Abstract
Microarray hybridization was used to assess acclimation responses to four UV regimes by near isogenic maize (Zea mays) lines varying in flavonoid content. We found that 355 of the 2,500 cDNAs tested were regulated by UV radiation in at least one genotype. Among these, 232 transcripts are assigned putative functions, whereas 123 encode unknown proteins. UV-B increased expression of stress response and ribosomal protein genes, whereas photosynthesis-associated genes were down-regulated; lines lacking UV-absorbing pigments had more dramatic responses than did lines with these pigments, confirming the shielding role of these compounds. Sunlight filtered to remove UV-B or UV-B plus UV-A resulted in significant expression changes in many genes not previously associated with UV responses. Some pathways regulated by UV radiation are shared with defense, salt, and oxidative stresses; however, UV-B radiation can activate additional pathways not shared with other stresses.
UV radiation is divided into three classes: UV-C, UV-B, and UV-A. Highly energetic UV-C (wavelengths ≤280 nm) is strongly absorbed by oxygen and ozone in the stratosphere such that none of this sterilizing radiation is present in terrestrial sunlight. Potentially harmful UV-B (280–315 nm) is strongly absorbed by atmospheric ozone, but approximately 4% of terrestrial radiation is UV-B from 290 to 315 nm. UV-A (315–400 nm) is not attenuated by atmospheric ozone, and this less damaging radiation is an important photomorphogenic signal in plant development (Bjorn, 1994). Chlorofluorocarbons and other pollutants have catalyzed depletion of the stratospheric ozone layer; consequently, terrestrial levels of UV-B are increasing with potentially deleterious consequences for all living organisms, particularly for plant development and physiology (Ballaré et al., 2001; Searles et al., 2001; Paul and Gwynn-Jones, 2003).
UV-B radiation induces diverse morphological and physiological responses in plants (for reviews, see Ballaré et al., 2001; Searles et al., 2001; Paul and Gwynn-Jones, 2003), but the underlying mechanisms governing these integrated responses are unknown. Concurrent with exposure, UV-B photons cause cellular damage by generating photoproducts in DNA (Britt, 1996) and direct damage to proteins (Gerhardt et al., 1999). In response to the inevitable exposure to damaging UV-B radiation, plants have evolved UV-induced mechanisms of protection and repair, such as accumulation of UV-absorbing pigments (Stapleton and Walbot, 1994; Mazza et al., 2000; Bieza and Lois, 2001) and use of UV-A photons to repair most UV-B-induced DNA damage (Britt, 1996). Because of its absorption spectrum, DNA is a major target of UV-B damage; even low doses of radiation can kill mutants lacking specific DNA repair pathways (Britt et al., 1993; Britt, 1996; Landry et al., 1997). In flowering plants, flavonoids, including anthocyanins, accumulate in the vacuoles of epidermal cells where they attenuate the UV component of sunlight with minimal absorption of photosynthetically active radiation (Stapleton and Walbot, 1994; Landry et al., 1995). In particular, the role of anthocyanins in UV protection has been investigated (Gould et al., 2002), these authors have reported that purified anthocyanin extracts show strong antioxidant properties in vitro, and they can also scavenge reactive oxygen in living cells. By real-time imaging of H2O2 in cells after mechanical injury, they found that anthocyanins, among various flavonoids, were the only molecules suitably located to account for the enhanced rates of H2O2 scavenging, suggesting that anthocyanins have elevated antioxidant capabilities in vivo (Gould et al., 2002). Therefore, the mechanism by which anthocyanins confer UV protection may involve UV absorption or scavenging of reactive oxygen species (ROS), or both. In Arabidopsis, sinapate esters also provide UV-B attenuation (Sheahan, 1996), but this biosynthetic pathway is not present in corn (Zea mays). In addition, cuticular waxes and lignins may also serve protective roles by absorbing UV radiation (Caldwell et al., 1983).
UV-B also stimulates production of ROS (Arnots and Murphy, 1991; Dai et al., 1997) and antioxidant defenses (Rozema et al., 1997; Jansen et al., 1998). It has been proposed that ROS not only function as destructive radicals, but also as signaling molecules during UV-B responses (Green and Fluhr, 1995; Mackerness et al., 1997, 2001; Mackerness and Jordan, 1999). In mammalian cells, ligand-independent activation of some membrane receptors is caused by oxidation of receptor-directed protein-Tyr phosphatases (Gross et al., 1999). Because no receptor Tyr kinase is known in plants, the corresponding phosphatases are also unlikely to exist in plants. However, parallel mechanisms may apply to other plant kinases. Receptor activation by UV was suggested for the systemin receptor in wild tomato (Lycopersicon peruvianum; Yalamanchili and Stratmann, 2002). Signaling cascades elicited by the initial products of DNA damage or by UV-B activation of receptors most likely govern the diverse physiological and developmental responses caused by exposure to this radiation. For example, in green tissues, UV-B exposure decreases the expression of photosynthetic proteins such as Rubisco, the D1 protein (psbA) of photosystem II, and the chlorophyll a/b binding protein (Lhcb; Jordan, 1996; Mackerness et al., 1997). Some UV-activated signaling components have been identified (Mackerness and Jordan, 1999; Stratmann et al., 2000; Mackerness et al., 2001; Miles et al., 2002); however, the actual signal transduction pathways activated by UV-B radiation are not yet well defined.
