.
Schematic depiction of α-, β-, and γ-chains of chicken (Ch) and human (Hu) fibrinogens showing missing repeat region (10 × 13 resolution, stippled) from chicken α-chains. Dark shading, homologous central domains and coiled-coil regions. Light shading, homologous βC and γC domains. Asterisks (*) denote locations of disulfide ring cysteines; scale shows chain lengths in residues. (Inset) SDS/5% polyacrylamide gels of human (H) and chicken (C) fibrinogens. Lanes 1 and 2, unreduced; lanes 3 and 4, reduced. Note that chicken α-chains are only slightly larger than β-chains.
Chicken Fibrinogen.
Fresh blood was collected at Wing Lee Poultry (San Diego) into approximately 1/10 volumes of cold 0.1 M trisodium citrate and centrifuged to remove cells. Crude fibrinogen was prepared by a modified cold ethanol method, after which the preparation was passed first over a Gly-Pro-Arg-Pro affinity column (13, 14) and then lysine-Sepharose (15). The material was concentrated to 6 mg/ml in 0.15 M NaCl/0.05 M imidazole, pH 7.0 and stored in small aliquots at −70°C. Unreduced preparations gave single bands on SDS gel electrophoresis (Fig. 1). Amino-terminal sequencing (University of California San Diego Sequencing Facility) gave a single amino-terminal sequence (YIATREN), both the α- and β-chains being blocked by cyclized terminal glutamines.
Crystallization.
Crystals were obtained by vapor diffusion from sitting drops at room temperature. A large range of conditions was explored. The quality of these crystals was improved significantly when peptide ligands corresponding to the A and B “knobs” were present (16, 17). In the end, fibrinogen solutions (6 mg/ml in 0.15 M NaCl/0.05 M imidazole, pH 7.0) containing 2 mM Gly-Pro-Arg-Pro-amide and Gly-His-Arg-Pro-amide were mixed volume for volume with well solutions containing 3–4% (vol/vol) polyethylene glycol (molecular weight = 3,350), 0.05 M imidazole (pH 6.5), 2 mM CaCl2, and 1 mM sodium azide. Crystals suitable for x-ray diffraction grow over the course of 1–2 weeks. Some crystals were treated with 20 mM glutaraldehyde (1:10 vol/vol) for 6–14 h just before mounting in an effort to stabilize them and protect them from radiation damage during data collection at room temperature (18).
Data Collection and Processing.
Diffraction data were collected both at the Stanford Synchrotron Radiation Laboratory (beamline 9-1) and at the National Synchrotron Light Source (Brookhaven, beamline X12C). Initial experiments at the Stanford Synchrotron Radiation Laboratory revealed that room temperature data were of higher quality than those obtained from flash-frozen crystals at 100 K, the latter being highly mosaic. Accordingly, advantage was taken of the very fast data collection possible with charge-coupled device detectors at the National Synchrotron Light Source, which allowed us to collect the better room temperature data. Treatment with glutaraldehyde was also helpful in obtaining crystals that survived for longer periods in the beam at room temperature.
At room temperature, the crystals of the chicken ternary fibrinogen-peptide complex diffracted asymmetrically to 4.0 Å in the a* and c* directions but to only 5.5 Å in the b* direction (Table 1). The observed anisotropic diffraction may be a consequence of the strong end-to-end packing that occurs around the terminal domains (corresponding to fragments D) and weak interactions involving the more centrally located entities, including the αC regions. Data processing with denzo and scalepack (19) indicated that the crystals belong to space group P21; comparison of the cell parameters of the native and glutaraldehyde-treated crystals showed only marginal differences, all within 3% (Table 1). There is one molecule in the asymmetric unit; the solvent content exceeds 65%. The glutaraldehyde-treated crystals proved sturdier and allowed the collection of a complete data set on a single crystal (Table 1).
Table 1.
