Abstract
The PII protein is Escherichia coli's cognate transducer of the nitrogen signal to the NRII (NtrB)/NRI (NtrC) two-component system and to adenylyltransferase. Through these two routes, PII regulates both amount and activity of glutamine synthetase. GlnK is the recently discovered paralogue of PII, with a similar trimeric x-ray structure. Here we show that PII and GlnK form heterotrimers, in E. coli grown in nitrogen-poor medium. In vitro, fully uridylylated heterotrimers of the two proteins stimulated the deadenylylation activity of adenylyltransferase, albeit to a lower extent than homotrimeric PII-UMP. Fully uridylylated GlnK did not stimulate, or hardly stimulated, the deadenylylation activity. We propose that uridylylated PII/GlnK heterotrimers fine-regulate the activation of glutamine synthetase. The PII/GlnK couple is a first example of prokaryotic signal transducer that can form heterotrimers. Advantages of hetero-oligomer formation as molecular mechanism for fine-regulation of signal transduction are discussed.
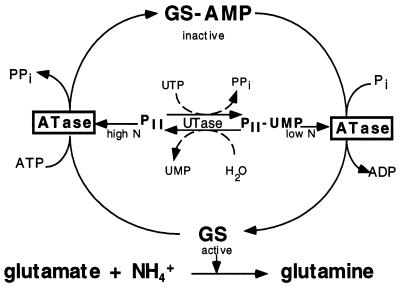
In enteric bacteria, the PII protein occupies a pivotal position in the network regulating the activity of glutamine synthetase (GS), itself a key enzyme in the assimilation of ammonia. The nitrogen status of the cell, as sensed by uridylyltransferase (UTase), is transferred to PII by adjusting the degree of uridylylation of the latter. Native PII signals a nitrogen-rich status whereas PII-UMP flags a nitrogen-poor status. From PII, the signal is transferred to two targets, i.e., adenylyltransferase (ATase) and NRII (NtrB), involved in the covalent modification of GS and in the transcriptional regulation of the gene encoding GS, respectively (refs. 1 and 2; Fig. 1).
Figure 1.
The adenylylation/deadenylylation cycle that controls the activity of glutamine synthetase. The enzyme UTase signals high N status through PII and low N status through PII-UMP by stimulating ATase to inactivate or activate, respectively, GS so that ATP can be used to ligate ammonia to glutamate to form glutamine. The paralogue GlnK that is expressed during low N and uridylylated at the same position as PII is not shown (see text for details); nor is the transcription regulation of the GS level through the two component system indicated.
Escherichia coli can also contain a PII-like protein: i.e., GlnK (3, 4). PII and GlnK are similar, both structurally and functionally. The two proteins are each composed of 112 amino acid residues that are 67% identical and have virtually the same trimeric crystal structure (5–7). Each protein has been purified as homotrimer (5, 8). Both PII and GlnK can be modified by UTase at tyrosine-51 (4, 9–12). Functionally, PII and GlnK can address the same targets; i.e., in vitro, they can both regulate the activities of ATase and NRII (4, 11). However, the in vivo response of either target to PII exceeds that to GlnK. This may in part be because of a difference in the effect of the small molecule 2-oxoglutarate on the two signal proteins (11, 13).
One functional aspect differs drastically between PII and GlnK; i.e., the expression of the glnB gene (which encodes PII) is constitutive with respect to the intracellular nitrogen status (14) whereas the expression of the glnK gene is regulated by it (4, 15). There is little or no GlnK after growth in nitrogen-rich conditions.
This report describes the identification of PII/GlnK-heterotrimers in E. coli after growing the cells in nitrogen-poor media and also shows that in vitro generated heterotrimers have lower activity than PII homotrimers. It suggests that GlnK may moderate the regulation of ammonia assimilation by replacing PII monomers in the regulatory trimer. This finding adds an additional level of complexity to this already sophisticated regulatory system.
Materials and Methods
Bacterial Strains and Media.
E. coli strains and plasmids used in this study are listed in Table 1. Minimal Mops medium has been described previously (4, 21) and contained 14 mM l-glutamine plus 14 mM NH4Cl, 14 mM l-glutamine or 14 mM l-arginine as nitrogen source. Adapted cultures of strains YMC10 (wild-type), WCH30 (glnK−), and RB9060 (glnB−) were grown at 37°C to an OD600 nm of 0.3–0.5, or to an OD600 nm of 0.1–0.15 for the strains grown in medium containing arginine as N source. RB9040 containing pBluescript II SK(+) (Stratagene) derived plasmids were grown at 37°C in Luria broth (LB) plus 14 mM l-glutamine to stationary phase. To RB9040 containing pWVH102, 200 μM IPTG was added to induce the glnB gene from an artificial IPTG-inducible promoter (20, 22). IPTG was not added to RB9040 containing pWVH149 or pWVH180. In these cases the expression of glnK resulted from leakiness of lacP/O, producing only native GlnK (4, 11, 18). Antibiotics were used at the following final concentrations: gentamycin, 15 μg/ml; ampicillin, 100 μg/ml; and tetracycline, 15 μg/ml.
Table 1.
