Abstract
The cAMP response element binding protein (CREB)-binding protein (CBP)/p300 family of coactivator proteins regulates gene transcription through the integration of multiple signal transduction pathways. Here, we show that induction of tumor necrosis factor α (TNF-α) gene expression in T cells stimulated by engagement of the T cell receptor (TCR) or by virus infection requires CBP/p300. Strikingly, in mice lacking one copy of the CBP gene, TNF-α gene induction by TCR activation is inhibited, whereas virus induction of the TNF-α gene is not affected. Consistent with these findings, the transcriptional activity of CBP is strongly potentiated by TCR activation but not by virus infection of T cells. Thus, CBP gene dosage and transcriptional activity are critical in TCR-dependent TNF-α gene expression, demonstrating a stimulus-specific requirement for CBP in the regulation of a specific gene.
The tumor necrosis factor α (TNF-α) gene encodes a pleiotropic cytokine, which plays a critical role in basic immunological processes as well as in a variety of infectious and autoimmune diseases (1). TNF-α is expressed in multiple cell types including lymphocytes, monocytes, mast cells, and fibroblasts and is induced by a wide range of extracellular stimuli (1). In T cells activated through their antigen receptor, TNF-α is expressed as an immediate early gene product (2). Transcriptional induction of the gene does not require de novo protein synthesis and depends on the recruitment of the transcription factors NFATp (nuclear factor of activated T cells) and ATF-2/Jun to cognate sites in the TNF-α promoter (2–4). Moreover, T cell production of TNF-α is critical in a variety of autoimmune diseases (1), as well as T cell-mediated lethal shock (5), and in the induction of HIV-1 replication in chronically HIV-1-infected cells (6). Thus, insights into the regulation of TNF-α in T cells may suggest therapeutic strategies to manipulate TNF-α levels in immunopathological processes in which TNF-α plays a destructive role.
Regulation of TNF-α gene expression is cell type-specific (4, 7). In T cells, binding of NFAT to both the κ3-NFAT and −76-NFAT TNF-α promoter sites is required for transcriptional induction of the gene, whereas in B cells, binding to the κ3-NFAT site is dispensable (4). In recent studies, we have demonstrated that regulation of TNF-α gene transcription is also stimulus-specific. In T cells, distinct enhancer complexes form on the TNF-α gene promoter in response to different extracellular stimuli. Virus infection of T cells leads to the recruitment of NFAT, ATF-2/Jun, and Sp1 proteins to the TNF-α promoter in vivo, whereas in response to calcium flux, the transcription factors NFAT and ATF-2/Jun, but not Sp1, are recruited to the promoter (8). Thus, in TNF-α gene regulation, different stimuli result in the assembly of distinct enhancer complexes, which use specific sets of binding sites and cognate proteins.
Here, we show that activation of TNF-α gene expression in T cells by TCR engagement, virus infection, and calcium influx depends on cAMP response element binding protein (CREB)-binding protein (CBP)/p300 coactivator proteins. Furthermore, we show that the composite TNF-α CRE/κ3 site, which binds ATF-2/Jun and NFAT proteins and is required for induction of the TNF-α gene by TCR engagement, virus infection, and calcium influx, is a CBP/p300-dependent element. Using mice lacking one allele of CBP, we show that gene dosage of CBP is selectively required for induction of TNF-α transcription by TCR engagement but not by virus infection. Moreover, TCR activation, but not virus infection, potentiates CBP transactivation in T cells. Thus, CBP is selectively required for TCR-mediated TNF-α gene transcription, demonstrating a novel stimulus-specific role for CBP in gene expression.
Materials and Methods
Plasmids.