Microarray technology has rapidly become an important tool for the simultaneous measurement of thousands of gene expression patterns after a change in exogenous conditions (for review, see Schaffer et al., 2000). In Arabidopsis, microarrays have been used to identify genes involved in responses to drought and cold (Seki et al., 2001), disease (Schenk et al., 2000) and systemic acquired resistance (Maleck et al., 2000), wounding and insect feeding (Reymond et al., 2000), oxidative stress (Desikan et al., 2001), and iron deficiency (Thimm et al., 2001). Salt stress responses have also been documented in rice (Oryza sativa; Kawasaki et al., 2001). A small-scale study of Arabidopsis response to UV-B was recently published (Brosche et al., 2002). Here, transcripts from plants after a 3-h UV-B exposure were compared with transcripts in control plants without UV-B on arrays with 5,000 genes. With the goal to better understand the processes involved in UV acclimation responses in plants, we have investigated gene expression in leaves of corn grown in UV exclusion and supplementation using microarrays containing approximately 2,500 maize cDNA clones. Because most UV photobiology studies in plants have been carried out in controlled environments using irradiation protocols of doubtful ecological significance, we accomplished UV exclusion experiments of natural sunlight in the field. Thus, one of our purposes was to compare transcripts affected by natural UV levels with those after UV-B irradiation in the greenhouse using UV lamps and to determine the degree of overlap in these responses. Moreover, we investigated differences in gene expression after exclusion of natural sunlight UV-B and UV-A+B radiation. The second aim of this work was to evaluate how UV-absorbing pigments modulate plant response to UV-B radiation. For this reason, we used four near-isogenic maize lines that differ in flavonoid and anthocyanin sunscreen compounds. Temporal and spatial patterns of flavonoid production are specified by the requirement for two transcription factors that act together to activate genes encoding biosynthetic enzymes. In maize leaves, the B and Pl genes encode for a basic helix-loop-helix and myb-domain transcription factors, respectively, that regulate the expression of flavonoid compounds (Winkel-Shirley, 2001). The maize inbred W23 with the functional B and Pl regulatory alleles has high levels of flavonoids; the near-isogenic b, pl W23 line is deficient in flavonoid accumulation (Dooner, 1983). The bronze2 (bz2), weak B, pl W23 line accumulates low levels of flavonoids including some anthocyanin. However, pigment is oxidized to a brown compound in the cytoplasm of bz2 mutants (Alfenito et al., 1998). A second bz2, weak B, pl line was also examined to analyze the interaction between radiation and pre-existing DNA damage. This W23 Mutator line has numerous MuDR/Mu transposons and as a consequence is under constant “genotoxic stress” from the DNA breakage associated with transposition reactions. Our final aim was to identify new pathways affected by UV radiation. Transcriptome profiling permitted identification of novel genes regulated by UV radiation, and an advantage of using large-scale methods is that coordinate regulation of genes acting in the same or different pathways can be analyzed.
RESULTS
UV-B Treatments of Maize
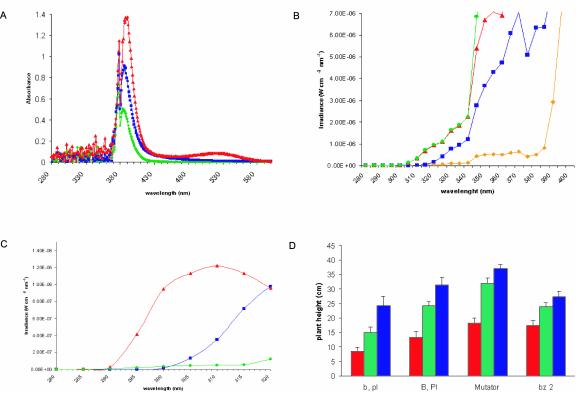
In our experiments, we used four near-isogenic maize lines that differ in flavonoid sunscreen compounds. Figure 1A shows that adult leaves of B, Pl plants accumulate high levels of purple anthocyanins (the peak at 530 nm corresponds to these pigments). Anthocyanins are localized in the vacuole and in maize are a mixture of at least five different compounds with partially overlapping and broad UV absorption properties (Harborne and Grayer, 1988). As shown in Figure 1A, bz2 plants contained more pigments than the green b, pl plants but only 30% the level of UV-absorbing pigments as the purple B, Pl genotype. If flavonoid sunscreens modulate UV-B responses, then we would expect the bz2 lines to be intermediate in response between the purple and green lines.
Figure 1.
A, Absorption spectra of extracted flavonoids in acidic methanol. Extract from B, Pl plants, ▴ in red; extract from Mutator plants, ▪ in blue; extract from bz2 plants, ♦ in green. The b, pl green plants were used as the blank, to normalize spectra among the samples. B, UV transmittance spectra of the filters and comparison with the solar spectrum. UV solar spectrum, • in green; CA filter (full sunlight), ▴ in red; PE filter (no UV-B), ▪ in blue; LE filter (no UV), ♦ in yellow. C, Irradiance of the supplementary UV-B lamps and comparison with the UV-B solar spectrum and with the UV-B spectrum of the UV-B lamps covered with PE. The supplementary lamps were used in a greenhouse, which has about 20% of the summer solar irradiation. Solar spectrum, ♦ in blue; UV-B lamps, ▴ in red; UV-B lamps covered with PE (no UV-B), • in green. D, Plant height after the different UV treatments. The values are the average of plants from the different plots after 3 weeks under the CA filter (six plots, green), PE filter (six plots, blue), or LE filter (four plots, red). Error bars are ses. There is a statistically significant difference between the values of plants under full UV-B compared with values under no UV-B or no UV-A+B for all of the lines, independently of the line used (two-way factorial ANOVA test, significance level P = 0.05; PT (effect of the treatment) < 0.05, PG (effect of the genotype) > 0.50; PT × G > 0.25).
To analyze steady-state transcript levels in response to UV exclusion, four near-isogenic maize lines that differ in flavonoid sunscreen compounds were grown in the field for 3 weeks under plastics designed to transmit full sunlight UV (cellulose acetate [CA]), no UV-B (polyester [PE]), or neither UV-A nor UV-B (lexan [LE]; exclusion experiments; Fig. 1B; see “Materials and Methods”). To study acclimation responses to UV-B, we also compared transcript levels in plants that were grown under full sunlight for 3 weeks with those in plants that were grown 3 weeks without UV-B and then exposed to natural UV-B levels in sunlight for 1 d (restoration experiments). Field experiments were done twice during the summer of 2001 outdoors at the Stanford Plant Growth Facility, using several plots for each treatment and genotype (six plots for CA and PE covered plants and four plots for LE covered plants) to control spatial variations, such as soil fertility and moisture. A diagram of the experimental layout is presented as supplementary material (Supplementary Fig. 1; they can be viewed at http://www.plantphysiol.org). Additionally, to study effects of UV-B at higher levels than present in field sunlight and to compare UV-B responses in controlled versus natural environments, we tested the effect of supplementary UV-B radiation with an intensity about 10-fold higher than is present in sunlight by giving greenhouse-grown plants an 8-h treatment (supplementation experiments, Fig. 1C). After all treatments, plants looked healthy, independent of the intensity of UV-B they received. The only noticeable phenotype was that plants grown without UV-B were taller than the sunlight control, whereas plants grown without any UV were shorter than the sunlight control (Fig. 1D). Height measurements in Figure 1D are the average from the different plots after 3 weeks under filters; the UV effects were statistically significant (P = 0.05), and were similar in all four lines (P > 0.25 for the UV × line interaction term for all lines). A decrease in height after total UV exclusion has not been reported for maize or any other plant; this observation could be a particular response of the W23 inbred or a more general morphogenic effect of UV radiation in certain conditions and treatments. The basis of this observation remains to be investigated more thoroughly in future studies.
