Abstract
We have found that the bacterial transposon Tn7 can recognize and preferentially insert adjacent to triple-helical nucleic acid structures. Both synthetic intermolecular triplexes, formed through the pairing of a short triplex-forming oligonucleotide on a plasmid DNA, and naturally occurring mirror repeat sequences known to form intramolecular triplexes or H-form DNA are preferential targets for Tn7 insertion in vitro. This target site selectivity depends upon the recognition of the triplex region by a Tn7-encoded ATP-using protein, TnsC, which controls Tn7 target site selection: the interaction of TnsC with the triplex region results in recruitment and activation of the Tn7 transposase. Recognition of a nucleic acid structural motif provides both new information into the factors that influence Tn7's target site selection and broadens its targeting capabilities.
There is much interest in gaining a better understanding of the factors that affect the target site selection of mobile DNAs. As an underlying process central to a diverse range of biological functions, selection of an insertion site can dramatically influence genome alteration, viral integration, and propagation of antibiotic resistance genes (1). Transposons typically show relatively little preference in selection of insertion sites (2), a property that has been widely exploited for their use in molecular genetics as mutagens, as “reporters” of gene expression, and for manipulating chromosome structure (3). However, many elements can respond to signals such as accessory proteins or structural features of target DNAs, dramatically altering their insertion profiles. For example, bent or cruciform DNA has been shown to be a hot-spot for HIV-1 integration (4, 5). Interestingly, we have found that triple-helical DNA structures can also influence the target site selection of the bacterial transposon Tn7. This discovery gives new insight into the factors that influence Tn7's target site selection. Tn7 is an excellent candidate to gain further insights into the targeting preferences of mobile genetic elements (6, 7), as it is distinguished by its ability to move with various degrees of target site selectivity (8–11). The ability to recognize and direct insertions adjacent to triplex DNA also allows a greater level of selectivity for Tn7, increasing its potential for use as a sequence-specific DNA delivery tool.
Five element-encoded proteins (TnsA, TnsB, TnsC, TnsD, and TnsE) mediate Tn7 transposition (7), promoting two distinct pathways that differ in their target site selectivity. The TnsABC + D pathway results in high-frequency transposition into a single site in the Escherichia coli chromosome called attTn7 (12, 13), whereas the TnsABC + E pathway yields low-frequency insertions into non-attTn7 sites (11). Little recombination to any site is observed with just TnsABC; thus TnsD and TnsE serve as both target site selectors and activators of the common TnsABC machinery.
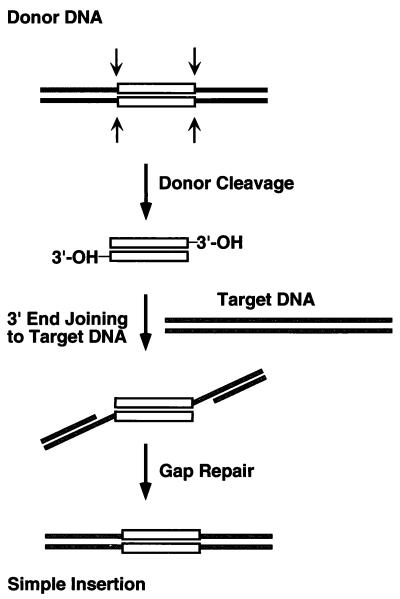
Reconstitution of a cell-free TnsABC + D transposition system (14) has allowed a molecular definition of the Tn7 transposition pathway and of the specific roles of each of these Tn7-encoded proteins. Tn7 moves through a cut-and-paste mechanism (Fig. 1) in which the element is first excised from the donor site by double-strand breaks at each end of the transposon, followed by the joining of these transposon ends to the target DNA. These DNA breakage and joining activities are executed by TnsA and TnsB, which collaborate to form the transposase (15, 16). TnsC plays a key role in the regulation of transposition and in target site selection. TnsC, a nonspecific DNA-binding protein and ATPase, evaluates and responds to potential target signals and also interacts with the TnsAB transposase to provoke its DNA breakage and joining activities (17, 18). In the presence of TnsC and TnsD, which binds to attTn7, a TnsC–TnsD target complex is formed that likely contains TnsC in an active ATP-bound state that interacts with transposase to stimulate transposition (17).
Figure 1.
Cut-and-paste transposition. Recombination initiates by means of double-strand breaks at either end of the transposon (white rectangle) from donor DNA (black lines), exposing terminal 3′-OH groups. Once cut from donor DNA, this excised linear transposon intermediate is joined to the target DNA (gray lines). The 5′ transposon ends are flanked by short gaps; upon repair, a simple insertion product is generated.
By binding to attTn7, TnsD promotes transposition into attTn7; little recombination is observed in the absence of TnsD. Analysis of this low level of recombination by TnsABC reveals that Tn7 inserts into many different target sites (14). Transposition by TnsABCA225V, a TnsC gain-of-function allele that provides for recombination in the absence of either TnsD or TnsE, occurs at much higher frequency, approaching the level of TnsABC + D recombination (18). Using TnsABCA225V and TnsABCwt, we have found that Tn7 is able to recognize and preferentially direct Tn7 insertions adjacent to triplex DNA.