The −200 TNF-α Luc, pcDNA3-NFATp S>A, E1A 12S, and E1A 12S Δ2–36 constructs have been described (8–10). The −39 TNF-α (CRE/κ3-NFAT)2 Luc reporter was created by subcloning the SmaI–HincII fragment of −39 TNF-α (CRE/κ3-NFAT)2 chloramphenicol acetyltransferase (CAT) (3) into the SmaI site of pGL3-Basic (Promega). The −40 IFN-β (CRE/κ3-NFAT)2 reporter was created by subcloning two copies of the double-stranded oligonucleotide κ3(L) (3) in the sense orientation into the BamHI site of −40 IFN-β CAT (11). To create −105 IFN-β Luc, the BamHI–XbaI fragment of −105 IFN-β CAT first was subcloned into pBluescript (Stratagene), and the KpnI–XbaI fragment then was subcloned into the KpnI–NheI sites of pGL3-Basic. The Gal4 × 5-Luc reporter was created by subcloning the PvuII–SmaI fragment of Gal4 × 5 E1bTATA-CAT (12) into the SmaI site of pGL3-Basic. The Gal4-CBP expression vector was a generous gift of Dimitris Thanos (Columbia University, New York).
Cell Culture and Transfection.
The T cell hybridoma 68–41 (a gift from Masato Kubo, Science University of Tokyo, Tokyo, Japan) and the Ar-5 T cell clone were grown, and transfections were performed by using DEAE-Dextran, as described (2, 13). Thirty-six hours after transfection, cells were activated with Sendai virus (Spafas, Cantell strain) at a final concentration of 300 hemagglutinin (HA) units/ml, with 1 μM ionomycin (Calbiochem), or with 1 μg/ml anti-CD3 (PharMingen) and harvested approximately 16 h later.
HeLa cells were maintained at 37°C, 5% CO2, in DMEM supplemented with penicillin, streptomycin, 10% FCS, and 2 mM l-glutamine. Transfections in HeLa cells were performed by using Lipofectamine (Life Technologies, Rockville, MD) at 25 μg/ml according to the manufacturer's protocol. Cells were incubated with DNA/Lipofectamine complexes in OptiMEM (Life Technologies) for 24 h posttransfection, after which medium was removed and replaced with complete DMEM. Cells were harvested after an additional 24 h.
CAT assays or luciferase assays were performed as described (2, 10). As a transfection control for the CAT assays and luciferase assays, the β-galactosidase control plasmid pCMVβ (CLONTECH) and the Renilla luciferase control plasmid pRL-TK (Promega), respectively, were cotransfected in all experiments. Transfections were normalized to β-galactosidase or Renilla luciferase as indicated in the figure legends.
RNA Analysis.
Spleen cells were prepared from CBP+/− mice and their wild-type littermates (14). Cells were stimulated with anti-CD3 or Sendai virus as indicated in the figure legends. 32P-labeled RNA probes were prepared from SP6 γ-actin and a murine TNF-α or IFN-β probe, and RNase protection assays were performed as described (2). Percentage of T and B cells was determined in a FACS analysis according to standard techniques by using antibodies to CD5 and B220 (Cappel), respectively.
Results
To assess the role of CBP/p300 proteins in TNF-α gene regulation, we performed cotransfection studies with the adenovirus E1A 12S protein, which inhibits CBP/p300 function (15). As a control, we used a mutant form of the protein (E1A 12S Δ2–36), which lacks the CBP/p300 interaction domain and fails to inhibit CBP/p300 activity (16). As a reporter, we used the region of the human TNF-α promoter containing 200 nt upstream of the transcriptional start site fused to the luciferase reporter gene. This region is sufficient for maximal transcriptional induction of the human TNF-α gene in response to multiple signaling pathways, including antigen receptor activation, calcium flux, and virus infection (2, 17).
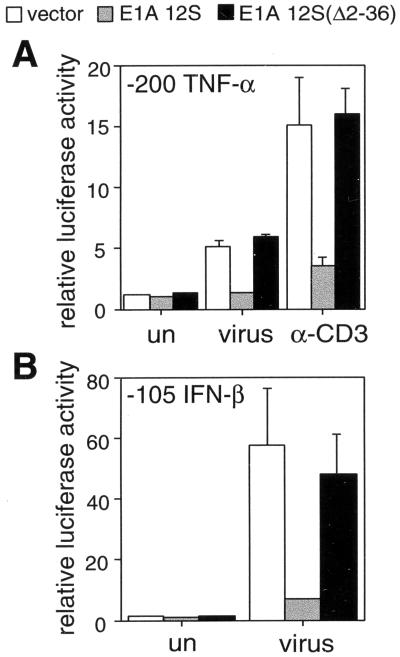
As shown in Fig. 1, activation of the TNF-α reporter gene upon stimulation of 68–41 T cells with virus or by crosslinking the TCR with antibodies directed toward the CD3 complex (anti-CD3) was strongly inhibited by E1A 12S but not by E1A 12S Δ2–36 (Fig. 1A). A similar result was observed in the T cell clone Ar-5 and by using calcium ionophore as the stimulus in the two cell types (data not shown). As a control we used the IFN-β gene, the activation of which by virus depends on CBP/p300 (18, 19). Virus induction of a IFN-β reporter in 68–41 T cells (Fig. 1B) and Ar-5 T cells (data not shown) also was abrogated by E1A 12S but not by E1A 12S Δ2–36, consistent with results obtained in HeLa cells (18).