E. coli strains and plasmids used in this study
| Strains/plasmids | Genotype/description | Ref. |
|---|---|---|
| Strain | ||
| YMC10 | endA1 thi-1 hsdR17 supE44 ΔlacU169 hutC (wt) | 16 |
| RB9040 | As YMC10, but glnD99∷Tn10 | 17 |
| RB9060 | As YMC10, but glnB2306 | 17 |
| WCH30 | As YMC10, but Ω(GmR)-ΔglnK1 | 18 |
| UNF3435 | As YMC10, but Ω(GmR)-ΔglnK1, ΔglnB2306 | 18 |
| Plasmid | ||
| pRJ1 | E. coli glnB expressed from λ PR-PL in pND707 | 9, 19 |
| pWVH102 | E. coli glnB expressed from PA1LacO-1 (22) in pBluescript II SK(+) (pBSK) | 20 |
| pWVH149 | E. coli glnK expressed from Plac in pBSK | 4 |
| pWVH180 | E. coli glnK glnB operon expressed from Plac in pBSK | This work |
Plasmid Constructions.
The operon containing the E. coli glnK and glnB genes was constructed by inserting the glnB gene, as a 1.4-kb EcoNI (blunted with DNA polymerase Klenow fragment)/SmaI fragment from pRJ1 (9) into pWVH149 (4) digested with BstXI (blunted), resulting in pWVH180.
Preparation of Cell Extracts to Study in Vivo Heterotrimer Formation.
Five-milliliter LB cultures or fifty-milliliter Mops cultures were chilled on ice for approximately 20 min and were centrifuged at 4°C. Cell pellets were resuspended in 500 μl of cold lysis buffer (50 mM Tris⋅HCl, pH 8.0/25% (wt/vol) sucrose/1 mM EDTA) containing 0.1 mg/ml lysozyme and were incubated for 15 min on ice. Cells were frozen in liquid nitrogen and subsequently were incubated at 65°C and regularly vortexed until completely thawed [at which temperature uridylyltransferase is inactivated (23)], were subjected to three additional freeze-thaw cycles and to an extra 10 min at 65°C, and then were chilled on ice. The resulting viscous solution was incubated with DNase I (0.1 mg/ml) at 37°C for 30 min after adding MgCl2 and MnCl2 to a final concentration of 10 mM and 1 mM, respectively. The solution was centrifuged (15,800 × g, 10 min, 4°C) and the supernatant was transferred to a new Eppendorf tube. After adding glycerol to a final concentration of 5%, cell extracts were frozen in liquid nitrogen and were stored at −20°C. Protein concentration was measured with the Bradford's protein reagent kit and BSA as the standard.
Gel Electrophoresis and Western Blot Analysis.
Cell extracts of RB9040 overexpressing glnB, glnK, or the glnK glnB operon, equivalent to 2 μg of total protein, or cell extracts of YMC10, WCH30, and RB9060, equivalent to 10 μg of total protein, were mixed with sample buffer (50% glycerol/0.2% bromophenol blue/0.3 M Tris) and were loaded without prior boiling onto a 8% nondenaturing acrylamide gel essentially as described before (9, 24). After electrophoresis, the gel was blotted onto nitrocellulose and was probed with polyclonal rabbit antiserum directed against E. coli PII (4) or monoclonal mouse antiserum 19G4 or 24H2 directed against E. coli PII (P.C. and S.G.V., unpublished work). Antiserum 19G4 can detect PII, PII-UMP, GlnK, and GlnK-UMP (Fig. 3A) whereas antiserum 24H2 can only detect PII and PII-UMP (Fig. 3B). The bands of the primary antibodies were probed with alkaline phosphatase-conjugated secondary antibodies and were visualized after exposure to AttoPhos detection reagent (Amersham), by scanning with a Storm PhosphorImager (Molecular Dynamics) or by staining with a mixture of nitro-blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate. Purified proteins were used as standards: 5 ng (PII)3, 5 ng (PII-UMP)3, 50 ng (GlnK)3, or 50 ng (GlnK-UMP)3 diluted in cell extract of the glnK glnB double mutant UNF3435 (10 μg protein). PII(-UMP) like bands in strain UNF3435 were not detectable (data not shown).
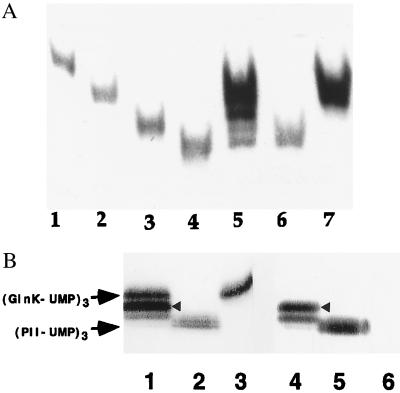
Figure 3.
(A) Formation of PII and GlnK heterotrimers in wild-type E. coli grown in “nitrogen-poor” medium. Western blot of nondenaturing gels of cell extracts detected with monoclonal PII antibody 19G4. Lanes: 1, 2, 3, and 4, purified homotrimeric proteins (GlnK)3, (GlnK-UMP)3, (PII)3, and (PII-UMP)3, respectively; 5, wild type-strain YMC10 (wt); 6, in-frame glnK deletion strain WCH30 (glnK−); 7, glnB deletion strain RB9060 (glnB−); 5–7, cells grown in minimal medium containing glutamine as nitrogen-source. (B) Detection of heterotrimers in wild-type cells grown in arginine medium using monoclonal PII antibody 19G4 (cross reacts with GlnK) (lanes 1–3) and PII monoclonal antibody 24H2 (lanes 4–6), which does not cross-react with GlnK or its uridylylated forms. Lanes: 1 and 4, strain YMC10; 2 and 5, strain WCH30; 3 and 6, strain RB9060. The different intensity of detection of second upper band in lanes 1 and 4 (highlighted with arrowhead) suggests that the identity of that band is most probably (GlnK-UMP)2/PII-UMP (see text). There is almost no PII-UMP homotrimer detected by 19G4 in lane 1 or lane 4, thus suggesting that proportions of homotrimers and heterotrimers vary according to the N source (arginine is a poorer source of N than glutamine).