Microarray Hybridization Reliability and Experimental Design
mRNA extractions were done using at least six leaves from each radiation treatment and genotype and from different plots to produce samples for microarray hybridizations. The array slides contained 2,500 maize cDNAs printed in triplicate spots; a triplicate set for nearly all cDNAs was printed in at least one additional location on each slide, so there is a minimum of six spots for each cDNA per slide (for more information, see http://gremlin3.zool.iastate.edu/zmdb/microarray/). The transcript levels for each cDNA were calculated as an average of the signal intensity of each spot within the same and duplicate slides.
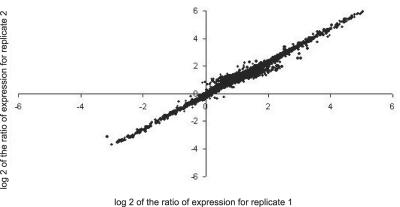
For the comparisons of greatest interest, direct pair wise hybridizations were done. In these cases, dye swap comparisons were also completed with different biological samples using mRNA from different extractions and experiments as described in “Materials and Methods.” Figure 2 shows an example of a dye-swapping experiment. mRNA from b, pl plants grown under sunlight and b, pl plants without UV-B were labeled separately with Cy3-dUTP or Cy5-dUTP. A Cy3-labeled mRNA from b, pl sunlight plants and Cy5-labeled mRNA b, pl no UV-B plants in one experiment were used for hybridization of a microarray slide. Reciprocally labeled samples from a second experiment (biological replicate) were used for hybridization to another slide. The comparison of signal ratios between the two channels produced after this dye-swapping protocol generated highly reproducible results (Fig. 2). The ratios of the signal intensities for each fluorescence channel for all of the cDNAs were linearly correlated with a correlation coefficient of 0.976. The same dye-swapping protocol was performed for all of the pair wise comparisons, using the b, pl sunlight treatment as the reference (Supplementary Fig. 2). In all cases, the correlation coefficients of the ratios were >0.95. These dye-swapping experiments also provided a repetition of each comparison, and the data were averaged and used for comparisons. Therefore, calculation of gene induction and repression was reproducible within the window of resolution of the microarray hybridization method independent of the dye used for sample labeling. For other comparisons, loop designs were used (Churchill, 2002; Yang and Speed, 2002). In these comparisons, samples were compared one to another in a daisy-chain fashion. These designs, combining loops with reference designs improved efficiency and robustness of the analysis by creating multiple links among samples (Churchill, 2002; Yang and Speed, 2002). Evaluations between samples not compared directly are computationally derived (Churchill, 2002).
Figure 2.
Scatter plot comparing ratios of signal values from two replicates on microarray hybridizations with mRNA from leaves of b, pl plants under full sunlight and under no UV-B labeled with Cy3-dUTP and Cy5-dUTP. Data from images of dye-swapping experiments were plotted as the mean intensity after normalization of expressed sequence tags (ESTs) spotted in triplicate. Additional examples are provided in Supplementary Figure 2.
Effects of UV-B Exclusion or Supplementation on Gene Expression in Different Maize Lines: Flavonoids and Anthocyanins Act as Sunscreens Mimicking the Effect of UV-B-Absorbing Plastics
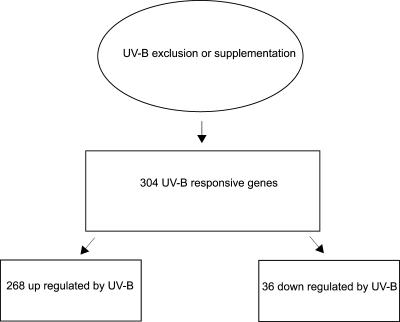
As described in the introduction, the role of flavonoids in UV protection can be vital. In the case of anthocyanins, this protection could be mediated both by UV absorption and via scavenging of ROS (Gould et al., 2002). To compare UV-B effects in maize lines with very low and varying contents of UV-absorbing pigments, RNA samples extracted from the purple (B, Pl genotype), bronze (bz2 genotypes), and green (b, pl genotype) plants after the diverse radiation treatments were analyzed by microarray experiments. For each pair wise comparison, the ratios of the fluorescence intensities of the two probes were calculated, and the number of clones showing a ratio difference greater than 2-fold was determined. This cutoff value was chosen so as to exclude the possibility of a gene being inappropriately designated as UV responsive as a result of a technical error. At this criterion, after analysis of exclusion and supplementation experiments for the four genotypes and restoration of solar UV-B for green b, pl and purple B, Pl plants, we identified 304 genes that were responsive to UV-B radiation, corresponding to 12% of the total probe sets (Fig. 3). Analysis of these data indicated that 268 gene transcripts had a significant increase in abundance after UV-B exposure in at least one of the treatments or genotypes. In contrast, only 36 genes were down-regulated (see Supplementary Tables I–III for complete listings of genes with putative known functions responding to specific treatments in each genotype). Many of these genes have not previously been shown to be regulated by UV treatment, and many of the genes in these groups have no assigned function; therefore microarrays are a powerful tool for gene discovery.
Figure 3.
Summary of the number of ESTs responsive to UV-B exclusion or supplementation.
The first goal of our study was to study maize leaf responses to natural UV levels. In addition, we studied leaf responses after UV-B irradiation in the greenhouse using UV lamps. Even if these treatments are not symmetrical because the UV-B exposure used in the greenhouse was of high irradiance and during a short period of time, we determined the degree of overlap in these different types of responses. As shown in Figure 4A, many more genes were upregulated by UV-B in the green (b, pl) plants compared with the purple (B, Pl) plants. This was true after exclusion of ambient solar UV-B (175 genes compared with 17) and after UV-B supplementation experiments in the greenhouse (122 genes affected in b, pl compared with 22 in B, Pl plants). The same pattern was observed for down-regulated ESTs. We found that even if there is some degree of overlap between treatments, many genes are regulated differentially either by exclusion of natural UV-B in the field or by UV-B supplementation in the greenhouse (Fig. 4). In this way, as shown in the intersections of the Venn diagrams in Figure 4A, 49 ESTs were upregulated and 17 were down-regulated by these two UV-B conditions in b, pl plants. Only in exclusion experiments, 126 were up-regulated and 14 were down-regulated by UV-B; and 72 were up-regulated and five were down-regulated only after UV-B supplementation in the greenhouse. For the B, Pl genotype, six of the up-regulated ESTs showed increased levels after both treatments, whereas five were down-regulated by both treatments. These results suggest that deviation from ambient conditions in sunlight is a significant variable for plants rather than simply a response to dosage per se.