Figure 1.
CBP/p300 is involved in multiple signaling pathways leading to the induction of TNF-α transcription. The −200 TNF-α Luc (A) or −105 IFN-β Luc (B) reporter plasmids were cotransfected with an expression vector for wild-type E1A 12S (E1A 12S) or mutant E1A 12S (E1AΔ2–36) protein or the expression vector alone (vector) in 68–41 T cells. Histograms of luciferase activity are shown, demonstrating inhibition of TNF-α transcription upon induction by anti-CD3 or virus (A) or inhibition of IFN-β transcription upon virus induction (B) by wild-type E1A 12S. The results are an average of three independent experiments, and the error bars represent SEM. Extracts were normalized to Renilla luciferase activity as described in Materials and Methods.
The composite TNF-α promoter element CRE/κ3 binds members of the ATF-2/Jun and NFAT transcription factor families (3). The CRE/κ3 site is critical in the regulation of TNF-α and can confer induction by anti-CD3, virus, and ionomycin upon a truncated TNF-α promoter (3, 8). Given that NFAT and ATF-2/Jun proteins can interact with CBP/p300 (20–22), we speculated that the integrative function of the CRE/κ3 site in TNF-α gene regulation might involve a functional association with CBP/p300 proteins.
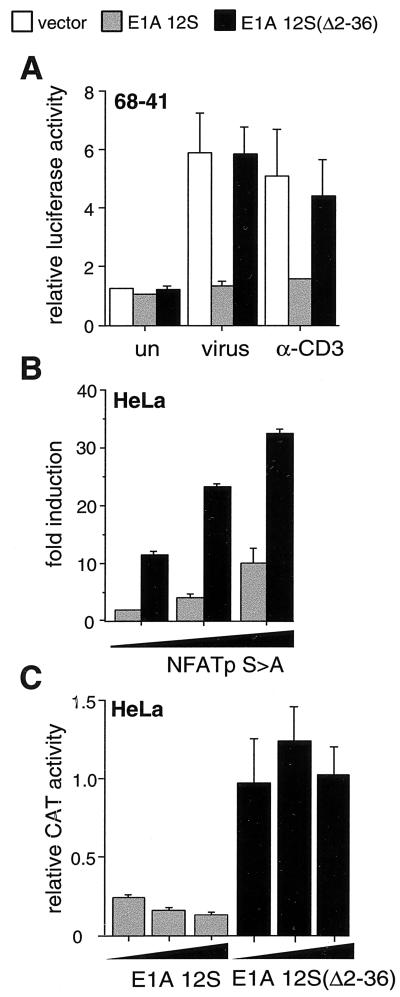
As shown in Fig. 2, we tested the effect of E1A 12S and E1A 12S Δ2–36 on the induction of a synthetic promoter construct, (CRE/κ3)2, containing two copies of the CRE/κ3 site fused to a truncated −39 TNF-α promoter. E1A 12S, but not E1A 12S Δ2–36, inhibited induction of the (CRE/κ3)2 reporter construct in T cells stimulated by anti-CD3 or virus (Fig. 2A). Thus, the CRE/κ3 element is sufficient for functional interaction with the coactivator proteins CBP/p300, consistent with its role of integrating diverse signal transduction pathways at the TNF-α promoter.
Figure 2.