Materials and Methods
Triplex Annealing and Crosslinking.
Triplex-forming oligomers (TFOs) were made by standard phosphoramidite synthesis or were purchased from Trilink Biotechnologies (San Diego). Annealing and crosslinking of triplexes was done at 0°C, pH 7.0, in triplex-annealing buffer, which contained a final concentration of 50 mM Mops, 20 mM MgCl2, and 10 mM NaCl. Crosslinking was carried out with 365-nm UV light for 15 min, 0°C. For both triplex sequences shown, >80% mono adduct between plasmid and TFO was confirmed by denaturing gel electrophoresis. pRAO1dup contains a polypurine tract (PPT) in the plasmid pUC-19. The TFO used for pRAO1dup has the sequence 5′-CTTTTTCCTTCTC-3′. The TFO sequence used to recognize pRAO2dup is 5′-TAACTAACTTTTTCT-3′. The modified bases 8-oxoadenine and 5-methylcytidine are underlined. Note these modifications allow a nontraditional Hoogsteen triad with the C⋅G base pair. The PPT used in pRAO2dup is a naturally occurring PPT found in the human collagenase I gene. This PPT was cloned from oligomers into the polylinker of plasmid pGEM-3Z.
In Vitro Transposition Reactions.
Each in vitro transposition reaction mixture (100 μl final volume) contained 25 mM Hepes (pH 7.6), 2.5 mM Tris⋅HCl (pH 7.6), 50 μg/ml BSA, 2 mM ATP, 2 mM DTT, 15 mM Mg(OAc)2, 600 ng (≈0.5 pmol of plasmid, or 1.4 nmol of nucleotides) of target plasmid DNA, 80 ng (0.038 pmol of plasmid) of mTn7-containing donor DNA, 1.2 pmol (39 ng) of TnsA, 0.3 pmol (25 ng) of TnsB, and 1.0 pmol (60 ng) of TnsCA225V. All of the components except TnsA, TnsB, donor plasmid, and Mg(OAc)2 are incubated at 30°C for 20 min to allow target assembly before the reaction. The remaining components are added and allowed to react at 30°C for 20 min. Extraction with phenol/chloroform and precipitation with ethanol were done before restriction digestion and Southern blotting.
Filter Binding.
Each of the binding reaction mixtures (25 μl final volume) contained 25 mM Hepes (pH 7.6), 2.5 mM Tris⋅HCl (pH 7.6), 50 μg/ml BSA, 2 mM ATP, 2 mM DTT, 1.8 μg (≈1.0 pmol of plasmid or 2.8 nmol of base pairs) of unlabeled competitor plasmid DNA, and ≈0.01 pmol (fragment) of 32P-labeled 110-bp target DNA fragment excised from either pRAO1dup for the duplex fragment or pRAO1tpx for the triplex fragment. For both duplex and triplex samples the total concentration of DNA was identical and held constant. Also, both the pmol and the total counts per reaction (50,000 cpm) of labeled DNA were identical for duplex and triplex samples. For the TnsA + TnsB reaction, standard in vitro amounts were used: 1.2 pmol (39 ng) of TnsA and 0.3 pmol (25 ng) of TnsB per reaction (a titration of TnsA and TnsB gave similar results; data not shown). Each reaction was incubated at 30°C for 20 min before loading onto a nitrocellulose filter preequilibrated in a buffer identical to the binding buffer with 7% sucrose. Samples were washed four times each with 1 ml of binding buffer before being dried. Radioactivity from individual filters was quantitated by scintillation counting and is listed as the percent bound of the total radiolabeled probe (100% = 50,000 cpm) by TnsCA225V (qualitatively similar preferences for triplex DNA were found with the much less active TnsCwt).
Dimethyl Sulfate (DMS) Footprinting.
Singly 5′-32P-labeled duplex and crosslinked triplex substrates were isolated by gel electrophoresis and resuspended under in vitro transposition conditions (above). Reaction mixtures (25 μl) were treated with 1 μl of a 1:40 dilution of DMS in dimethyl sulfoxide for 3 min at 0°C. Reactions were stopped by precipitation with ethanol. For both DNA cleavage of DMS-treated substrates and removal of psoralen crosslink, pellets were dried and resuspended in 100 μl of 0.1 M NaOH and heated to 90°C for 1 h. After an additional precipitation, samples were resuspended in 5 μl of loading buffer and loaded on an 8 M urea/8% polyacrylamide gel and electrophoresed under denaturing conditions. The gel was dried and exposed to x-ray film. The autoradiographs were scanned with a Molecular Dynamics scanner and the bands were quantified with imagequant and excel 5 software.
Sequence Analysis.