Data collection and processing statistics
| Data statistics | Native | Glutaraldehyde-treated |
|---|---|---|
| Space group | P21 | P21 |
| Unit cell dimensions | a = 114.6 | 114.7 |
| b = 104.8 | 101.8 | |
| c = 207.2 | 210.3 | |
| β = 105.9° | 106.7° | |
| Matthews coefficient | 3.9 | 3.8 |
| Percentage solvent | 67 | 66 |
| Molecules/asym. unit | 1 | 1 |
| Number of crystals used | 2 | 1 |
| Mosaicity | 0.58 | 0.53 |
| Highest resolution, Å | 5.0 | 5.0 |
| Observations, no. | 44,029 | 45,940 |
| Unique reflections, no. | 15,526 | 16,412 |
| Mean redundancy | 2.9 | 2.8 |
| Completeness, % | 73.9 | 80.0 |
| Completeness outer shell (5.9-5.5) | 52.3 | 65.4 |
| Rsym (I)* | 0.11 | 0.09 |
*Rsym (I) = ∑ | I − 〈I〉 |/∑ | I |.
Structure Determination.
Molecular replacement was carried out with amore (20) on the two sets of data (native and glutaraldehyde-treated) in the resolution range of 15–6 Å. A single D domain from a 2.3-Å structure of crosslinked fragments D (PDB code = 1FZC) was used as a search model. Residues of human fragment D that differ from the chicken sequence were replaced by alanines or, where appropriate, glycines. For both data sets, solutions were found that corresponded to the two fragments D in each fibrinogen molecule, but as a result of ambiguity in the choice of unit cell, the solutions for the native and glutaraldehyde-treated data were unique but equivalent. In the case of the native data, the correlation coefficient was 59.5 and the R factor 0.47. The corresponding values for the glutaraldehyde data were 58.2 and 0.45.
Initial 2Fo − Fc and Fo − Fc difference electron density maps were calculated at 5.5 Å by using the fragment D phases with SIGMAA weighting (21). A suitable mask was constructed, after which phases were improved by density modification involving solvent flattening and histogram mapping as provided in the ccp4 package (22). At this point, tubes of density corresponding to the full extent of the coiled coils were clearly evident. Model building of the regions not present in fragment D was conducted with the program package o (23). The extension of the model greatly improved the phases, to the point where the mask was not used, and the disordered regions outside the initial envelope appeared. In some cases, the glutaraldehyde data were used exclusively.
Results
General Structure.
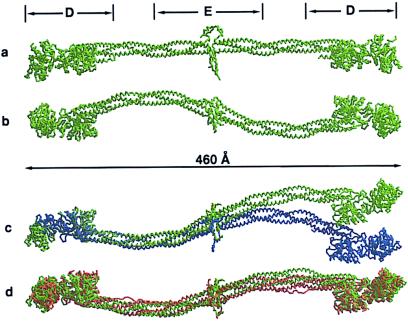
The mainframe structure of chicken fibrinogen, not including the flexible αC domains, is sigmoidal in shape (Fig. 2); it measures 460 Å in its longest dimension. The structure bears a remarkable resemblance to images obtained by negative-staining electron microscopy (2) and is virtually superimposable on the recently reported bovine structure (Fig. 2d). The coiled-coil connections are significantly bent, over and beyond what is expected for a three-stranded coiled coil.
Figure 2.
Mainframe αC backbone structures of full-length chicken fibrinogen. D and E denote regions of major core fragments. (a) Top view. (b) Side view. (c) Demonstration of pseudosymmetry of fibrinogen dimer; the γC domain from one end of structure (chain C; green) was superimposed on the γC domain from the other (chain F; blue). (d) Superposition of chicken model (green) on that recently reported for modified bovine fibrinogen (brown; ref. 8). Figures were prepared with raster3d (32, 33) and xtalview (34).
The central domain has the shape of an oblate disk defined by sets of radial spokes on each side of the covalent dimer. When viewed along an axial projection from either coiled coil, the disulfide rings have a trigonal aspect, the γ-, β-, and α-chains emerging along the three legs of a Y. The two γ-chain segments are opposite each other, and as a result, the comparable α- and β-chain segments are juxtaposed, allowing disulfide bonds between α36 and β65 from opposing sides of the dimer. The situation with regard to the chain segments on the amino-terminal sides of those bonds is less clear. The α-chain portions are constrained by a disulfide bond between the two cysteines at α28 and must end up diametrically opposite to where the two γ-chains are bridged. The sidedness is reminiscent of the distinct dorsal–ventral aspect of the D dimer structure (5); it is essential that the A knobs be near each other to pin the two halves of the D dimer together. The amino-terminal segments of the α- and β-chains, which constitute the fibrinopeptides A and B, respectively, were not apparent in the electron density maps, and these regions must be highly flexible.