Purification of Proteins, Denaturation and Renaturation of PII/GlnK Mixture, and in Vitro Assays.
The preparations of PII (25), GlnK (8), uridylylated PII (9), uridylylated GlnK (12), ATase (26), UTase (26), GS (20), and GS-AMP (20) were carried as described. To allow heterotrimers to form, PII, PII-UMP, GlnK, and GlnK-UMP were mixed at various ratios (1:1, 1:2, 2:1), to a final concentration of 5 mM in 50 mM Tris⋅HCl (pH 7.5) and 8 M urea (27), and were incubated at 45°C for 1 h. Renaturation was achieved by dialyzing samples against 50 mM Tris⋅HCl (pH 7.5) (three changes) at 4°C overnight. The different molecular forms [e.g., homotrimer of PII-UMP indicated as (PII-UMP)3] of the signal proteins made during the renaturation were separated on a 8% nondenaturing PA gel, were stained with Coomassie blue, were destained and the respective proportions quantitated (Table 2), with a Fluor-S MultiImager (Bio-Rad) using the quantity one 4.0.1 software version. The adenylylation and deadenylylation assays of GS and GS-AMP, respectively, were carried out with the renatured mix as described (26, 28).
Table 2.
Proportion of heterotrimers in renatured mixtures
| 1:1 | 1:2 | 2:1 | |
|---|---|---|---|
| PII:GlnK mixture | |||
| GlnK3 (%) | 12 (12) | 28 (30) | 6 (4) |
| PII1GlnK2 (%) | 34 (37) | 40 (44) | 18 (22) |
| PII2GlnK1 (%) | 43 (37) | 27 (22) | 47 (44) |
| PII3 (%) | 11 (12) | 5 (4) | 29 (30) |
| PII-UMP:GlnK-UMP mixture | |||
| GlnK-UMP3, % | 30 | 45 | 16 |
| PII-UMP1GlnK-UMP2, % | 28 | 27 | 24 |
| PII-UMP2GlnK-UMP1, % | 29 | 20 | 35 |
| PII-UMP3, % | 13 | 8 | 25 |
| PII-UMP:GlnK mixture | |||
| GlnK3, % | 14 | 22 | 5 |
| PII-UMP1GlnK2, % | 34 | 34 | 16 |
| PII-UMP2GlnK1, % | 41 | 35 | 46 |
| PII-UMP3, % | 11 | 9 | 33 |
Proportions of various species separated on native 8% polyacrylamide gel were quantified as in Materials and Methods. In PII:GlnK section, between parentheses, the theoretical fractions expected for an unbiased binomial distribution are given. The same numbers should have applied to the PII-UMP:GlnK-UMP section had their distributions been unbiased. In this section, the bias observed is slight and in favor of a stronger attractive interaction between uridylylated GlnKs than between uridylylated PIIs. The PII-UMP:GlnK section also shows a distribution close to the theoretically expected values.
Results
Heterotrimer Formation in Vivo, After Overexpressing a glnK-glnB Operon.
In preliminary experiments, purified GlnK and PII, and their covalently modified forms containing one, two, or three UMP groups, ran on a native gel as eight discrete bands (12). To simplify the interpretation of the native gels, the issue of heterotrimer formation was addressed in the absence of uridylylation. As glnK is not expressed in a UTase-deficient strain (4, 15), plasmid pWVH180 containing a glnK-glnB operon expressed from a leaky lacP/O (18) was derived from pWVH149 by inserting the glnB gene downstream of the glnK gene.
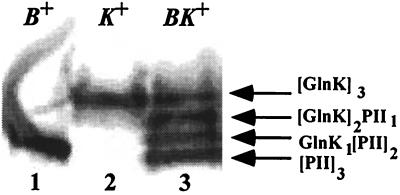
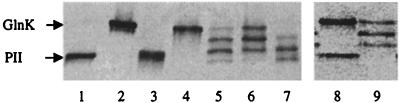
The plasmids containing the glnB gene, the glnK gene, or the glnK-glnB operon were grown in RB9040 (UTase-deficient strain) overnight in rich medium, and cell extracts of these cultures were separated on a native gel. A Western blot of the native gel, using monoclonal PII antiserum 19G4 [which cross-reacts with GlnK (Fig. 2)] or polyclonal PII antiserum (data not shown), revealed that PII and GlnK were separated by the gel electrophoresis under nondenaturing conditions and that PII migrated faster than GlnK. Strain RB9040 overexpressing the glnK-glnB operon from pWVH180 showed not only PII and GlnK bands, but also two bands that migrated at positions in between PII and GlnK (Fig. 2, lane 3). The detection of these two additional bands by the PII monoclonal antibody, as well as their relative mobility in the native gel, suggests that they constitute heterotrimers of GlnK and PII. From comparison of their relative mobility with that of the homotrimers of PII and GlnK (Fig. 2, lanes 1 and 2, respectively), we propose that the subunit compositions amount to (GlnK)2/PII for the more slowly migrating heterotrimer and GlnK/(PII)2 for the faster migrating heterotrimer. A mixture of purified PII and GlnK separated as two bands with mobilities of homotrimeric PII and GlnK respectively, excluding the possibility that the PII/GlnK heterotrimers were formed in the stacking gel of the nondenaturing acrylamide gel (Fig. 4, lane 8).