In vitro transposition reaction products were precipitated with ethanol and digested with EcoRV to linearize donor plasmid before transformation into E. coli. Cultures were grown under conditions selecting for appropriate antibiotic resistance and plated, and colonies were individually isolated to obtain a single target plasmid containing a unique mini-Tn7 insertion.
RNase Hydrolysis of r-TFO Substrates.
Once annealed and crosslinked, 1.0 μg of plasmid DNA containing either d-TFO (pRAO1tpx) or r-TFO (pRAO1tpx) triplexes (d- indicates all deoxyribonucleotides and r- indicates the presence of a single ribonucleotide) were treated with 1 μl of a 40 ng/μl RNase solution in a final volume of 50 μl at 37°C for 1 h. As a control, pRAO1dup was also treated. Reaction mixtures were extracted three times with phenol/chloroform, and products were precipitated and brought into the in vitro transposition reaction as pRAO1pso. To confirm hydrolysis, a separate assay was used: by excising a 100-bp region of the triplex DNA from each of the substrates above, end labeling, and running the products out on both 6% native and denaturing acrylamide gels (data not shown), mobility shift confirmed the hydrolysis of the third strand.
Results
Targeting the Triplex Region by Tn7.
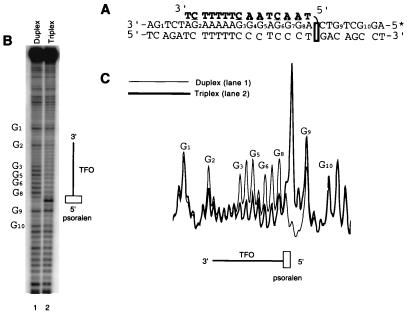
The reconstitution of Tn7's transposition in vitro has allowed us to directly examine the effects of individual nucleic acid structural motifs on target site selection. To generate transposition targets containing triplex DNA, several intermolecular triplex targets were constructed by annealing short (13–15 bp) TFOs to plasmids ranging in size from 2.5 to 5.2 kb. To increase stability of these pyrimidine-motif (i.e., Y⋅R⋅Y) triplexes under in vitro transposition reaction conditions (14), TFOs contained nucleoside analogs 5-methylcytosine (19, 20) and 8-oxoadenine (21) to negate the low pH requirement of Hoogsteen hydrogen bond formation (22, 23), and they also had a psoralen intercalator (24) to allow covalent attachment to the plasmid by photoactivated crosslinking. Once annealed and crosslinked, nearly 80% of the duplex plasmids contained a TFO attached by mono adduct. The stability of the triplex within these plasmids was assayed to verify proper annealing of the third strand throughout the transposition reactions. DMS footprinting confirms stable triplex formation under in vitro transposition conditions (Fig. 2). A comparison of the cleavage pattern within the triplex binding site in pRAO1dup (Fig. 2B, lane 1) to that of pRAO1tpx (Fig. 2B, lane 2) illustrates protection by the TFO. Line graph analysis further illustrates the magnitude of cleavage and protection of the target duplex and triplex regions (Fig. 2C). These triplex-containing plasmids and their parent duplex plasmids were used in the in vitro transposition reaction as targets where the effects of the triplex region on target site selection were measured. Because the increased activity of TnsABCA225V allows easier visualization and quantification of reaction products, experiments using a TnsABCA225V system are shown here; qualitatively similar results have been obtained with TnsABCwt (J.E.R. and N.L.C., unpublished results).
Figure 2.
Chemical footprinting of the duplex and intermolecular triplex found in pRAO1dup and pRAO1tpx, respectively, under in vitro transposition conditions. DMS methylation followed by NaOH treatment of the 5′-end-labeled purine strand generates a guanine and, to a lesser extent, adenine cleavage pattern. (A) The sequence of the triplex region. (B and C) Sequencing gel (B) and line graph (C) analysis of the cleavage pattern reveals the presence of the 15-bp TFO in the major groove; triplex formation results in a corresponding protection from DMS methylation and subsequent cleavage (lane 2) compared with duplex (lane 1). Hypercleavage at the site of psoralen intercalation and crosslinking is seen at the 5′ triplex-duplex junction.