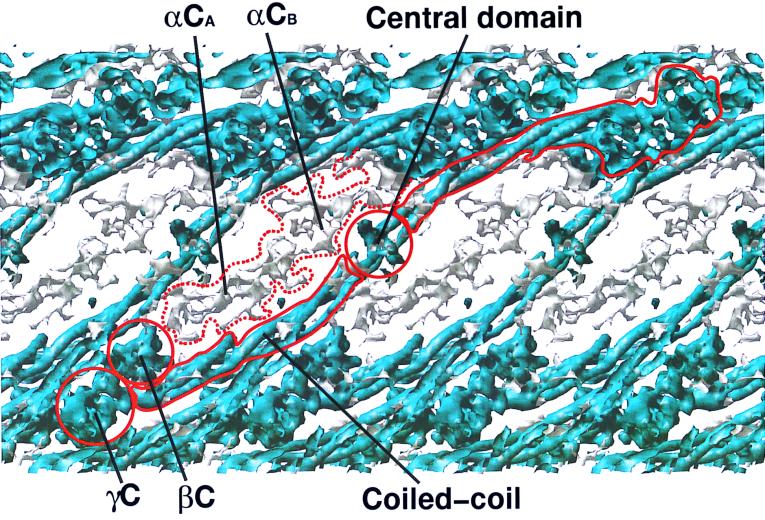
The electron density corresponding to the flexible αC domains is weak (Fig. 3), and at the present resolution, it was not possible to trace the individual chains. Indeed, it was not even possible to assign the αC domains with certainty to particular molecules. Our preliminary interpretation is that, in the crystal, an interaction occurs between αC domains of adjacent molecules, rather than within the same molecule.
Figure 3.
Section of an electron density map calculated at 8 Å showing native chicken fibrinogen mainframe backbone density (blue) and disordered regions of αC domains (gray). The figure was prepared with the program ribbon (35).
Packing Considerations.
As in other fibrinogen structures, the molecules are packed end-to-end in the same way that occurs at the interface of the γC domains in fibrin (5). The side-by-side packing primarily involves a looser interaction between the βC and γC domains of neighboring molecules. These two sets of interactions account for the fundamental layering in the crystals. The layers themselves are accommodated by interactions between the spines (mid points) of the coiled coils packing into the groove between βC and γC domains. Additional interactions involve the central domain, one side of which is packed against a neighboring βC domain, and the other of which faces the coiled coil.
Central Domain.
The electron density was clearer on one side of the central domain dimer than on the other, a likely result of crystal packing. In this regard, one side of the domain packs against the globular region of the β-domain, whereas the other is situated vis-à-vis the disordered αC domain. Accordingly, some features of the weaker side were modeled on the basis of expected symmetry. Generally, maps were interpretable beginning at residue α34, β63, and γ1, although the model could have been extended into a region of weaker density in the case of the α-chain on the basis of an anticipated disulfide bond at α28. Given the uncertainty of the course of the nearby β-chain in the segment preceding the disulfide bond between β65 and α36, we have not included this region in the model.
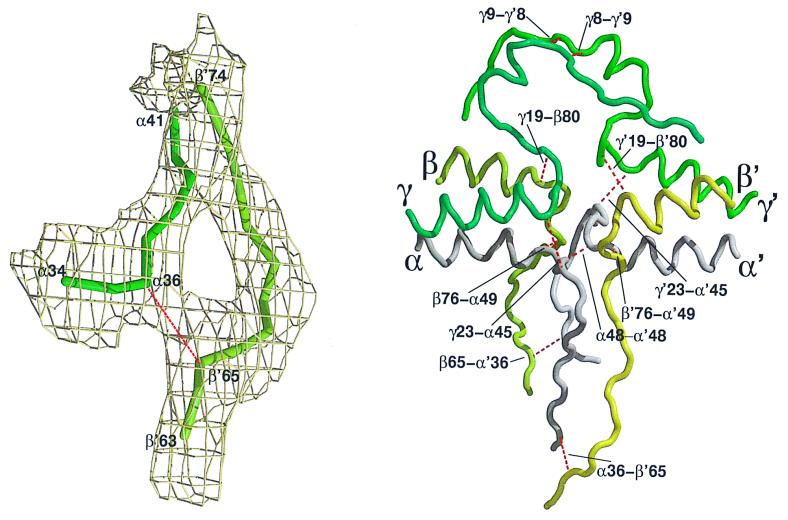
The disulfide connections include the three disulfide ring junctions between cysteines at β76 and α49, α45 and γ23, and γ19 and β80 (Fig. 4). The interhalf dimer connections include α36 and β65, the partners for which arise on opposite sides of the dimeric interface. There are also antiparallel connections between the two sets of cysteines at γ8–γ9. Also, biochemical experiments require that there be an interdimeric bond between the two α28 residues (25). Finally, chicken fibrinogen has an additional cysteine in each half of the dimeric molecule at α48, and a disulfide bond clearly connects them across the dyad axis (Fig. 4).