Figure 2.
Heterotrimer formation after over-expressing a glnK-glnB operon in a UTase negative strain. Shown is a Western blot of a native gel of cell extracts detected with monoclonal PII antibody 19G4 that cross-reacts with GlnK. Lanes: 1, PII encoded by pWVH102 (B+); 2, GlnK encoded by pWVH149 (K+); 3, expression of the glnK-glnB operon from pWVH180 (KB+). Two heterotrimers, (GlnK)2PII and GlnK(PII)2, in lane 3 migrate in between the homotrimeric PII and GlnK.
Figure 4.
Formation of PII/GlnK heterotrimers in vitro by denaturation-renaturation procedure. Samples of pure PII or GlnK or mixtures of the two were either untreated or treated with a denaturation-renaturation procedure (see Materials and Methods). Samples corresponding to 2 μg of protein were applied to each lane and were separated on a 8% native gel followed by staining Coomassie brilliant blue to visualize the protein bands. Lanes: 1, untreated PII; 2, untreated GlnK; 3, treated PII; 4, treated GlnK; 5–7, PII/GlnK mixtures in 1:1, 1:2, and 2:1 ratios, respectively, that were treated. Lane 8 shows a similar 8% native gel of untreated 1:2 mixture of PII:GlnK separating as homotrimers compared with a treated sample (lane 9), indicating that heterotrimers are not formed in the stacking gel.
Heterotrimer Formation of PII and GlnK in Wild-Type E. coli.
The above results indicate that heterotrimers of PII and GlnK can form in vivo, when both proteins are overexpressed from a plasmid based operon in an UTase negative strain. The question remained whether heterotrimers are formed in wild-type E. coli grown in physiological conditions in which both the glnB and glnK genes are expressed. To address this, three different strains, wild-type YMC10, the glnB mutant strain RB9060, and the glnK deletion mutant strain WCH30, were grown in nitrogen-poor medium, and cell extracts were prepared as above. Samples were again electrophoresed through a native gel and were subjected to Western blot analysis. Fig. 3A shows that the cell extracts of the glnB mutant and of the glnK mutant, when grown in nitrogen-poor medium, contained GlnK (Fig. 3A, lane 7) and PII (lane 6) trimers, respectively, which migrated as if they were fully uridylylated. The presence of fractions of PII (and GlnK, respectively) that were incompletely uridylylated cannot be excluded with the available resolution. The most important point of the experiment was, however, the result obtained with wild-type cells grown in nitrogen-poor medium. Here, the gel does display bands with the mobility of (GlnK-UMP)3 and (PII-UMP)3, but also two bands migrating with intermediate mobilities (Fig. 3A, lane 5). These additional bands comigrated neither with native PII nor with native GlnK. In fact, the second upper band in Fig. 3A, lane 5 migrated more slowly than did native PII (lane 3), hence more slowly than any of the PII trimers, but faster than (GlnK-UMP)3 (lane 2), and hence faster than any of the GlnK trimers. The third upper band in lane 5 of Fig. 3A migrated more slowly than (PII-UMP)3 but faster than native PII. Because only four equally spaced discrete bands are discernible in lane 5 of Fig. 3A, two of which comigrate with (GlnK-UMP)3 or (PII-UMP)3, we presume that the heterotrimer bands are uniformly and probably fully uridylylated, as perhaps expected because these wild type cells were grown in nitrogen-poor medium. Had the heterotrimers not been uniformly uridylylated, more bands should have been detected that migrated with unequal spacing between the various hybrids. The appearance and mobility of the two extra bands in lane 5 of Fig. 3A are consistent with (GlnK-UMP)2/PII-UMP for the more slowly migrating heterotrimer (second upper band) and GlnK-UMP/(PII-UMP)2 for the faster migrating heterotrimer (third upper band).