CBP/p300 is involved in induction of the CRE/κ3-NFAT element in response to multiple stimuli or by exogenous NFATp. The −39 TNF-α (CRE/κ3-NFAT)2 Luc (A) or −40 IFN-β (CRE/κ3-NFAT)2 CAT (B and C) reporter plasmids were cotransfected with an expression vector for wild-type E1A 12S (E1A 12S) or mutant E1A 12S (E1AΔ2–36) protein or the expression vector alone (vector) into 68–41 T cells (A) or HeLa cells (B and C). (A) Histograms of luciferase activity upon induction by anti-CD3 or virus (V) demonstrating inhibition of CRE/κ3-NFAT reporter activity by wild-type E1A 12S. (B and C) Histograms of CAT activity in response to increasing amounts of exogenous NFATp (30, 100, or 300 ng) in the presence of fixed amounts of wild-type or mutant E1A 12S (0.1 μg) (B) or in response to increasing amounts of wild-type or mutant E1A 12S (0.1, 0.3, or 1.0 μg) in the presence of fixed amounts of exogenous NFATp (100 ng) (C), demonstrating inhibition of NFATp-mediated CRE/κ3-NFAT reporter activity by wild-type E1A 12S. The results in A—C are an average of three independent experiments, and the error bars represent SEM. For luciferase assays, extracts were normalized to Renilla luciferase activity; for CAT assays, extracts were normalized to β-galactosidase activity.
Two molecules of NFAT bind to the κ3 site of the CRE/κ3 composite element in activated T cells (2, 23). As shown in Fig. 2B, transfection of increasing amounts of a constitutively nuclear form of NFATp (NFATp S>A) (8) into HeLa cells, which lack NFAT (24), activates the (CRE/κ3)2 reporter construct. To investigate the role of CBP/p300 in activation of the (CRE/κ3)2 reporter construct by NFATp, we tested the effect of cotransfection of E1A. NFATp-driven induction of the (CRE/κ3)2 reporter construct was strongly inhibited by a constant amount of E1A but not by E1A Δ2–36 (Fig. 2B). In the reciprocal experiment, increasing amounts of E1A but not E1A Δ2–36 inhibited induction of the reporter by a fixed amount of NFATp (Fig. 2C). We note that vector alone did not influence NFATp-dependent activation of the (CRE/κ3)2 reporter (data not shown). Thus, CBP/p300 functionally cooperates with NFATp at the CRE/κ3 site.
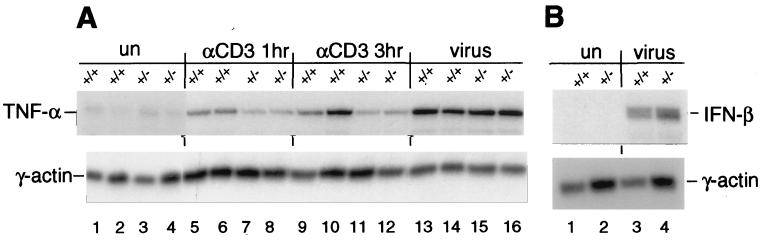
We next examined the role of CBP in the induction of the endogenous TNF-α gene in spleen cells derived from mice lacking one copy of the CBP gene (CBP+/−) (14). Using anti-CD3, we selectively stimulated splenic T cells in CBP+/− mice and wild-type littermates. The CBP protein levels in the spleens from CBP+/− mice are concordant with gene dosage (ref. 14 and data not shown). Strikingly, in T cells from the CBP+/− mice, induction of TNF-α gene transcription by treatment with anti-CD3 was inhibited, whereas virus infection of spleen cells from the same CBP+/− mice resulted in equivalent levels of TNF-α mRNA relative to wild-type littermates (Fig. 3A). Similarly, virus induction of the IFN-β gene was not impaired by the loss of one CBP allele (Fig. 3B). We note that IFN-β is not activated by anti-CD3 (data not shown) and is selectively induced by virus (19). Taken together, these results demonstrate an inducer-specific requirement for CBP in anti-CD3-stimulated TNF-α gene induction.
Figure 3.