Figure 4.
Structural arrangement of central domain. (Left) Final 2Fo − Fc electron density map of α-chain residues 34–41 and β′-chain residues 63–74. This map was calculated at 5.5 Å with data from the glutaraldehyde-treated crystal. (Right) Backbone structure of dimeric central domain encompassed by residues α-chain 34–60 (purple), β-chain 63–90 (yellow), and γ-chain 1–35 (green). Disulfide bonds (n = 11) are shown (red broken lines). The figures were prepared with raster3d (32, 33), xtalview (34), and molscript (36).
The regions of the α- and β-chains nearer to the amino termini, including the fibrinopeptides, are disordered, and it was not possible to identify the likely locations of the A and B knobs. Nonetheless, the α-chains are restricted by the disulfide bond connecting the residues Cysα28 and Cysα′28 and must reside relatively near each other on the side of the protein diametrically opposite to the disulfide-bonded γ-chain terminal segments. The β-chains are not so encumbered, and the B knobs may be widely separated.
Discussion
Crystal Packing and Symmetry.
Although fibrinogen is a genuine covalent dimer, the two halves of which have identical sequences, packing constraints make the two halves distinctive. In particular, the end-to-end packing between abutting γ-chains necessitates an accommodating twist, and as in the case of the crosslinked D dimer (5), the two halves of each molecule are defined arbitrarily as either A (made of the A, B, and C chains) or B (made of the D, E, and F chains) according to the interfacial partners of Tyrγ275 (5). The extent and nature of the required twist is demonstrated easily by superposing one end of a fibrinogen molecule on its other (Fig. 2c). It is conceivable that interactions between the two halves of the central domain may also contribute to the twist and resulting pseudosymmetry, but that is a matter that can be ascertained only on molecules or fragments that are not packed end-to-end. The fact that the end-to-end packing is exactly the same in D dimer (5), in which the central domain is not present, makes such a contribution seem unlikely, however.
Flexible αC Domains.
The pseudosymmetry is also evident in the cases of the flexible αC domains, the two versions of which seem to have settled into somewhat different forms in the crystal (Fig. 3). It is likely that, under physiological conditions, this region of the molecule is extremely flexible, the zones occupied in the crystal being favored for reasons of packing. On the other hand, the fact that the observed end-to-end packing has a physiological counterpart (5) suggests that these conformations may also occur under natural circumstances, an aspect relevant to proposals that implicate αC domains in the lateral association of protofibrils (24).
Central Domain.
Because of its great functional importance, the central domain has been a subject of great biochemical scrutiny over the years, and the disulfide connections in this region have been the subject of many past experiments. Initially, the 11 (12 in chicken) disulfide bonds in this central core were divided into two categories: those that bound the various chains within a given half of the molecule and those that joined the two halves together (25). Three of the bonds in each half of the protein involved braces of cysteines separated by three residues in each of the three nonidentical chains. Although it was never possible to assign the connections by direct experiment, it was proposed on evolutionary grounds that they must be in the form of rings (9, 26). Because the three nonidentical chains are homologous, it was reasoned, the original molecule was likely a homotrimer. The only way three chains can be bound by such braces of identically situated cysteines is to have the first of one set bond to the second of another and the same again and still again until the ring is closed. In fact, the observed structure conforms exactly to that reasoning (Fig. 4). The evolutionary homology is reflected further in the trigonally disposed spokes that emerge.
With regard to the interhalf attachments, it should be noted that chicken α-chains have an additional cysteine (α48, human numbering) within the sequence that contributes to the disulfide ring. We had supposed it would of necessity have to cross the dyad between the two half molecules, and it does. Additionally, γ-chain residues 8 and 9 were known to form interhalf attachments (25). The nature of the attachment is antiparallel (residue 8 of one side to residue 9 of the other and vice versa), just as had been shown previously for human fibrinogen by a chemical approach (27). The bond between α-chain residue Cys-36 and β-chain residue Cys-65 was thought originally to be within the half molecule (25), but site-directed mutagenesis studies subsequently showed that the bond must be holding half molecules together (28, 29). The x-ray structure verifies that observation.