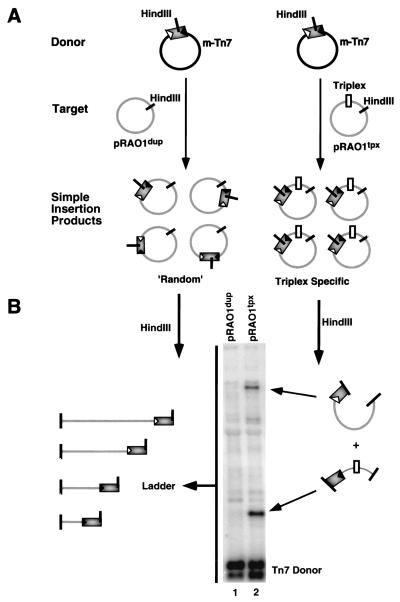
A direct comparison of Tn7's targeting preferences between a representative intermolecular triplex-containing plasmid, pRAO1tpx, and its parental duplex, pRAO1dup, was made by gel electrophoresis and Southern blotting of the in vitro transposition reaction products (Fig. 3). The location of the insertions into target plasmids were evaluated by restriction digestions at unique sites within both the target backbone and the transposon itself (Fig. 3A). On gel electrophoresis, the resulting distribution patterns for a population of insertions were visualized by hybridization of a transposon-specific probe. TnsABCA225V reaction mixtures containing the pRAO1dup target generated a ladder of bands corresponding to a “random” distribution throughout the target plasmid (Fig. 3B, lane 1). However, reaction mixtures that contained pRAO1tpx target plasmid displayed two discrete bands corresponding to an accumulation of insertions to a specific region of the target (Fig. 3B, lane 2). The size of these fragments coincided with the insertions falling in a region of the plasmid containing the 13-bp triplex.
Figure 3.
Assay for target specificity. (A) The TnsABCA225V in vitro transposition reaction generates a population of insertion products in which Tn7 is excised from the donor plasmid (mTn7) and inserted “randomly” into a duplex target plasmid (pRAO1dup). Target plasmids containing triplex (pRAO1tpx) generate specific insertion products adjacent to the triplex motif. (B) Unique HindIII sites located inside the transposon and in the target are used to digest reaction products, allowing random and specific insertions to be distinguished. On gel electrophoresis, the population of “random” inserts into pRAO1dup generates a ladder of products which are visualized by Southern blotting (lane 1). In contrast, an accumulation of specific insertions adjacent to triplex yields two specific products upon digestion (lane 2).
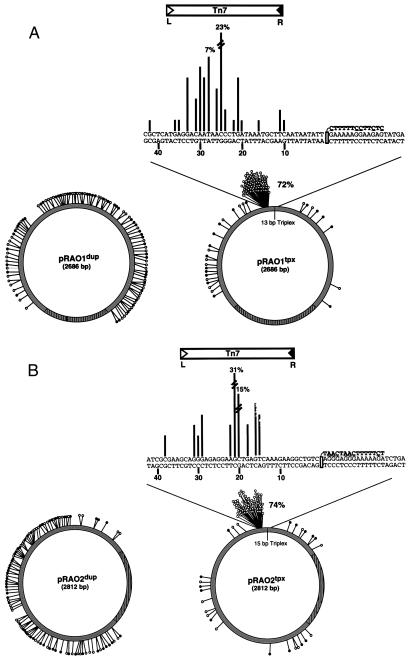
To determine the precise position of these pRAO1tpx insertions and the insertions into pRAO1dup, 100 individual transposition products from reaction mixtures containing each target were isolated by transformation and sequenced. The distribution profiles were generated by plotting the position and number of insertion events into each target DNA (Fig. 4A). To assay the contributions of target and TFO sequence on the insertion profile, reaction products from a second representative Y⋅R⋅Y triplex-containing target, pRAO2tpx, and its parent duplex, pRAO2dup were also sequenced (Fig. 4B). The profiles obtained from both triplex-containing target plasmids confirm the accumulation of insertions seen by gel electrophoresis (Fig. 3B, lane 2), with >70% of all insertions sequenced falling within a 40-bp region upstream of the 5′ end of the triplex (Fig. 4). This regional hot-spot for each triplex target is focused between 21 and 25 bp upstream from the 5′ end of the TFO. In contrast, the insertion products isolated from reactions of pRAO1dup and pRAO2dup show a “random” distribution pattern throughout the duplex targets. These results show that the presence of a 13- to 15-bp triplex strongly influences Tn7 target site selection, with >70% of all insertions occurring near the triplex.
Figure 4.
Comparison of insertion profiles of duplex- and triplex-containing targets. (A) Distribution of 100 insertions into pRAO1dup and pRAO1tpx. These target plasmids are identical except for a 13-bp TFO annealed and crosslinked to pRAO1tpx. (B) A second set of targets, pRAO2dup and pRAO2tpx, with varied sequence for both plasmid and TFO is shown. For the distribution patterns into the duplex plasmids, each line represents a single insertion; an open circle corresponds to a left-to-right orientation for Tn7, while a closed circle is right-to-left. The origin of replication is drawn in hatched lines. For the triplex-containing plasmids, the nucleotide sequence surrounding the triplex is shown; the number of insertions at each nucleotide is proportional to the length of the lines drawn. More than 70% of all insertions are found within a 40-bp region of each TFO. For these insertions, nearly all (>98%) occurred such that the right end of Tn7 element lands nearest the 5′ end of the triplex. A Tn7 element is drawn above the regional hot-spot to illustrate this orientation bias. Underlined nucleotides within the TFO sequence denote the use of modified bases.