Gene dosage-dependent requirement for CBP in TNF-α gene transcription in response to activation via the T cell receptor. An autoradiograph of RNase protection assays is shown. Levels of endogenous TNF-α (A) and IFN-β (B) mRNA induced by various stimuli from wild-type mice (+/+) or from heterozygous littermates (+/−) lacking one copy of the CBP allele were measured. The γ-actin gene was used as a control for RNA loading and processing. (A) In CBP heterozygous mice, induction of TNF-α mRNA upon stimulation by α-CD3 after 1 or 3 h is inhibited relative to levels observed in wild-type mice, whereas induction of TNF-α mRNA upon stimulation by virus remains unaffected. Results from two CBP+/− and two wild-type CBP+/+ littermates are shown. (B) Virus induction of IFN-β mRNA in CBP+/− mice is not reduced relative to levels observed in wild-type littermates. In the experiment displayed, when total mRNA levels are normalized to γ-actin, the relative levels of virus-inducible IFN-β mRNA derived from the CBP+/− mouse relative to the levels observed in the wild-type littermate are equivalent. The percentage of T and B cells in spleens from CBP+/− and wild-type CBP+/+ littermates was equivalent as determined by FACS analysis (data not shown). We note that the CBP+/− mice used in the experiments displayed were in a 129/Sv background and are representative of three individual CBP+/− and wild-type CBP+/+ littermates. When the mice were backcrossed into a BL6 background, inhibition of anti-CD3 induction of TNF-α was attenuated (data not shown), suggesting a strain-specific modifier. It should be noted that such a strain-specific backcrossing effect previously has been observed with regard to heterozygote lethality in p300+/− mice (25).
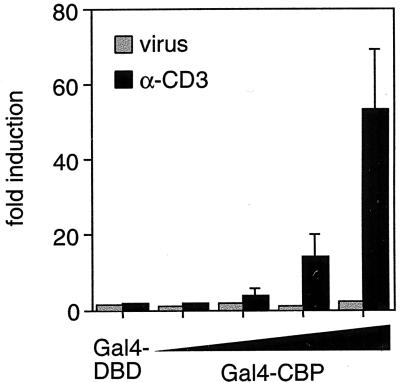
TCR engagement- and NFAT-dependent transcription depends on calcium flux (26, 27). In neuronal cells, the transcriptional activity of CBP is potentiated by calcium flux (28). We therefore next investigated whether engagement of the TCR and/or virus infection could potentiate the transcriptional activity of CBP in T cells. We cotransfected increasing amounts of full-length CBP fused to the DNA-binding domain of Gal4 with a Gal4-dependent reporter into T cells. Strikingly, Gal4-CBP was strongly activated in response to TCR activation, but not in response to virus infection (Fig. 4). These results thus are consistent with the selective requirement for wild-type CBP levels in the transcriptional activation of TNF-α in T cells stimulated by anti-CD3.
Figure 4.
Transcriptional activation of CBP in response to α-CD3 and virus stimulation of T cells. A histogram of the fold induction of luciferase activity of a minimal promoter containing five copies of the Gal4-binding site fused to luciferase (1 μg) is shown. Increasing amounts (130 ng, 400 ng, 1.3 μg, and 4 μg) of vector expressing the Gal4 DNA-binding domain fused to full-length CBP (Gal4-CBP) or 4 μg of vector expressing the Gal4 DNA-binding domain (Gal4-DBD) was transfected into 68–41 T cells and stimulated with α-CD3 or virus as indicated. Because Gal4-CBP expression depends on the simian virus 40 (SV40) promoter, and the SV40 promoter-driven expression of exogenous β-galactosidase is not potentiated by anti-CD3 or virus (data not shown), we conclude that the observed effects on Gal4-CBP transcriptional activity are posttranslational. The results are an average of three independent experiments, and the error bars represent SEM. Extracts were normalized to Renilla luciferase activity.
Discussion
CBP and p300 both function as histone acetyltransferases and interact with a broad range of transcription factors and coactivators. Although these proteins are functionally interchangeable in many in vitro and in vivo assays, CBP and p300 do not have completely overlapping biological functions (29, 30). For example, mutant forms of the HTLV-I Tax protein that selectively interact with either CBP or p300 activate transcription from ATF/CREB- or NF-κB-dependent pathways, respectively, in transformed hamster (BHK21) cells (31). Reduction of CBP or p300 protein levels using ribozyme technology also has suggested a specific role for p300 in retinoic acid-induced differentiation of murine embryonal carcinoma F9 cells (32) and in the accumulation of p53 in response to DNA damage in human breast cancer (MCF-7) cells (33).