Figure 4.
Venn diagrams of comparisons between UV-B-responsive genes in lines with different levels of flavonoids and anthocyanins. A, Venn diagrams of comparisons between UV-B-responsive genes in b, pl (green) and B, Pl (purple) plants in UV-B exclusion experiments for 3 weeks in the field (Exclusion, SL UV-B/no SL UV-B); 1 d after UV-B restoration in plants grown for 3 weeks without UV-B in the field (Restoration, 1 d SL UV-B after 3 weeks without UV-B/SL UV-B); and after 10-fold UV-B supplementation for 8 h in the greenhouse (Supplementation, 10-fold UV-B for 8 h in the greenhouse/no UV-B in the greenhouse). B, Venn diagrams of comparisons between UV-B-responsive genes in bz2 (beige) and Mutator (pink) plants in UV-B exclusion experiments for 3 weeks in the field (Exclusion, SL UV-B/no SL UV-B); and after 10-fold UV-B supplementation for 8 h in the greenhouse (Supplementation, 10-fold UV-B for 8 h in the greenhouse/no UV-B in the greenhouse). Up-regulated genes are in red; down-regulated genes are in green. Sets of genes were selected using the criteria described in “Materials and Methods.”
Restoration of ambient solar UV-B after 3 weeks of exclusion showed that a subset of UV-B responses is readily reversible. Among the genes with altered expression after solar UV-B exclusion in green b, pl plants, 98.6% showed similar expression to the reference (continuous sunlight b, pl sample) after 1 d of ambient UV-B in sunlight (Fig. 4A). Consequently, for these genes, there is a rapid return to the expression level conditioned by solar UV-B. On the contrary, for purple B, Pl plants, the same was observed for only 39% of the ESTs. In 61% of the cases, gene expression after 1 d in solar UV-B was different from the reference (Fig. 4A). The B, Pl plants, after being depleted of solar UV-B for 3 weeks, have lower levels of UV-B inducible flavonoids and anthocyanins than in B, Pl plants grown under full UV-B sunlight (data not shown). From the perspective of gene expression, B, Pl plants in a depleted UV-B environment are hypersensitive to solar UV-B; exposure to UV-B elicits a “shock” response that requires adjustment to the new environmental conditions.
As shown in Figure 4B, UV-B exclusion and supplementation experiments were also done with bz2 weak B, pl genotypes, with intermediate levels of flavonoids. Again, we found that some ESTs are regulated by UV-B in both treatments, whereas many are only affected by one of the treatments. In addition, we found that even modest levels of flavonoid screening as found in the bz2 weak B, pl genotype suffices to moderate plant responses. Figure 4B shows that the number of ESTs changed after experiments of UV-B exclusion and supplementation in these genotypes is intermediate between the number of ESTs changed in b, pl compared with B, Pl plants. In all genotypes, a much smaller number of genes were down-regulated by UV-B exposure compared with the number up-regulated (Table I). Collectively, these observations confirm the protective role of flavonoids and anthocyanins as natural sunscreens in plants: Plants with lower levels of these compounds are more responsive to UV-B, in this case showing greater changes in transcript levels for some genes than plants with higher levels of flavonoids.
Table I.
Summary of significant gene expression changes by UV-B and/or UV-A radiation in b, pl, B, Pl, bz2, and Mutator plants
A description of genes with putative function showing a significant change (>2-fold) is provided in supplementary Tables I, II and III.
| Response
|
Affected Genes
|
Process (No. of genes)
|
Example of Genes
|
Detected by
|
||||
|---|---|---|---|---|---|---|---|---|
| Total | Known | Unknown | Exclusion | Restoration | Supplementation | |||
| Decrease by UV-B | 36 | 17 | 19 | Photosynthesis (7) | Rubisco small and large subunits | X | — | X |
| Detoxification (3) | Multidrug resistance associated proteins | X | — | — | ||||
| Increase by UV-B | 268 | 187 | 81 | Protein synthesis (41) | Ribosomal proteins, translation factors | X | X | X |
| Cytoskeleton (6) | α-Tubulins, actin | X | X | X | ||||
| Protein degradation (12) | Ubiquitin, proteosome subunits | X | X | X | ||||
| DNA metabolism (12) | Histones, DNA-binding proteins | X | X | X | ||||
| Stress response and antioxidant responses (31) | Chaperons, thioredoxins, detoxifying enzymes, Cys proteinase | X | X | X | ||||
| Signal transduction and transcription factors (19) | Receptor protein kinases, GTP-binding proteins, Ca2+-binding proteins, myb-related transcription factor | X | X | X | ||||
| Non-photosynthetic energy metabolism (25) | Respiration, glycolisis, lipid biosynthesis, S-adenosyl-Met metabolism | X | X | X | ||||
| Cell wall biosynthesis (5) | Xyloglucan endotransglycosylase, chorismate mutase | X | — | X | ||||
| Decrease by UV-A + B | 25 | 14 | 11 | Various (16) | Protein phosphatases, ubiquitin | |||
| Increase by UV-A + B | 26 | 14 | 12 | Photosynthesis (4) | Rubisco small and large subunits | |||
| Flavonoid/ anthocyanin (3) | Chalcone synthase, Bz1 | |||||||
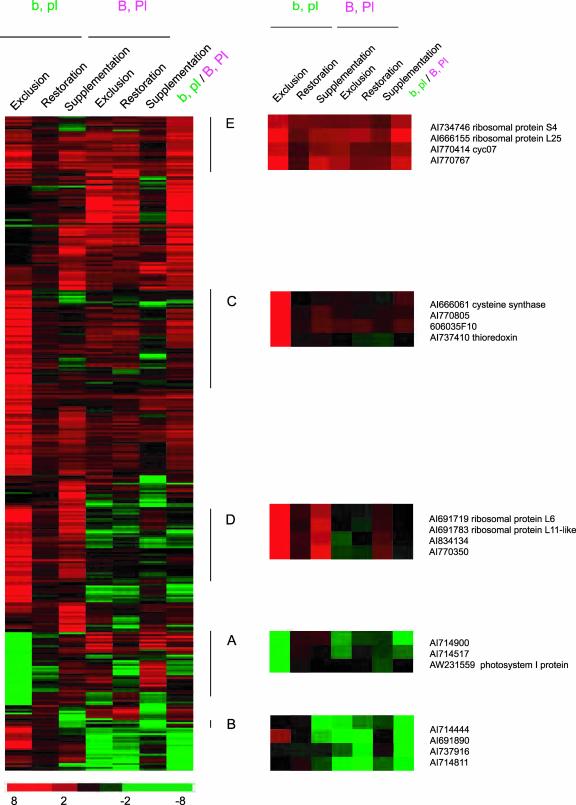
To further evaluate how UV-absorbing pigments can modulate plant responses to UV-B radiation, additional analyses were done with b, pl and B, Pl genotypes under different UV-B conditions as presented in Figure 5. Hierarchical clustering of the UV-B-regulated genes in purple B, Pl and green b, pl plants reveals that blocks of genes were regulated in several patterns. For example, ESTs in group a are down-regulated by solar UV-B only in b, pl plants (exclusion column, b, pl plants). This down-regulation seems to reflect a long-term effect on the plants, because supplementary UV-B for only 8 h does not change the levels of these transcripts. Moreover, these transcripts were unchanged in B, Pl plants, confirming again the filtering role of flavonoids and anthocyanins. Furthermore, when comparing transcript levels of these genes in purple and green plants in the sun (last column, b,pl/B,Pl), we found that some show lower levels in b, pl plants than in B, Pl plants. This demonstrates that endogenous flavonoid sunscreens parallel the effect of the plastic filter absorbing UV-B radiation. Examples of ESTs in this group are shown on the right of Figure 5, including transcripts for photosynthetic proteins such as a photosystem I protein and granule-bound starch synthase.