Not only did the locations of the insertions vary greatly between pRAOdup and pRAOtpx targets, there is a striking bias in the orientation of the insertions as well. Tn7 is structurally asymmetric—i.e., Tn7L is distinct from Tn7R (25)—so that Tn7 insertion can occur in two distinct orientations. For example, insertions into attTn7 almost exclusively occur such that the right end of the element, Tn7R, is closest to the TnsD binding site (14). Interestingly, of the insertions landing adjacent to the triplex, >98% occurred with Tn7R closest to the triplex, whereas insertions away from the triplex and in pRAOdup displayed little preference in orientation (Fig. 4).
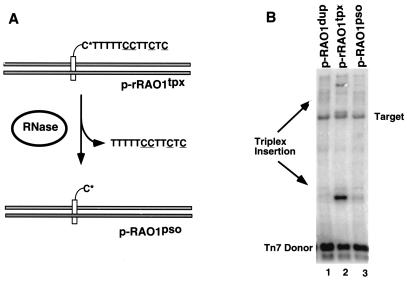
What feature of the triplex attracts Tn7? As we rely on the psoralen crosslink to maintain triplex formation under in vitro transposition reaction conditions, one possibility was that the transposon recognized the psoralen moiety. To better understand the recruitment signal, the contribution of the psoralen anchor was individually examined. A TFO was synthesized in which the deoxyribonucleotide nearest the psoralen on the 5′ end was replaced with a ribonucleotide, allowing removal of the TFO via RNase hydrolysis (Fig. 5A). Other than the replacement of a single deoxyribonucleotide, this TFO (r-TFO) was identical in sequence and its ability to target Tn7 transposition to the original all-deoxyribonucleotide TFO in pRAO1tpx (data not shown). Hydrolysis of this r-TFO triplex (p-rRAO1tpx) after annealing and crosslinking gave a target plasmid with psoralen alone (p-RAO1pso) (Fig. 5A) which was used in vitro to evaluate the effects of psoralen on Tn7 targeting. The insertion profile seen for the p-RAO1pso target (Fig. 5B, lane 3) resembles that of pRAO1dup (Fig. 4B, lane 1), with a “random” distribution pattern showing little accumulation of inserts at or near the original triplex binding site. Thus removal of the TFO resulted in a corresponding loss of insertion specificity. Only when the triplex is present (Fig. 5B, lane 2) is Tn7 stimulated to specifically insert adjacent to this site, making this targeting pathway truly triplex dependent.
Figure 5.
Effect of TFO removal on targeting. (A) Schematic illustrating removal of ribonucleotide-containing TFO upon hydrolysis. C* denotes a ribonucleotide cytosine, which is susceptible to hydrolysis by RNase treatment. Upon hydrolysis, a target substrate with psoralen still crosslinked (p-rRAO1pso) is generated. These pRAO1dup, p-rRAO1tpx, and p-rRAO1pso plasmids were then brought into the in vitro transposition reaction and assayed for target specificity. (B) A Southern blot using Tn7-specific probes illustrates the loss of targeting adjacent to the triplex upon removal of the TFO. Although the psoralen is still attached, no accumulation of triplex specific inserts was observed in reactions with the p-rRAO1pso target plasmid (lane 3). The band labeled “Target” is due to background cross-hybridization to linearized target DNA.
TnsC Preferentially Binds Triplex DNA.
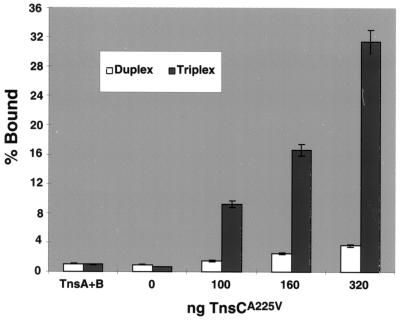
Which of the Tns proteins is responsible for triplex recognition? The transposase TnsAB? The ATP-dependent regulator TnsC? Filter binding was used to determine which transposition protein(s) was responsible for recognition of the triplex target (Fig. 6). TnsCA225V showed a significant preference for triplex DNA under the in vitro transposition reaction conditions. A combination of TnsA + TnsB showed no binding preference for triplex over duplex. These data reveal that triplex recognition is mediated by TnsC. We also note that recognition and insertion adjacent to triplex DNA by TnsC depended on the presence of TnsC's ATP cofactor (J.E.R. and N.L.C., unpublished observations). Thus, as has been seen for other targeting routes for Tn7, the ATP state of TnsC was critical to its activities (17, 26).
Figure 6.
Binding activity of transposition proteins: A graph showing relative affinities of TnsA, TnsB, and TnsCA225V for duplex and triplex DNA. Under in vitro reaction conditions, a titration of TnsCA225V binding shows a distinct preference for triplex over duplex DNA, whereas TnsA + TnsB shows little binding to either substrate. The percent bound reflects the fraction of the labeled substrate bound by TnsCA225V.
Taken together, our results suggest strongly that Tn7's ability to select triplex DNA as a target is derived from a specific interaction between TnsC and the triplex.