The critical requirement for the correct dosage of CBP and its lack of complete functional redundancy with other CBP/p300 proteins is illustrated further by Rubinstein–Taybi Syndrome. The genetic basis of Rubinstein–Taybi Syndrome is a mutation in a CBP allele (34), which results in growth and mental retardation and craniofacial, skeletal, and heart defects in affected patients (30). A similar phenotype is observed in mice that lack one allele of CBP (14, 35) or express a truncated form of CBP protein (36, 37). Mice lacking one p300 allele exhibit defects in heart development and cell proliferation (25). These defects undoubtedly result from perturbations in the cell- and inducer-specific regulation of genes and signal transduction pathways that selectively depend on CBP or p300.
Here, we have shown that activation of TNF-α gene transcription by multiple stimuli, including TCR engagement and virus infection, depends on CBP/p300. TNF-α gene transcription induced by TCR engagement, however, is inhibited in cells from mice lacking one CBP allele, whereas transcription of the gene in response to virus infection is not compromised. Thus, whereas CBP/p300 proteins appear to be generally required for induction of TNF-α gene transcription, at least one family member, CBP, is specifically required for TNF-α gene induction by a distinct stimulus, TCR engagement. This specificity may involve the intrinsic ability of CBP to activate transcription in the context of the TNF-α enhancer complex; this hypothesis would be consistent with our demonstration that TCR engagement, but not virus infection, is a potent effector of the intrinsic transactivation function of CBP.
Interestingly, distinct sets of TNF-α promoter elements are required for induction of TNF-α gene transcription by TCR stimulation and calcium influx or by virus infection (3, 8). Whereas calcium influx results in the recruitment of ATF-2/c-jun and NFATp to the TNF-α promoter, in response to virus infection, Sp1 is also recruited (8). Thus, it is possible that the presence of an additional factor, Sp1, bound to the TNF-α promoter after virus infection creates a more extensive surface for protein—protein interactions. This surface may strengthen the recruitment of CBP to the TNF-α enhancer complex, thereby permitting transcription to proceed in the absence of wild-type levels of CBP protein. In the case of IFN-β, virus infection leads to the cooperative assembly of transcription factors and CBP/p300 at the IFN-β promoter to form a stereospecific, higher-order complex, the enhanceosome (19). Wild-type levels of CBP also are not required for activation of IFN-β transcription by virus, consistent with efficient recruitment of CBP to this highly cooperative complex. Thus, the group of transcription factors recruited to the TNF-α and IFN-β promoters by virus infection may have a relatively stronger interaction with the CBP-containing complex than the groups of factors assembled on the TNF-α promoter in response to TCR activation.
Alternatively, in the case of virus-inducible transcription of both the TNF-α and IFN-β genes, wild-type levels of p300 may compensate for reduced levels of CBP. In this case, recruitment of either CBP or p300 to the TNF-α and IFN-β enhancer complexes would be sufficient for expression of either gene in response to virus infection. We note that both CBP and p300 have been detected as components of the IFN-β enhanceosome (19). Although in the context of the TCR-mediated TNF-α enhancer complex, the intrinsic transactivation potential of CBP may be important, in the context of the virus-induced TNF-α and IFN-β enhancer complexes, CBP may function primarily as a scaffold for the complex. This idea is consistent with our observation that CBP-mediated transcription is not potentiated by virus infection.
The selective role of CBP in TCR-mediated TNF-α expression also may be an underlying factor in the phenotype of mice lacking one allele of CBP. A full complement of CBP is required for normal hematopoietic differentiation and hematopoietic tumor suppression in aging mice (14). Given that TNF-α is involved in the generation of follicular dendritic cells (38) and splenic germinal centers (39, 40) and is an autocrine growth factor for human splenic B cells (41), it is interesting to speculate that dysregulation of TNF-α in CBP+/− mice may contribute to this phenotype.
In summary, these results lend insight into the general question of how specificity in transcriptional activation is achieved. The requirement for, and activation of, CBP in TCR-mediated TNF-α gene expression illustrates the role of a specific coactivator protein in the transduction of a stimulus originating at the cell surface, which culminates in a distinct pattern of gene expression in the nucleus.