Implications for Fibrin Formation.
The crystal structure reveals that the knobs for the knob–hole interaction involved in protofibril formation must be extended well away from the central portion of the molecule, giving rise to a very open structure for fibrin. Calorimetric studies long ago showed the independent melting of the D and E domains in both fibrinogen and fibrin (30), suggesting that the intermolecular contacts between units in fibrin must be few in number; it is not an intimate association.
In this regard, the flexible nature of two α-chains near their amino termini makes it likely that the A knobs can readily adapt to the offset nature of the γ-chain holes in abutting fibrin units (5). On the other hand, it would be expected that B knobs should be widely separated if they play a role in the lateral association of protofibrils; the necessary flexibility may be reflected in the lack of defined density corresponding to residues β1 to β60. Finally, we should mention in passing that chicken fibrinogen crystals are greatly affected by the presence of the synthetic peptide Gly-His-Arg-Pro-amide, a surrogate of mammalian B knobs, even though in chicken fibrinogen, the β-chain of fibrin ends with the sequence Ala-His-Arg-Pro (31). The strong sequence resemblance between chicken and mammalian fibrinogens in the vicinity of the β-chain hole indicates that the chicken protein should bind Gly-His-Arg-Pro-amide just as the human protein does (6, 7).
Comparison with Bovine Structure.
Chicken fibrinogen β- and γ-chains are 65–68% identical with their mammalian counterparts (Fig. 1); there are no significant structural differences apparent in the mainframe regions available for comparison (Fig. 2d). The sequence resemblances are stronger in the globular regions than in the coiled coils, but even in the coiled-coil region, the three-dimensional structures are virtually indistinguishable. In the case of α-chains, the obvious homology between chicken and mammals extends to human α245 (bovine α248), at which point the 13-residue repeats begin in mammals; the homology resumes at human residue α395. In this regard, it is noteworthy that the folded back portion of the α-chain extends significantly further along the coiled coil in the bovine case than in the chicken. Thus, in the bovine structure, the so-called “fourth helix” (5) extends to residue α224 (bovine numbering, ref. 8; α221 human numbering), whereas in the chicken, the fourth helix could be traced only to α200 (human numbering). It may be that the absence of the αC domain in the bovine preparation allows more of the α-chain to fall back against the coiled coil. Alternatively, there may be a real structural difference between the two fibrinogens in this region.
Acknowledgments
We are grateful to Justin Kollman for assistance with some of the data collection, and we thank G. Brown and C. Cohen for providing the backbone coordinates of the structure for the bovine modified fibrinogen before publication. We also thank the staffs at the Stanford Synchrotron Radiation Laboratory and the Brookhaven National Laboratory for their assistance during our visits. This research was supported by National Institutes of Health Grant HL26873. The Brookhaven National Synchrotron Light Source is supported by the U.S. Department of Energy and the National Science Foundation. The Stanford Synchrotron Radiation Laboratory Biotechnology Program is supported by the National Institutes of Health National Center for Research Resources Biomedical Technology Program and by the Department of Energy.