Tn7 Targets H-DNA, a Naturally Occurring Triplex Structure.
Another class of naturally occurring triplexes, known as H-DNA, form from polypurine⋅polypyrimidine mirror repeats and are believed to be involved in many biological processes (27), including homologous recombination (28–32). These sequences are found throughout the genomes of many organisms and have been implicated in playing a role in vivo (33). A particularly well characterized example of such a structure comes from the human PKD1 gene, which contains a 2.5-kb polypurine⋅polypyrimidine tract (34). It has been suggested that the ability to form higher-order DNA structures within this region may cause its characteristically high mutation frequency, ultimately contributing to autosomal dominant polycystic kidney disease (ADPKD). Interestingly, a 46-bp perfect mirror repeat taken from within intron 21 of this gene and cloned into pUC19 has recently been shown to readily form an H-DNA pyrimidine motif triplex (H-y3 isomer) at acidic pH in vitro (35).
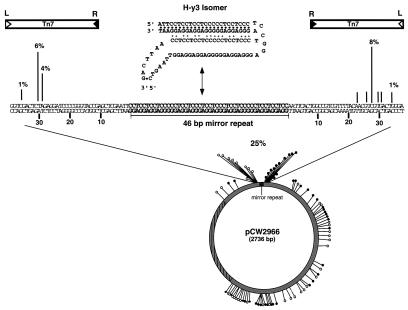
The plasmid containing this 46-bp mirror repeat sequence known to form H-DNA, pCW2966, was used as a target in our in vitro transposition reaction to assay Tn7's ability to recognize a naturally occurring intramolecular triplex structure (Fig. 7). At pH 8.0, little or no targeting was seen by Southern blotting or sequencing to the mirror repeat sequence (data not shown). However, reactions at pH 6.5 show a significant accumulation of insertions adjacent to the sequence known to form triplex (Fig. 7). Again, sequencing was used to view the insertion profile at nucleotide resolution (Fig. 7). Of 85 insertions sequenced, 25% fell within 50 bp of the mirror repeat sequence at pH 6.5.
Figure 7.
H-DNA target and Tn7 insertion profile. The 46-bp mirror repeat sequence capable of forming an intramolecular triplex or H-DNA (H-y3 isomer) is illustrated. Distribution of 85 independent insertions isolated by means of transformation were mapped onto pCW2966. An accumulation of insertions is found on either side of the 46-bp mirror repeat sequence; >98% of these insertions occur such that Tn7's right end is closest to the mirror repeat.
In comparison to the intermolecular triplex insertion profiles (Fig. 4), the pCW2966 insertions are focused on both the 5′ and 3′ ends of the repeat sequence, whereas nearly all of the intermolecular directed insertions occurred on the 5′ end (Fig. 4). An orientation bias is again observed, with 99% of the mirror-repeat insertions having Tn7's right end adjacent to the triplex moiety (Fig. 7). The disparity in target specificity between intermolecular (>70%) and the intramolecular triplex targets (25%) is likely due to the availability of target plasmids in the H-y3 triplex conformation. While crosslinking of the modified TFOs ensures that >80% of the population of plasmids contain a stable triplex, the pyrimidine motif H-DNA triplex structure is favored at more acidic pH values (35), and thus less H-y3 triplex isomer is likely available as substrate for site-specific recognition and transposition than for the crosslinked triplex plasmids.
Discussion
How does Tn7 preferentially recognize and direct insertions adjacent to triplex DNA? One possibility is the conformation of the triplex target site. The structural consequences of intermolecular triplex formation are found mostly at the triplex–duplex junctions, where bends and unwinding of duplex generate subtle asymmetric distortions (36–39). These conformational changes may provide a recognition signal for TnsC, which in turn stimulates the activities of the transposase, TnsA + TnsB. Tn7's wild-type target site selector, TnsD, has recently been found to induce conformational changes on its target, attTn7 (P. N. Kuduvalli and N.L.C., unpublished observations), suggesting this triplex-induced structural signal mimics the signals normally recognized by Tn7. The ability of TnsC to recognize and direct transposition adjacent to a nucleic acid structural motif, such as triplex DNA, raises the possibility that the recognition signal used in a TnsD-directed insertion into attTn7 also relies on a conformational change in the target DNA.
Structural features of target DNA have also been shown to influence target site selection in a variety of other recombination systems. Retroviral integrases and other nonretroviral transposition systems, for example, prefer to integrate into bent or cruciform target sites (5, 40). Many of these systems, however, rely on direct interaction between the integrase or transposase with target DNA, whereas our findings reveal that for Tn7, positioning of the transposase on target DNA through the use of an accessory protein, TnsC, is critical to target site selection. This strategy has been seen in other systems such as the Ty elements, which display a range of target site selectivity believed to be influenced by collaborations between host-encoded and Ty-encoded proteins. Highly specific integration of the retroelement Ty3, for example, has been shown to be positioned by the cellular transcription factors TFIIIB and TFIIIC (41).