Acknowledgments
We are indebted to Tom Maniatis for his support and critical comments on the manuscript. We also thank David Livingston for his support of this work. We are grateful to D. Thanos for the gift of Gal4-CBP plasmid, T. Collins for the gift of the E1A constructs, T. Kawakami for the gift of the −200 TNF-α luciferase construct, and M. Kubo for the gift of the 68–41 cells. We are also grateful to A. Uglialoro for laboratory assistance, K. Bloch for critical reading of the manuscript, and anonymous reviewers for helpful comments. This work was supported by a grant from the National Institutes of Health (GM-56492) and an Established Investigator Award from the American Heart Association (to A.E.G.) and by a grant from the Howard Hughes Medical Institute (to A.L.K.).
Abbreviations
- CBP
cAMP response element binding protein (CREB)-binding protein
- TNF
tumor necrosis factor
- TCR
T cell receptor
- NFAT
nuclear factor of activated T cells
- CAT
chloramphenicol acetyltransferase
References
- 1.Aggarwal B B, Puri R K. Human Cytokines: Their Role in Disease and Therapy. Cambridge, MA: Blackwell Science; 1995. [Google Scholar]
- 2.Goldfeld A E, McCaffrey P G, Strominger J L, Rao A. J Exp Med. 1993;178:1365–1379. doi: 10.1084/jem.178.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsai E Y, Jain J, Pesavento P A, Rao A, Goldfeld A E. Mol Cell Biol. 1996;16:459–467. doi: 10.1128/mcb.16.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsai E Y, Yie J, Thanos D, Goldfeld A E. Mol Cell Biol. 1996;16:5232–5244. doi: 10.1128/mcb.16.10.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miethke T, Wahl C, Heeg K, Echtenacher B, Krammer P H, Wagner H. J Exp Med. 1992;175:91–98. doi: 10.1084/jem.175.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fauci A. Nature (London) 1996;384:529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- 7.Goldfeld A E, Maniatis T. Proc Natl Acad Sci USA. 1989;86:1490–1494. doi: 10.1073/pnas.86.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falvo J V, Uglialoro A M, Brinkman B N M, Merika M, Parekh B S, King H C, Tsai E Y, Morielli A D, Peralta E G, Maniatis T, et al. Mol Cell Biol. 2000;20:2239–2247. doi: 10.1128/mcb.20.6.2239-2247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerritsen M E, Williams A J, Neish A S, Moore S, Shi Y, Collins T. Proc Natl Acad Sci USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hata D, Kawakami Y, Inagaki N, Lantz C S, Kitamura T, Khan W N, Maeda-Yamamoto M, Miura T, Han W, Hartman S E, et al. J Exp Med. 1998;187:1235–1247. doi: 10.1084/jem.187.8.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du W, Maniatis T. Proc Natl Acad Sci USA. 1992;89:2150–2154. doi: 10.1073/pnas.89.6.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lillie J W, Green M R. Nature (London) 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- 13.Kubo M, Ransom J, Webb D, Hashimoto Y, Tada T, Nakayama T. EMBO J. 1997;16:4007–4020. doi: 10.1093/emboj/16.13.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kung A L, Rebel V I, Bronson R T, Ch'ng L-E, Sieff C A, Livingston D M, Yao T-P. Genes Dev. 2000;14:272–277. [PMC free article] [PubMed] [Google Scholar]