Figure 5.
Cluster analysis of transcripts from b, pl (green) and B, Pl (purple) plants after 3 weeks under UV-B exclusion in the field, 1 d after UV-B restoration in plants grown for 3 weeks without UV-B in the field, and after 10-fold UV-B supplementation for 8 h in the greenhouse. Clustering was performed according to Eisen et al. (1998). The color saturation reflects the magnitude of the log2 expression ratio for each transcript. Transcripts are grouped into patterns A to E accordingto their expression profiles. b, pl (in green), Low flavonoid plants; B, Pl (in purple), high flavonoid and anthocyanin plants. Exclusion, Plants grown under full UV-B sunlight/plants grown without UV-B in the field; Restoration, plants 1 d after removing the PE filters/plants grown under full sunlight; Supplementation, plants with supplementary UV-B for 8 h in the greenhouse/plants grown without UV-B in the greenhouse. Last column, b, pl/B, Pl under full UV-B sunlight. Red, Higher transcript levels than the reference; green, lower transcript levels than the reference. Gray, Flagged ESTs that had signals similar to the background in some condition and were eliminated during the analysis. The color log2 scale is provided at the bottom of the Figure. For clustering analysis, the same data from microarray experiments shown in Venn diagrams in Figure 4A were used.
Group b depicts genes that are down-regulated by solar UV-B in both green and purple genotypes, and some are also repressed by supplementary UV-B for 8 h or after restoration of sunlight UV-B. This group of genes seems to be affected by both long- and short-term UV-B treatments, and they are down-regulated even in plants with protective UV-B-absorbing compounds. Examples of genes in this cluster are shown in Figure 5 and include the photosynthesis-related Rubisco small and large subunits and a chlorophyll a/b-binding protein.
Transcripts in groups c, d, and e have higher expression in the presence of UV-B. Cluster c shows genes in the b, pl genotype whose normal expression depends on long-term effects of UV-B levels present in sunlight but are not changed after supplementary UV-B; cluster c genes are mostly unaffected in the B, Pl genotype. Transcripts for translation factor 4A2 and a homolog to transcription factor scarecrow-5, as well as a thioredoxin and a glutathione S-transferase, are found in this cluster. Cluster d contains genes up-regulated by UV-B by both long-term effects in the sunlight and the short-term 8-h UV-B supplementation in the greenhouse; again, cluster d changes are only observed in the green b, pl plants. Therefore, these genes are highly responsive to different UV-B treatments but through mechanisms that can be significantly attenuated by sunscreen pigments. Some ribosomal protein transcripts belong to this group, such as S8, L6, and L11.
Finally, cluster e shows that there are genes that respond to UV-B to some extent in all genotypes, independent of flavonoid content and of the UV-B treatment used for the experiments. Again, as shown for group a, when comparing transcript levels of these genes in purple and green plants in the sun (last column, b,pl/B,Pl), we found that some show higher levels in b, pl plants than in B, Pl plants, confirming again that anthocyanin and flavonoids parallel the effect of the plastic filter absorbing this radiation. Transcripts for several ribosomal proteins such as S4, S15, and L25, together with transcripts for histones and chaperones, are examples of genes up-regulated by UV-B in this cluster.
A central aim of this work was to identify new pathways affected by UV radiation and to find novel genes that are regulated by UV-B only in certain conditions, in this case after different UV-B treatments and in different genotypes. Table I summarizes the key findings by the plant processes affected after the different UV-B treatments, based on identification of representative responding genes. Apparently, genes involved in the same function can either be regulated by different mechanisms after UV-B radiation or respond differentially to treatment severity. For example, in b, pl plants, some photosynthesis-related genes are down-regulated by UV-B only in solar UV-B exclusion experiments (for example, a photosystem I protein; Supplementary Table I), whereas others are decreased by 10-fold supplementary UV-B in the greenhouse (such as a photosystem II protein), and some by both treatments (such as Rubisco large subunit). Therefore, the use of different UV-B treatments allowed us to identify transcripts regulated by UV-B in different ways, which would not have been possible using only one UV-B treatment. The description of all of the ESTs with a putative function showing a significant change (greater than 2-fold) in any treatment is provided in Supplementary Tables I through III. We found that some transcripts that are regulated by UV radiation are shared with other stresses, such as thioredoxin in cluster c of Figure 5. More importantly, we found that UV-B radiation can regulate the expression of additional genes that are not affected by other stresses, such as the genes encoding ribosomal proteins. A more extensive description of transcripts regulated by UV-B is discussed later in the paper. To validate our experiments, transcriptome profiling identified UV-regulated genes for proteins involved in pathways already known to be affected by UV-B, such as photosynthesis and antioxidant responses. Moreover, we demonstrated that flavonoids and anthocyanins have a similar shielding effect as the UV-B-absorbing plastics, because as many genes were similarly expressed in purple plants under full sunlight UV-B as in green plants under the plastic.