Extra evidence for heterotrimers in the wild-type cells was obtained by probing extracts of wild-type or mutant cells grown in nitrogen “very poor” medium (arginine) with monoclonal PII antibody 19G4 (Fig 3B, lanes 1–3) or 24H2 (lanes 4–6). As previously noted, 19G4 antiserum detected a band with electrophoretic mobility similar to that of (PII-UMP)3 in Fig. 3B, lane 2 in an extract from the glnK− strain, and one with similar mobility to (GlnK-UMP)3 in lane 3 in the extract from the glnB− strain. Notably, the PII specific monoclonal antibody 24H2 that cross reacts neither with GlnK nor with any of its uridylylated forms did not detect (GlnK-UMP)3 in Fig. 3B, lane 6 (same sample as in lane 3) but did reveal the band corresponding to (PII-UMP)3 in lane 5 (same sample as in lane 2). In wild-type cells grown in arginine, only three distinct bands were detected with antibody 19G4. The band corresponding to (PII-UMP)3 was hardly detectable. The slowest migrating band corresponded to (GlnK-UMP)3. The second upper band (indicated by an arrowhead in Fig. 3B, lanes 1 and 4) is presumably (GlnK-UMP)2/PII-UMP whereas the other band is GlnK-UMP/(PII-UMP)2. The two heterotrimers formed in the wild-type cells appeared to be present in different quantities, the presumed (GlnK-UMP)2/PII-UMP exceeding GlnK-UMP/(PII-UMP)2 (Fig. 3B, lane 1). The most important results here are that (i) the PII-specific monoclonal antibody 24H2 (Fig. 3B, lane 4) detected the two presumed heterotrimers bands and (ii) the upper band of the two presumed heterotrimers bands migrated faster than (GlnK-UMP)3 but more slowly than unmodified or modified PII homotrimer; i.e., that band was retarded compared with PII3 or even (PII-UMP)3 but still cross-reacted with the PII-specific antibody 24H2. This is good evidence that the latter band corresponds to a mixed oligomer. In addition, the intensity of the presumed heterotrimer (GlnK-UMP)2/PII-UMP band probed with antibody 24H2 was strongly reduced compared with the same band (indicated by arrowhead in Fig. 3B) probed with antibody 19G4.
Heterotrimer Formation in Vitro with Purified Proteins.
To examine the biochemistry of heterotrimers, purified PII or GlnK were denatured in 8 M urea and were renatured as described in Materials and Methods (27). Separation on an 8% native gel demonstrated that PII and GlnK can each be denatured and renatured to form homotrimers on their own (Fig. 4). When the two proteins were mixed at various ratios and subjected to the denaturation-renaturation protocol, the native gel analysis (Fig. 4, lanes 5–7) revealed mixed oligomers of the two proteins, migrating to positions between homotrimeric GlnK and homotrimeric PII. Probably the slower migrating of the two extra bands in Fig. 4, lanes 5–7 corresponds to (GlnK)2/PII, and the other one is GlnK/(PII)2. As with the in vivo experiment, the level of each of the heterotrimer species depended on the ratio of PII to GlnK in the peptide mixture used in the renaturation procedure. The proportions of the various species in the denatured-renatured mixtures were not far from that expected for an unbiased binomial distribution (Table 2). This suggests that PII and GlnK monomers had the same affinities for their own kind as for their paralogues. Thus, heterotrimer formation is not dominant. This agrees with the structural data: the critical trimer contact residues such as Val 30, Lys 34, and Glu32 are identical for the two paralogues (5–7). Heterotrimers of mixtures of purified PII-UMP/GlnK-UMP (Table 2) and PII-UMP/GlnK (Table 2) were produced by using the same protocol as above.
Function of Homo- and Heterotrimers in GS Activation (Deadenylylation).
Because the expression of GlnK is nitrogen-regulated, and the present study indicates that this protein is presumably fully uridylylated in cells grown in poor nitrogen medium, we chose to examine the ability of the uridylylated PII/GlnK heterotrimers as well as that of the uridylylated PII and GlnK homotrimers to stimulate the deadenylylation activity of ATase in vitro. When GS has been adenylylated, it is unable to form γ-glutamyl hydroxymate (γ-GH). In the presence of fully uridylylated PII, ATase rapidly converts GS-AMP to GS, thereby increasing the rate of formation of γ-GH (Fig. 5; refs. 4 and 9). PII-UMP alone that had been subjected to the denaturation-renaturation protocol was almost as active as untreated PII-UMP in stimulating ATase deadenylylation activity. In fact, the initial rate for deadenylylation in the presence of the treated PII-UMP was similar to that with untreated PII-UMP whereas the extent of deadenylylation was a little lower compared with untreated PII-UMP (data not shown). Accordingly, we measured how quickly the rate of γ-GH formation increased after addition of untreated GlnK-UMP or uridylylated heterotrimers prepared by the denaturation-renaturation protocol. Surprisingly, fully uridylylated GlnK (as evidenced by native gel analysis) hardly stimulated ATase into deadenylylating GS-AMP (Fig. 5). This was observed under the conditions in which PII-UMP did stimulate the deadenylylation activity of ATase. In fact, the ultimate GS activity in the presence of GlnK-UMP was only marginally lower as compared with when PII-UMP was omitted from the assay mixture, as demonstrated by the almost negligible change in the rate of increase of γ-glutamyl hydroxymate formation for the two conditions (Fig. 5). PII-UMP stimulated the deadenylylation activity more, even when its concentration was reduced to 1/20th of that of 27 nM GlnK-UMP. One reason for the lack of activity of GlnK-UMP could be the absence of an essential cofactor from the deadenylylation mixture. This factor should then be unique to GlnK: i.e., unnecessary for the PII-UMP stimulated deadenylylation. Because an anion-binding site had been noted in the crystal structure of GlnK (7), the possibility that acetyl phosphate was involved in activation of GlnK was examined. However, neither GlnK nor GlnK-UMP was able to stimulate ATase to deadenylylate GS-AMP in the presence of 50 mM acetyl phosphate (data not shown). Unmodified GlnK either at the standard concentration (27 nM) or 10 times that concentration was also unable to activate deadenylylation, although 10 times the standard PII concentration activated deadenylylation somewhat (ref. 29; data not shown).
Figure 5.