- 15.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 16.Lee J S, Galvin K M, See R H, Eckner R, Livingston D, Moran E, Shi Y. Genes Dev. 1995;10:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- 17.Goldfeld A E, Doyle C, Maniatis T. Proc Natl Acad Sci USA. 1990;87:9769–9773. doi: 10.1073/pnas.87.24.9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merika M, Williams A J, Chen G, Collins T, Thanos D. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 19.Maniatis T, Falvo J V, Kim T H, Kim T K, Lin C H, Parekh B S, Wathelet M G. Cold Spring Harbor Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 20.Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Nature (London) 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 21.Kawasaki H, Song J, Eckner R, Ugai H, Chiu R, Taira K, Shi Y, Jones N, Yokoyama K K. Genes Dev. 1998;12:233–245. doi: 10.1101/gad.12.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Rodriguez C, Rao A. J Exp Med. 1998;187:2031–2036. doi: 10.1084/jem.187.12.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCaffrey P G, Goldfeld A E, Rao A. J Biol Chem. 1994;269:30445–30450. [PubMed] [Google Scholar]
- 24.Northrop J P, Ho S N, Chen L, Thomas D J, Timmerman L A, Nolan G P, Admon A, Crabtree G R. Nature (London) 1994;369:497–502. doi: 10.1038/369497a0. [DOI] [PubMed] [Google Scholar]
- 25.Yao T-P, Oh S P, Fuchs M, Zhou N D, Ch'ng L-E, Newsome D, Bronson R T, Li E, Livingston D M, Eckner R. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 26.Rao A, Luo C, Hogan P G. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 27.Crabtree G R. Cell. 1999;96:611–614. doi: 10.1016/s0092-8674(00)80571-1. [DOI] [PubMed] [Google Scholar]
- 28.Chawla S, Hardingham G E, Quinn D R, Bading H. Science. 1998;281:1505–1509. doi: 10.1126/science.281.5382.1505. [DOI] [PubMed] [Google Scholar]
- 29.Shikama N, Lyon J, La Thangue N B. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01149-5. [DOI] [PubMed] [Google Scholar]
- 30.Giles R H, Peters D J M, Breuning M H. Trends Genet. 1998;14:178–183. doi: 10.1016/s0168-9525(98)01438-3. [DOI] [PubMed] [Google Scholar]
- 31.Bex F, Yin M J, Burny A, Gaynor R B. Mol Cell Biol. 1998;18:2392–2405. doi: 10.1128/mcb.18.4.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawasaki H, Eckner R, Yao T-P, Taira K, Chiu R, Livingston D M, Yokoyama K K. Nature (London) 1998;393:284–289. doi: 10.1038/30538. [DOI] [PubMed] [Google Scholar]
- 33.Yuan Z M, Huang Y, Ishiko T, Nakada S, Utsugisawa T, Shioya H, Utsugisawa Y, Yokoyama K, Weichselbaum R, Shi Y, et al. J Biol Chem. 1999;274:1883–1886. doi: 10.1074/jbc.274.4.1883. [DOI] [PubMed] [Google Scholar]
- 34.Petrij F, Giles R H, Dauwerse H G, Saris J J, Hennekam R C, Masuno M, Tommerup N, van Ommen G-J B, Goodman R H, Peters D J M, et al. Nature (London) 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka Y, Naruse I, Maekawa T, Masuya H, Shiroishi T, Ishii S. Proc Natl Acad Sci USA. 1997;94:10215–10220. doi: 10.1073/pnas.94.19.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oike Y, Hata A, Mamiya T, Kaname T, Noda Y, Suzuki M, Yasue H, Nabeshima T, Araki K, Yamamura K. Hum Mol Genet. 1999;8:387–396. doi: 10.1093/hmg/8.3.387. [DOI] [PubMed] [Google Scholar]
- 37.Oike Y, Takakura N, Hata A, Kaname T, Akizuki M, Yamaguchi Y, Yasue H, Araki K, Yamamura K, Suda T. Blood. 1999;93:2771–2779. [PubMed] [Google Scholar]
- 38.Le Hir M, Bluethmann H, Kosco-Vilbois M H, Muller M, di Padova F, Moore M, Ryffel B, Eugster H P. J Exp Med. 1996;183:2367–2372. doi: 10.1084/jem.183.5.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marino M W, Dunn A, Grail D, Inglese M, Noguchi Y, Richards E, Jungbluth A, Wada H, Moore M, Williamson B, et al. Proc Natl Acad Sci USA. 1997;94:8093–8098. doi: 10.1073/pnas.94.15.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cook M C, Korner H, Riminton D S, Lemckert F A, Hasbold J, Amesbury M, Hodgkin P D, Cyster J G, Sedgwick J D, Basten A. J Exp Med. 1998;188:1503–1510. doi: 10.1084/jem.188.8.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boussiotis V A, Nadler L M, Strominger J L, Goldfeld A E. Proc Natl Acad Sci USA. 1994;91:7007–7011. doi: 10.1073/pnas.91.15.7007. [DOI] [PMC free article] [PubMed] [Google Scholar]