Transposition directed to a specific target site in the absence of Tn7's wild-type targeting proteins has yielded both insights into the targeting signals normally recognized by Tn7 and novel targeting routes. A triplex-directed pathway may broaden the current applications of both Tn7 and TFOs as therapeutics; as Tn7 can be designed to carry a gene of interest, one can now envision DNA delivery to a location that has the ability to form triplex. Although target sequences are currently limited to polypurine⋅polypyrimidine tracts, TFOs offer an attractive alternative to integration systems that rely on protein chimeras to generate novel binding activities (42–44). Moreover, the interaction of TnsC with triplex DNA motifs may uncover further information protein recognition of triple-helical nucleic acids, providing new evidence of their potential biological roles. Although many useful applications for triplex DNA have been found over recent years (27, 45–47), their biological role remains to be demonstrated.
Acknowledgments
We are grateful to Dr. John Bissler for supplying pCW2966; P. Eckhoff for help in the preparation of this manuscript; members of the Craig lab for critical reading of this manuscript; members of the Miller and Bryant labs for helpful discussion; Bob Sarnovsky, Dr. Prasad Kuduvalli, Dr. David Noll, and Dr. Joseph Peters for expert advice and insightful discussion; and Dr. Tom Tullius for guidance. This work was supported by the National Institutes of Health under Grant GM53824 to N.L.C. N.L.C. is an investigator with Howard Hughes Medical Institute.
Abbreviations
- TFO
triplex-forming oligomer
- r-TFO
TFO containing a single ribonucleotide
- DMS
dimethyl sulfate
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.080061497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.080061497
References
- 1.Berg D E, Howe M M, editors. Mobile DNA. Washington, DC: Am. Soc. Microbiol.; 1989. [Google Scholar]
- 2.Craig N L. Annu Rev Biochem. 1997;66:437–474. doi: 10.1146/annurev.biochem.66.1.437. [DOI] [PubMed] [Google Scholar]
- 3.Berg C M, Berg E. In: Mobile Genetic Elements. Sherratt D J, editor. Oxford: IRL; 1995. pp. 38–68. [Google Scholar]
- 4.Katz R A, Gravuer K, Skalka A M. J Biol Chem. 1998;273:24190–24195. doi: 10.1074/jbc.273.37.24190. [DOI] [PubMed] [Google Scholar]
- 5.Pryciak P M, Varmus H E. Cell. 1992;69:769–780. doi: 10.1016/0092-8674(92)90289-o. [DOI] [PubMed] [Google Scholar]
- 6.Barth P T, Datta N, Hedges R W, Grinter N J. J Bacteriol. 1976;125:800–810. doi: 10.1128/jb.125.3.800-810.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craig N L. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd Ed. Neidhardt F C, Curtiss R I, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2339–2362. [Google Scholar]
- 8.Rogers M, Ekaterinaki N, Nimmo E, Sherratt D. Mol Gen Genet. 1986;205:550–556. doi: 10.1007/BF00338097. [DOI] [PubMed] [Google Scholar]
- 9.Waddell C S, Craig N L. Genes Dev. 1988;2:137–149. doi: 10.1101/gad.2.2.137. [DOI] [PubMed] [Google Scholar]
- 10.Kubo K M, Craig N L. J Bacteriol. 1990;172:2774–2778. doi: 10.1128/jb.172.5.2774-2778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolkow C A, DeBoy R T, Craig N L. Genes Dev. 1996;10:2145–2157. doi: 10.1101/gad.10.17.2145. [DOI] [PubMed] [Google Scholar]
- 12.Lichtenstein C, Brenner S. Nature (London) 1982;297:601–603. doi: 10.1038/297601a0. [DOI] [PubMed] [Google Scholar]
- 13.Craig N L. Mol Microbiol. 1991;5:2569–2573. doi: 10.1111/j.1365-2958.1991.tb01964.x. [DOI] [PubMed] [Google Scholar]
- 14.Bainton R J, Kubo K M, Feng J-N, Craig N L. Cell. 1993;72:931–943. doi: 10.1016/0092-8674(93)90581-a. [DOI] [PubMed] [Google Scholar]
- 15.Sarnovsky R, May E W, Craig N L. EMBO J. 1996;15:6348–6361. [PMC free article] [PubMed] [Google Scholar]
- 16.May E W, Craig N L. Science. 1996;272:401–404. doi: 10.1126/science.272.5260.401. [DOI] [PubMed] [Google Scholar]