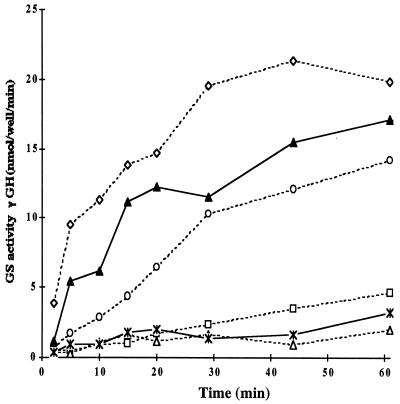
Uridylylated PII/GlnK heterotrimers can regulate ATase in vitro. The in vitro assay that monitors the rate of formation of γ-glutamyl hydroxamate (γ-GH) based on the γ-glutamyl transferase activity of non-adenylylated GS was used to monitor the effect of heterotrimers on ATase to deadenylylate GS-AMP and was carried out as described (26). The reaction mixture contained Hepes-HCl (100 mM, pH 7.6), BSA (2 mg/ml), potassium phosphate (50 mM), MgCl2 (10 mM), ATP (2 mM, pH 7.2), ATase (27 nM), 2-oxoglutartae (40 mM), GS-AMP (100 nM; n = 11), and PII-UMP, GlnK-UMP, or uridylylated heterotrimers as indicated. Deadenylylation stimulation by untreated PII-UMP (broken line) at 27 nM (open diamond); 6.75 nM (open circle); 1.35 nM (open square); and without PII-UMP (open triangle) are shown. The deadenylylation stimulation by 27 nM of treated mixtures (solid line) of PII-UMP/GlnK-UMP at 1:1 ratio (closed triangle) is presented. Based on proportions of homotrimer PII-UMP in the treated mixtures (see Table 2), it appears that at least the PII-UMP in heterotrimers are active in deadenylylation. Twenty-seven nanomolar purified untreated GlnK-UMP (asterisk connected by solid line) is almost inactive in the deadenylylation assay.
Next, the in vitro deadenylylation assay was used to examine the ability of pure PII-UMP/GlnK-UMP mixtures (1:1, 1:2, and 2:1) that had been treated with the denaturation-renaturation protocol, to stimulate deadenylylation activity of ATase (Fig. 5). When 27 nM 1:1 treated mixture (i.e., 13 nM of each homotrimer) was examined in the deadenylylation assay, the initial rate of γ-GH formation and the extent of deadenylylation was close to that for 13 nM (i.e., 50% of the amount in the standard assay) untreated PII-UMP (data not shown). Table 2 shows that, in this mixture, only around 13% (less than 4 nM) of the species is (PII-UMP)3, so that the observed stimulation of deadenylylation must reflect some contribution from the heterotrimers. Similarly, 27 nM 1:2 mixture showed a decrease in the initial deadenylylation rate as compared with 27 nM (PII-UMP)3 (data not shown), but the rate was still higher than that observed with 3.4 nM untreated (PII-UMP)3, although the amount of (PII-UMP)3 in the mixture was only 2 nM (8%) (Table2). Likewise, in the 2:1 PII-UMP/GlnK-UMP mixture, the initial rate of γ-GH formation exceeded that observed with untreated 50% PII-UMP, although the proportion of (PII-UMP)3 in the mixture was 25% (data not shown). Importantly, in all three cases, the activities of the heterotrimers were significantly higher than could be accounted for by the amount of PII-UMP trimer still present in the heterotrimer preparation. This demonstrates that the uridylylated heterotrimers do have activity, although less than PII-UMP homotrimers. A mixture of homotrimers PII-UMP and GlnK-UMP (not treated with the denaturation-renaturation protocol) showed similar stimulation of the deadenylylation activity as the untreated PII-UMP homotrimer on its own (data not shown). This result suggests that GlnK-UMP has no or little influence on the deadenylylation rate when it is not incorporated into heterotrimers, at least not under the conditions of the present in vitro assay in which PII-UMP is highly active.
Heterotrimers containing PII-UMP and non-uridylylated GlnK (Table 2) exhibited γ-GH formation rates that suggested that only the homotrimeric (PII-UMP)3 in this mixture was active in the deadenylylation reaction (data not shown). This suggests that heterotrimeric GlnK inhibited PII-UMP to a greater extent than did heterotrimeric GlnK-UMP when deadenylylation activities of heterotrimers of PII-UMP/GlnK-UMP were compared with those of PII-UMP/GlnK heterotrimers.
The PII/GlnK heterotrimers were similarly examined for their ability to stimulate adenylylation (inactivation) of GS. Here again, the results indicated that the treated mixtures stimulated adenylylation to a greater extent than that which can be accounted by the proportion of homotrimeric PII (data not shown). A PII monomer inserted in a heterotrimer with GlnK was more active in adenylylating GS than homotrimeric GlnK (data not shown), which was at least 40-fold less active than PII (4, 11).