Footnotes
Data deposition: The Cα atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1EI3).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.080065697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.080065697
References
- 1.Hall C E, Slayter H S. J Biophys Biochem Cytol. 1961;5:11–15. doi: 10.1083/jcb.5.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willams R C. J Mol Biol. 1981;150:399–408. doi: 10.1016/0022-2836(81)90555-6. [DOI] [PubMed] [Google Scholar]
- 3.Rao S P S, Poojary M D, Elliott B W, Jr, Melanson L A, Oriel B, Cohen C. J Mol Biol. 1991;222:89–98. doi: 10.1016/0022-2836(91)90739-s. [DOI] [PubMed] [Google Scholar]
- 4.Yee V C, Pratt K P, Cote H C, LeTrong I, Chung D W, Davie E W, Stenkamp R E, Teller D. Structure (London) 1997;5:125–138. doi: 10.1016/s0969-2126(97)00171-8. [DOI] [PubMed] [Google Scholar]
- 5.Spraggon G, Everse S J, Doolittle R F. Nature (London) 1997;389:455–462. doi: 10.1038/38947. [DOI] [PubMed] [Google Scholar]
- 6.Everse S J, Spraggon G, Veerapandian L, Riley M, Doolittle R F. Biochemistry. 1998;37:8637–8642. doi: 10.1021/bi9804129. [DOI] [PubMed] [Google Scholar]
- 7.Everse S J, Spraggon G, Veerapandian L, Doolittle R F. Biochemistry. 1999;38:2941–2946. doi: 10.1021/bi982626w. [DOI] [PubMed] [Google Scholar]
- 8.Brown J H, Volkmann N, Jun G, Henschen-Edman A H, Cohen C. Proc Natl Acad Sci USA. 2000;97:85–90. doi: 10.1073/pnas.97.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doolittle R F. Adv Protein Chem. 1973;27:1–109. doi: 10.1016/s0065-3233(08)60446-5. [DOI] [PubMed] [Google Scholar]
- 10.Doolittle R F, Watt K W K, Cottrell B A, Strong D D, Riley M. Nature (London) 1979;280:464–468. doi: 10.1038/280464a0. [DOI] [PubMed] [Google Scholar]
- 11.Murakawa M, Okamura T, Kamura T, Shibuya T, Harada M, Niho Y. Thromb Haemostasis. 1993;69:351–360. [PubMed] [Google Scholar]
- 12.Weissbach L, Grieninger G. Proc Natl Acad Sci USA. 1990;87:5198–5202. doi: 10.1073/pnas.87.13.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuyas C, Haeberli A, Walder P, Straub P W. Thromb Haemostasis. 1990;63:439–444. [PubMed] [Google Scholar]
- 14.Yamazumi K, Doolittle R F. Proc Natl Acad Sci USA. 1992;89:2893–2896. doi: 10.1073/pnas.89.7.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deutsch D G, Mertz E T. Science. 1970;170:1095–1096. doi: 10.1126/science.170.3962.1095. [DOI] [PubMed] [Google Scholar]
- 16.Laudano A P, Doolittle R F. Proc Natl Acad Sci USA. 1978;75:3085–3089. doi: 10.1073/pnas.75.7.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laudano A P, Doolittle R F. Biochemistry. 1980;19:1013–1019. doi: 10.1021/bi00546a028. [DOI] [PubMed] [Google Scholar]
- 18.Quiocho F A, Richards F M. Proc Natl Acad Sci USA. 1964;52:833–839. doi: 10.1073/pnas.52.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 21.Read R J. Acta Crystallogr A. 1986;42:140–149. [Google Scholar]
- 22.Collaborative Computing Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 23.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 24.Veklich Y I, Gorkun O V, Medved L V, Nieuwenhuizen W, Weisel J E. J Biol Chem. 1993;268:13577–13585. [PubMed] [Google Scholar]
- 25.Blomback B. In: Biochemical Evolution and the Origin of Life. Schoffeniels E, editor. Amsterdam: North–Holland; 1971. pp. 112–129. [Google Scholar]
- 26.Doolittle R F, Goldbaum D M, Doolittle L R. J Mol Biol. 1978;120:311–325. doi: 10.1016/0022-2836(78)90070-0. [DOI] [PubMed] [Google Scholar]
- 27.Hoeprich P D, Jr, Doolittle R F. Biochemistry. 1983;22:2049–2055. doi: 10.1021/bi00278a003. [DOI] [PubMed] [Google Scholar]
- 28.Huang S, Cao Z, Davie E W. Biochem Biophys Res Commun. 1993;190:488–495. doi: 10.1006/bbrc.1993.1074. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J-Z, Kudryk B, Redman C M. J Biol Chem. 1993;268:11278–11282. [PubMed] [Google Scholar]
- 30.Donovan J W, Mihalyi E. Proc Natl Acad Sci USA. 1974;71:4125–4128. doi: 10.1073/pnas.71.10.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weissbach L, Oddoux C, Procyk R, Grieninger G. Biochemistry. 1991;30:3290–3294. doi: 10.1021/bi00227a017. [DOI] [PubMed] [Google Scholar]
- 32.Bacon D J, Anderson W F. J Mol Graphics. 1988;6:219–220. [Google Scholar]
- 33.Merritt E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 34.McRee D E. J Mol Graphics. 1992;10:44–46. [Google Scholar]
- 35.Carson M. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]
- 36.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]