RNA Gel-Blot Analysis
To determine whether the transcript changes identified by microarray analysis were reliable, total RNA obtained from the same plants used for microarrays experiments was examined by RNA gel-blot analysis (Fig. 6). RNA samples for sunlight plants, UV-B exclusion for 3 weeks, and after 1 d of sunlight UV-B restoration were examined for both the green b, pl and purple B, Pl genotypes. Four genes (elongation factor 1α, ribosomal protein QM, MRP33, and MRP47) were selected as probes, because these genes exhibited altered mRNA levels by microarray analysis. The RNA-blot hybridization results correspond closely in magnitude and UV-B sensitivity to the microarray results for each of these genes (Fig. 6). The reference RNA in all comparisons is from plants that were grown under natural levels of UV-B (listed as 0 change), and log2 ratios were calculated in the same way as was done for microarray experiments (see Fig. 5 legend). For example, both elongation factor 1α (Fig. 5A) and ribosomal protein QM (Fig. 5B) decrease in abundance in the green plants when UV-B is filtered from sunlight as was found by microarray analysis. The purple B, Pl genotype is less responsive to removal or restoration of UV-B as found by microarray hybridization. Similarly, both MRP33 (Fig. 5C) and MRP47 (Fig. 5D) are induced more than 4-fold when UV-B is filtered from sunlight in b, pl plants, similar to the induction measured by microarray analysis.
Figure 6.
RNA gel-blot analysis to confirm microarray data. Lanes contained 10 μg of total RNA extracted from plants after different UV-B treatments. Several identical gels were prepared and blotted. Each blot was hybridized with 32P-labeled elongation factor 1α (A), ribosomal protein QM (B), MRP33 (C), or MRP47 (D) probes. b, pl, Low flavonoid plants; B, Pl, high flavonoid and anthocyanin plants. Exclusion, Plants grown without UV-B in the field; Restoration, plants 1 d after removing the PE filters; Sunlight, plants grown under full sunlight. Figure 4E shows an ethidium bromide-stained gel as a check for equal loading. The log2 ratio was calculated as for microarray experiments by quantification of hybridization signals and ethidium bromide-stained bands using Kodak ds 1D Digital Science, as described in “Materials and Methods,” and is provided at the bottom of each blot, using as a reference RNA from plants that were grown under natural levels of UV-B (listed as 0 change).
bz2 Mutator Plants Show Discrete Responses
As presented in the introduction, one of the maize lines used is a Mutator line with numerous MuDR/Mu transposons, and as a consequence, it is under constant genotoxic stress from the DNA breakage associated with transposition reactions. One of our questions was to determine whether there is any interaction between radiation and pre-existing DNA damage. Figure 7 is a cluster analysis showing gene expression differences in leaves of the active Mutator bz2-mu2, standard bz2, and purple B, Pl plants grown under full sunlight compared with green b, pl plants in sunlight. First, we found that 43 ESTs show differences in expression under the same levels of UV-B (Fig. 7). Again, the bz2 lines confirm the biological role of flavonoid sunscreens: The bz2, weak B, pl plants are intermediate in flavonoid content and show responses intermediate between b, pl and B, Pl genotypes. For example, under solar UV-B, transcripts levels for ribosomal protein L31 were 6.36 times lower in B, Pl plants relative to levels in b, pl plants, although only 2.73 times lower in bz2 plants and 1.40 times lower in bz2 Mutator plants.
Figure 7.
Cluster analysis of transcripts from Mutator (Mu, pink); bz2 (bz2, beige); and B, Pl (B, purple) plants in sunlight. RNA from b, pl green plants in sunlight was used as the reference for microarray hybridization. Clustering was performed according to Eisen et al. (1998). The color saturation reflects the magnitude of the log2 expression ratio for each transcript. Red color means higher transcript levels than the reference, whereas green means lower transcript levels than the reference. The log2 scale of color values is provided at the bottom of the figure.
Interestingly, we found 26 ESTs that are highly transcribed in Mutator plants with respect to b, pl plants and that were also increased in b, pl plants by UV-B exposure (Fig. 7). Active Mutator plants suffer continuous DNA damage during Mu transposition. Consequently, the up-regulation of these transcripts could reflect DNA damage and its consequences. Moreover, we found that the mudrA gene, which encodes the transposase-mobilizing Mu elements, was up-regulated in sunlight in Mutator plants (Supplementary Table III). In UV-B supplementation experiments in the greenhouse, the only transcript selectively up-regulated in these plants encodes MURB, the second protein encoded by the MuDR transposon (data not shown). These results provide a plausible mechanism of transcriptional activation to explain how UV-B radiation can reactivate cryptic Mu transposons (Walbot, 1999).
Modulation of Gene Expression in the Absence of UV-A and UV-B
Finally, we investigated differences in gene expression after exclusion of both UV-B and UV-A radiation, with the aim to identify genes regulated differentially by these two radiations. For this experiment, all four genotypes were grown under complete UV filters (Fig. 1B, LE). First, we observed that 81 transcripts altered by UV-B exclusion returned to “normal” levels when UV-A was also depleted (for genes with putative known functions in this group, see Supplementary Table III). Twenty-five transcripts were down-regulated by UV (UV-A + B), and these transcripts were not changed or were up-regulated by UV-B. This observation could reflect down-regulation in response to UV-A or counteracting effects of UV-B and UV-A (Table I; Supplementary Table III). It is important to note that as result of the properties of the plastics used, UV-A was manipulated only in the absence of UV-B. Thus, it cannot be ruled out that some of the contrasts between responses to UV-B and UV-A + B could be accounted for by nonlinear responses to overall UV fluence.