Discussion
The results presented in this study show that the simultaneous in vivo expression of glnB and glnK genes located on the haploid chromosome of E. coli results in the synthesis of two molecular species of heterotrimers as well as homotrimers of PII and GlnK. That the species on native gels other than homotrimers indeed correspond to heterotrimers was confirmed biochemically by mixing purified E. coli PII and GlnK under denaturation-renaturation conditions. In vitro, homotrimeric PII-UMP stimulated deadenylylation (activation) of GS-AMP whereas homotrimeric GlnK-UMP did not. We demonstrated that uridylylated heterotrimers retained the ability to stimulate deadenylylation of GS-AMP, although to a smaller extent than did homotrimeric PII-UMP. This observation suggests, in agreement with the in vivo data (4, 20), that at least one important function of the mixed oligomers is that of down-tuning the activity of PII-UMP in the deadenylylation reaction (Fig. 6). Our present demonstration that the signal transduction proteins PII and GlnK can form heterotrimers in vivo and that heterotrimers have different levels of activity in vitro is intriguing because it has not previously been seen in bacteria.
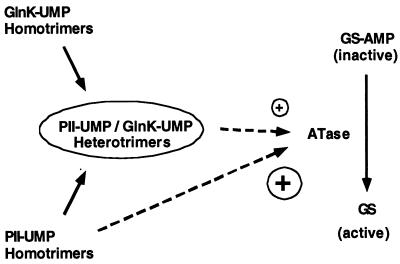
Figure 6.
The role of uridylylated PII and GlnK in stimulation of deadenylylation activity of ATase. Homotrimeric PII-UMP is a potent stimulant (signified by large plus) of the ATases deadenylylation activity whereas under the same in vitro conditions homotrimeric GlnK-UMP is almost completely inactive even when present in two-fold excess together with homotrimeric PII-UMP. The PII-UMP/GlnK-UMP heterotrimers appear to be able to stimulate the deadenylylation activity to lower extent (signified by small plus sign).
Mixed oligomers of PII and GlnK can down-tune the activity of PII in stimulating the adenylylation (inactivation) of GS (data not shown). Thus, fine-tuning the activity of PII by hetero-oligomers may even extend to control of transcription by NRII. However, the level of fine-tuning by mixed oligomers may be different for the various reactions. The upper and lower limits of regulation are set by homotrimeric PII (UMP) and GlnK (UMP), respectively. In this regard, our observation that the ratio of PII, GlnK, and the two heterotrimers varied according to the nitrogen status of the cell is important. Indeed, cells adapted to grow in a minimal medium with arginine (a poorer nitrogen source than glutamine) had hardly any detectable (PII-UMP)3 whereas the other three species, i.e., (GlnK-UMP)3, PII-UMP/(GlnK-UMP)2, and (PII-UMP)2/GlnK-UMP, could be detected (Fig. 3B).
Although we cannot assess whether GlnK-UMP in the heterotrimer has some activity, the results can be understood only if the PII-UMP monomers within these heterotrimers are active. Recently it has been demonstrated in vitro that a heterotrimer containing one wild-type E. coli PII monomer and two inactive PII mutant monomers with deleted T-loops was sufficient for productive interaction with its receptor (27). That study supports our hypothesis that heterotrimers of active PII-UMP and inactive GlnK-UMP may still function when bound to ATase. However, heterotrimer formation between plasmid encoded PII from the evolutionarily distinct Synechococcus (Sc PII) and E. coli PII or GlnK resulted in the formation of inactive complexes (30). The corollary in this case is our observation that PII-UMP appears to be inactive in a heterotrimer consisting of (unmodified) GlnK (data not shown).
What might be the function of the heterotrimerization? First, it might be more difficult to prevent heterotrimerization than to cause it, in view of the close similarity of PII and GlnK. Second, the heterotrimerization inhibition mechanism can provide a fine tuning control like a rheostat as opposed to the on/off switch mechanism as in competition between homotrimers. A GlnK (UMP) homotrimer with low affinity for ATase would not be able to compete with PII (UMP) homotrimers away from the ATase. Likewise, if homotrimeric GlnK-UMP had a much stronger affinity for ATase than PII-UMP, then the latter would be readily replaced by (inactive) GlnK-UMP, thereby compromising the regulatability of the GS cascade. Although the present data support a role for heterotrimers in fine regulation, we are currently trying to separate the various heterotrimers from each other and from homotrimers. This should allow quantitative assessment of the ability of each heterotrimer to stimulate the deadenylylation reaction.
Acknowledgments
We thank Sydney Kustu for valuable criticisms of the manuscript. This work was supported by the Australian Research Council (ARC) and the Netherlands Organization for Scientific Research (NWO). W.C.v.H. was supported by a joint-grant of the ARC and NWO.