- 17.Stellwagen A, Craig N L. Trends Biochem Sci. 1998;23:486–490. doi: 10.1016/s0968-0004(98)01325-5. [DOI] [PubMed] [Google Scholar]
- 18.Stellwagen A, Craig N L. EMBO J. 1997;16:6823–6834. doi: 10.1093/emboj/16.22.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Povsic T J, Dervan P B. J Am Chem Soc. 1989;111:3059–3061. [Google Scholar]
- 20.Lee J S, Woodsworth M L, Latimer L J, Morgan A R. Nucleic Acids Res. 1984;12:6603–6614. doi: 10.1093/nar/12.16.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller P S, Bi G, Kipp S A, Fok V, DeLong R K. Nucleic Acids Res. 1996;24:730–736. doi: 10.1093/nar/24.4.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Felsenfeld G, Davies D R, Rich A. J Am Chem Soc. 1957;79:2023–2024. [Google Scholar]
- 23.Hoogsteen K. Acta Crystallogr. 1959;12:822–824. [Google Scholar]
- 24.Takasugi M, Guendouz A, Chassignol M, Decout J L, Lhomme J, Thuong N T, Helene C. Proc Natl Acad Sci USA. 1991;88:5602–5606. doi: 10.1073/pnas.88.13.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arciszewska L K, Drake D, Craig N L. J Mol Biol. 1989;207:35–52. doi: 10.1016/0022-2836(89)90439-7. [DOI] [PubMed] [Google Scholar]
- 26.Bainton R, Gamas P, Craig N L. Cell. 1991;65:805–816. doi: 10.1016/0092-8674(91)90388-f. [DOI] [PubMed] [Google Scholar]
- 27.Soyfer V N, Potaman V N. Triple-Helical Nucleic Acids. New York: Springer; 1996. [Google Scholar]
- 28.Zhurkin V B, Raghunathan G, Ulyanov N B, Camerini-Otero R D, Jernigan R L. J Mol Biol. 1994;239:181–200. doi: 10.1006/jmbi.1994.1362. [DOI] [PubMed] [Google Scholar]
- 29.Rooney S M, Moore P D. Proc Natl Acad Sci USA. 1995;92:2141–2144. doi: 10.1073/pnas.92.6.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kowhi Y, Panchenko Y. Genes Dev. 1993;7:1766–1778. doi: 10.1101/gad.7.9.1766. [DOI] [PubMed] [Google Scholar]
- 31.Collier D A, Griffin J A, Wells R D. J Biol Chem. 1988;263:7397–7405. [PubMed] [Google Scholar]
- 32.Weinreb A, Collier D A, Birshtein B K, Wells R D. J Biol Chem. 1990;265:1352–1359. [PubMed] [Google Scholar]
- 33.Frank-Kamenetskii M D, Mirkin S M. Annu Rev Biochem. 1995;64:65–95. doi: 10.1146/annurev.bi.64.070195.000433. [DOI] [PubMed] [Google Scholar]
- 34.Watson M L, Torres V E. Polycystic Kidney Disease. New York: Oxford Univ. Press; 1996. [Google Scholar]
- 35.Blaszak R T, Potaman V V, Sinden R R, Bissler J J. Nucleic Acids Res. 1999;27:2610–2617. doi: 10.1093/nar/27.13.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asensio J L, Dosanjh H S, Jenkins T C, Lane A N. Biochemistry. 1998;37:15188–15198. doi: 10.1021/bi980057m. [DOI] [PubMed] [Google Scholar]
- 37.Rhee S, Han Z, Liu K, Miles H T, Davies D R. Biochemistry. 1999;38:16810–16815. doi: 10.1021/bi991811m. [DOI] [PubMed] [Google Scholar]
- 38.Stonehouse T J, Fox K R. Biochim Biophys Acta. 1994;1218:322–330. doi: 10.1016/0167-4781(94)90184-8. [DOI] [PubMed] [Google Scholar]
- 39.Chomilier J, Sun J-S, Collier D A, Garestier T, Helene C, Lavery R. Biophys Chem. 1992;45:143–152. doi: 10.1016/0301-4622(92)87006-5. [DOI] [PubMed] [Google Scholar]
- 40.Pruss D, Reeves R, Bushman F D, Wolffe A P. J Biol Chem. 1994;269:25031–25041. [PubMed] [Google Scholar]
- 41.Kirchner J, Connolly C M, Sandmeyer S B. Science. 1995;267:1488–1491. doi: 10.1126/science.7878467. [DOI] [PubMed] [Google Scholar]
- 42.Bushman F D, Craigie R. Proc Natl Acad Sci USA. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goulaouic H, Chow S A. J Virol. 1996;70:37–46. doi: 10.1128/jvi.70.1.37-46.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katz R A, Merkel G, Skalka A M. Virology. 1996;217:178–190. doi: 10.1006/viro.1996.0105. [DOI] [PubMed] [Google Scholar]
- 45.Moser H E, Dervan P B. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 46.Thuong N T, Helene C. Angew Chem Int Ed Engl. 1993;32:666–690. [Google Scholar]
- 47.Majumdar A, Khorlin A, Dyatkina N, Lin F L, Powell J, Liu J, Fei Z, Khripine Y, Watanabe K A, George J, et al. Nat Genet. 1998;20:212–214. doi: 10.1038/2530. [DOI] [PubMed] [Google Scholar]