Table I and Supplementary Table III also describe genes that are only up-regulated by UV-A + B, because transcript levels are not changed by UV-B exclusion. In this group, it is important to note that the transcripts for some photosynthetic genes down-regulated by UV-B, such as Rubisco and a photosystem II protein, are up-regulated when UV-B and UV-A are both depleted. Two anthocyanin biosynthetic genes are up-regulated by UV in sunlight: chalcone synthase and Bz1 (Table I). Exclusion of UV-B alone does not modulate the levels of transcripts for these genes, because UV-A and visible light are effective in maintaining these transcripts. Only when both UV-B and UV-A are depleted do transcripts levels decrease. Transcripts for chalcone synthase were not detected in b, pl plants, in agreement with previous data showing that expression of this gene in maize leaves requires the B and Pl transcription factors (Dooner, 1983).
DISCUSSION
Terrestrial levels of UV-B are increasing as stratospheric ozone is depleted. UV-B changes biomass accumulation, plant morphology, and relationships with herbivores (for reviews, see Ballaré et al., 2001; Searles et al., 2001; Paul and Gwynn-Jones, 2003). Using microarray hybridization we examined changes in transcript abundance for approximately 2,500 maize ESTs after several UV irradiation treatments in leaves of four maize genotypes that differ in flavonoid and anthocyanin content and in preexisting DNA damage levels. We found that 355 transcripts (14% of array elements) are regulated by UV in at least one of the genotypes studied; of these, 304 are regulated only by UV-B. Among these, 204 transcripts have putative identified functions, whereas 100 encode proteins of unknown function with homologies to others in GenBank or show no similarity to any known protein (Supplementary Fig. 3). With this information, new insights can be generated by identifying genes involved in the same biological pathway and by clustering genes with similar transcription profiles. Our findings are especially relevant because many of the genes characterized in this article have no assigned function.
First, we found that many transcripts are regulated in a different manner by natural UV-B levels than by UV-B irradiation in the greenhouse using UV lamps, even if there is some overlap and some transcripts levels are changed by both treatments. It is important again to note that the irradiation protocol used in the greenhouse delivered a very high UV-B irradiance over a short period; therefore, our supplementation and exclusion experiments were not symmetrical and cannot be directly compared. Nonetheless, a robust understanding of maize responses to UV-B requires multiple protocols to elucidate the diversity of transcript changes.
Second, we confirmed that flavonoids and anthocyanins have a similar shielding effect as the UV-B-absorbing plastics, because as many genes were similarly expressed in purple plants under full sunlight UV-B as in green plants under the plastic. We found that the Mutator line, which is under constant genotoxic stress from the DNA breakage, shows upregulation of some transcripts that suggests that some interaction between radiation and pre-existing DNA damage probably exists (Fig. 7). Of particular note, ambient UV-B results in higher mudrA transcript levels and the mudrB transcript is up-regulated by supplemental UV-B after 8 h. The rapidity of this response suggests that the ability of UV-B radiation to reactivate cryptic Mu transposons may result from transcriptional activation of MuDR (Walbot, 1999).
Finally, one key purpose of this work was to identify novel genes regulated by UV-B radiation. As shown in Table I and Supplementary Tables I through III, microarrays allowed us to study coordinate gene expression of enzymes participating in a variety of different pathways. Many of the genes identified in our study have not previously been shown to be regulated by UV-B, illustrating that microarrays are a powerful tool for the discovery of gene functions. Dozens of candidate maize genes and pathways were identified here that had not been directly associated with responses to UV-B radiation in plants. For example, the functional group with the largest number of genes up-regulated by UV-B is those encoding proteins involved in protein synthesis (41 transcripts; Table I; Supplementary Fig. 3). Examples include cytoplasmic ribosomal proteins, initiation and elongation factors, and poly(A+)-binding proteins. It is important to note that previous studies using microarrays to identify transcripts regulated by stress treatments did not detect the induction of any gene involved in protein biosynthesis. A major component of DNA damage is UV-B-induced base dimerization. In a similar way, UV-B could damage ribosomes by forming cross-links in ribosomal RNA or between mRNA, tRNA, rRNA, and proteins (Noah et al., 2000). As a consequence, coordinate up-regulation of ribosomal protein synthesis is likely to be important for the maintenance of this crucial cellular function. Our findings are similar to the report of Valéry et al. (2001) who conducted microarray analysis of cultured normal human melanocytes after UV-B exposure, by hybridization to arrays of human ESTs. They identified up-regulation of transcripts encoding ribosomal proteins after UV-B treatment, and these transcripts had been previously demonstrated to be highly expressed in melanomas, a skin cancer caused by UV-B. In this way, there are many parallels in gene regulation by UV-B in animal and plant cells, although tumor formation is not the end result in plants.
We found that transcripts encoding proteins related to photosynthesis and CO2 fixation, such as Rubisco and proteins of both photosystems I and II, were down-regulated by UV-B radiation. The transcripts for some photosynthetic genes that were down-regulated by UV-B were also up-regulated by UV-A, indicating that UV-A in natural sunlight may be necessary for the expression of these genes, probably by the action of one of the blue/UV-A photoreceptors. Down-regulation of photosynthetic proteins has been documented in pea (Pisum sativum) and wheat (Triticum aestivum; Jordan, 1996; Mackerness et al., 1997), but this is the first report for corn, a C4 plant adapted to a high-sunlight fluence rate. Our novel finding is that UV-B radiation increased the level of transcripts for some enzymes of the glycolytic pathway, fermentation, and respiration such as the pyrophosphate-dependent phosphofructokinase β-subunit, enolase, pyruvate kinase, and alcohol dehydrogenase (Supplementary Table II), particularly in b, pl plants, which are the most sensitive to UV-B. These alternative energy sources may be used in various situations of stress, such as UV-B exposure. Involvement of enzymes of primary metabolism in stress has been demonstrated by Logemann et al. (2000), who found UV-induction of enzymes that can provide carbon substrates for the shikimate pathway, but this is the first report, to our knowledge, of induction of enzymes that can also provide energy in the form of ATP for the synthesis of these and other molecules necessary for cell functions under UV-B stress.