Abbreviations
- GS
glutamine synthetase
- UTase
uridylyltransferase
- ATase
adenylyltransferase
- γ-GH
γ-glutamyl hydroxymate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Reitzer L J. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Low K B, Magasanik B, Reznikoff W, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 391–407. [Google Scholar]
- 2.Rhee S G, Chock P B, Stadtman E R. Adv Enzymol Relat Areas Mol Biol. 1989;62:37–92. doi: 10.1002/9780470123089.ch2. [DOI] [PubMed] [Google Scholar]
- 3.van Heeswijk W C, Stegeman B, Hoving S, Molenaar D, Kahn D, Westerhoff H V. FEMS Microbiol Lett. 1995;132:153–157. doi: 10.1111/j.1574-6968.1995.tb07825.x. [DOI] [PubMed] [Google Scholar]
- 4.van Heeswijk W C, Hoving S, Molenaar D, Stegeman B, Kahn D, Westerhoff H V. Mol Microbiol. 1996;21:133–146. doi: 10.1046/j.1365-2958.1996.6281349.x. [DOI] [PubMed] [Google Scholar]
- 5.Cheah E, Carr P D, Suffolk P M, Vasudevan S G, Dixon N E, Ollis D L. Structure (London) 1994;2:981–990. doi: 10.1016/s0969-2126(94)00100-6. [DOI] [PubMed] [Google Scholar]
- 6.Carr P D, Cheah E, Suffolk P M, Vasudevan S G, Dixon N E, Ollis D L. Acta Crystallogr D. 1996;52:93–104. doi: 10.1107/S0907444995007293. [DOI] [PubMed] [Google Scholar]
- 7.Xu Y, Cheah E, Carr P D, van Heeswijk W C, Westerhoff H V, Vasudevan S G, Ollis D L. J Mol Biol. 1998;282:149–165. doi: 10.1006/jmbi.1998.1979. [DOI] [PubMed] [Google Scholar]
- 8.Macpherson K H R, Xu Y, Cheah E, Carr P D, van Heeswijk W C, Westerhoff H V, Luque E, Vasudevan S G, Ollis D L. Acta Crystallogr D. 1998;54:996–998. doi: 10.1107/s0907444998001887. [DOI] [PubMed] [Google Scholar]
- 9.Jaggi R, Ybarlucea W, Cheah E, Carr P D, Edwards K J, Ollis D L, Vasudevan S G. FEBS Lett. 1996;391:223–228. doi: 10.1016/0014-5793(96)00737-5. [DOI] [PubMed] [Google Scholar]
- 10.Jiang P, Zucker P, Atkinson M R, Kamberov E S, Tirasophon W, Chandran P, Schefke B R, Ninfa A J. J Bacteriol. 1997;179:4342–4353. doi: 10.1128/jb.179.13.4342-4353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atkinson M R, Ninfa A J. Mol Microbiol. 1999;32:301–313. doi: 10.1046/j.1365-2958.1999.01349.x. [DOI] [PubMed] [Google Scholar]
- 12.Jaggi R. Ph.D. thesis. Townsville, Australia: James Cook Univ.; 1998. [Google Scholar]
- 13.Kamberov E S, Atkinson M R, Ninfa A J. J Biol Chem. 1995;270:17797–17807. doi: 10.1074/jbc.270.30.17797. [DOI] [PubMed] [Google Scholar]
- 14.van Heeswijk W C, Rabenberg M, Westerhoff H V, Kahn D. Mol Microbiol. 1993;9:443–457. doi: 10.1111/j.1365-2958.1993.tb01706.x. [DOI] [PubMed] [Google Scholar]
- 15.Atkinson M R, Ninfa A J. Mol Microbiol. 1998;29:431–447. doi: 10.1046/j.1365-2958.1998.00932.x. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y M, Backman K, Magasanik B. J Bacteriol. 1982;150:214–220. doi: 10.1128/jb.150.1.214-220.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bueno R, Pahel G, Magasanik B. J Bacteriol. 1985;164:816–822. doi: 10.1128/jb.164.2.816-822.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arcondéguy T, van Heeswijk W C, Merrick M. FEMS Microbiol Lett. 1999;180:263–270. doi: 10.1111/j.1574-6968.1999.tb08805.x. [DOI] [PubMed] [Google Scholar]
- 19.Love C A, Lilley P E, Dixon N E. Gene. 1996;176:49–53. doi: 10.1016/0378-1119(96)00208-9. [DOI] [PubMed] [Google Scholar]
- 20.van Heeswijk W C. Ph.D. thesis. Amsterdam: Univ. of Amsterdam; 1998. [Google Scholar]
- 21.Neidhardt F C, Bloch P L, Smith D F. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lutz R, Bujard H. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang P, Peliska J A, Ninfa A J. Biochemistry. 1998;37:12782–12794. doi: 10.1021/bi980667m. [DOI] [PubMed] [Google Scholar]
- 24.Forchhammer K, Tandeau de Marsac N. J Bacteriol. 1994;176:84–91. doi: 10.1128/jb.176.1.84-91.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vasudevan S G, Gedye C, Dixon N E, Cheah C, Carr P D, Suffolk P M, Ollis D L. FEBS Letts. 1994;337:255–258. doi: 10.1016/0014-5793(94)80203-3. [DOI] [PubMed] [Google Scholar]
- 26.Jaggi R, van Heeswijk W C, Westerhoff H V, Ollis D L, Vasudevan S G. EMBO J. 1997;16:5562–5571. doi: 10.1093/emboj/16.18.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang P, Zucker P, Ninfa A J. J Bacteriol. 1997;179:4354–4360. doi: 10.1128/jb.179.13.4354-4360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stadtman E R, Smyrniotis P Z, Davis J N, Wittenberger M E. Anal Biochem. 1979;95:275–285. doi: 10.1016/0003-2697(79)90217-3. [DOI] [PubMed] [Google Scholar]
- 29.Jiang P, Peliska J A, Ninfa A J. Biochemistry. 1998;37:12802–12810. doi: 10.1021/bi980666u. [DOI] [PubMed] [Google Scholar]
- 30.Forchammer K, Hedler A, Strobel H, Weiss V. Mol Microbiol. 1999;33:338–349. doi: 10.1046/j.1365-2958.1999.01477.x. [DOI] [PubMed] [Google Scholar]