In agreement with previous studies, we found that transcripts for a number of enzymes implicated in the oxidative burst and antioxidant responses are increased after UV-B exposure in b, pl plants (Table I; Supplementary Table II). Previous studies have shown that UV-B exposure increases ROS species generating oxidative stress (Arnots and Murphy, 1991; Dai et al., 1997). It was proposed that, in response to UV-B radiation, ROS function as destructive radicals and may also be components of signal cascades that change plant gene expression (Mackerness et al., 1997; Mackerness and Jordan, 1999). Genes implicated in oxidative stress such glutathione S-transferases and superoxide dismutases are increased in our experiments; they have also been detected by microarray hybridization analysis during elicitation of disease resistance in Arabidopsis (Schenk et al., 2000) and salt stress in rice (Kawasaki et al., 2001). These proteins help to maintain redox homeostasis in cells and also scavenge ROS (Edwards et al., 2000; Schurmann and Jacquot, 2000). Additional studies will be required to determine whether ROS or a direct effect of UV-B increases transcripts for gene encoding the antioxidant enzymes. The origin of ROS remains elusive; candidate enzymes are peroxidases, lipoxygenases, and oxalate oxidase (Wojtaszek, 1997). A membrane-bound NADPH-oxidase, analogous to the one present in mammalian cells, has been proposed as the ROS source in plants (Lamb and Dixon, 1997). The transcript for a NADPH-oxidase is up-regulated after UV-B exposure in b, pl, B, Pl, and Mutator plants (Supplementary Table III); this result can be very valuable as a platform for further studies to confirm its biochemical role.
In this paper, we confirm that UV-B is a potent inducer of protective mechanisms. Numerous transcripts that were up-regulated by UV-B as listed in Supplementary Table II encode proteins of unknown functions that are also induced by other stresses. These results demonstrate that some of the responses elicited by UV-B are shared by other acclimation responses to a fluctuating environment. Identification of the functions of these proteins in response to general stress can help to understand mechanisms of stress responses, and to use them as markers for stress-tolerant genotypes. An increase in transcripts whose products are involved in the biosynthesis of the plant cell wall and cuticular waxes was observed in b, pl plants, such as endoxyloglucan transferase, beta-ketoacyl-CoA synthase and acetyl-CoA acyltransferase. Both of these cellular components can protect against UV-B radiation (Caldwell et al., 1983), and wall alterations could modify leaf shape and size. Additionally, transcripts for chorismate mutase were increased; this enzyme participates in the synthesis of Phe and Tyr and therefore could increase precursors for both cell wall and UV-B-absorbing compounds.
Another important novel finding is the coordinated up-regulation by UV-B of transcripts for enzymes participating in S-adenosyl-Met metabolism. S-Adenosyl-Met is the major donor of methyl groups for transmethylation reactions in eukaryotic cells (Chiang et al., 1996). The concentration of this molecule and the diverse methyltransferases could modulate reactions that regulate many aspects of gene expression. In addition, transcripts for several transcription factors and components of signal transduction pathways were increased by UV-B (Table I), particularly in b, pl plants (Supplementary Table II); these proteins are probably involved in UV-B-induced signaling cascades that respond to the initial products of DNA damage, to ROS, or to UV-B interaction with receptors.
We also found that a group of transcripts regulated by UV-B are involved in genome integrity and cell cycle control. DNA is damaged by UV-B during the day in maize leaves, but detectable damage monitored using sensitive antibody tests for cyclobutane pyrimidine dimers is photoreactivated (Stapleton et al., 1997); damage in sunlight is matched by DNA repair to ensure genomic integrity. Transcripts for histones and other DNA-binding proteins are upregulated by UV-B. The high level of these transcripts could be an indirect effect after DNA damage caused by UV-B radiation. Interestingly, transcripts for highly abundant proteins that vary during the cell cycle including cytoskeletal proteins such as actins, tubulins, and ankyrins (Klessig et al., 2000; Catterou et al., 2001); thioredoxin h (Marty et al., 1993); proteins participating in ubiquitin-mediated protein degradation such as cdc20, ubiquitin, proteosome proteins, and cyclins (Mironov et al., 1999; Kramer et al., 2000); and S-adenosylhomo-Cys hydrolase and argininosuccinato synthase (Coller et al., 2000) are all up-regulated by UV-B in our experiments. Furthermore, cell cycle regulatory proteins are elevated, which may reflect tighter control of cell division when leaf cells incur DNA damage from UV-B radiation. In contrast with our results, Logemann et al. (1995) found that induction of flavonoid biosynthetic enzymes in UV-irradiated cell suspension cultures of parsley (Petroselinum crispum) shows an inverse correlation with the activities of several cell cycle-related genes. This opposite observation could depend on the different UV-B treatment used (field experiments versus UV-B controlled-environments) or the different species used (parsley cell suspension cultures versus maize leaves). Despite this, all of these results confirm that the cell cycle is affected by UV-B radiation and that this occurs under natural UV levels.
UV-B also causes cross-linking and oxidative damage to proteins (Gerhardt et al., 1999), and we found significant increases in the transcript levels of ubiquitin, ubiquitin-binding proteins, proteosome proteins, and proteinases, together with several chaperonins (Table I). The ubiquitin- and proteasome-dependent pathway is highly conserved in eukaryotes. Specificity is achieved through the diversity of conjugating enzymes, some of which recognize abnormal or damaged proteins and others that recognize specific transcription factors and cell cycle regulators (Ingvardsen and Veierskov, 2001). From our data, an enhanced capacity to repair and recycle damaged proteins is implicated as an acclimation response to UV-B in maize. Moreover, posttranslational processes may be very important as a mechanism of regulation of gene expression by UV-B.
CONCLUSIONS
In this paper, we report “discovery genes” that were not previously identified during studies of UV responses in plants. There have been several previous studies of responses to UV-B exclusion under field conditions, such as growth inhibition (Papadopoulos et al., 1999), sunscreen responses (Turunen et al., 1999), photosynthesis (Bischof et al., 2002), and insect-plant interactions (Zavala et al., 2001). This is the first large-scale study reporting global changes induced by UV radiation in gene expression patterns in experiments under field conditions. An important result is the significant activation of genes supporting translation. Experiments in progress have shown that UV-B radiation inhibits protein synthesis in maize leaves in vivo as damage accumulates in ribosomes by formation of cross-links between RNA and ribosomal proteins (P. Casati and V. Walbot, unpublished data). In this way, our study can provide markers for various physiological processes affected by UV-B suitable for more detailed studies of mechanism. Moreover, microarray data can be used to demonstrate particular roles in UV-B acclimation responses by use of mutants and transgenic plants and for the study of genetic networks through the analysis of epistatic relationships between mutant phenotypes. Microarray analysis confirmed literature reports on the impact of UV-B on photosynthetic genes and on flavonoid biosynthesis. Cluster analysis further demonstrated coordinate regulation of suites of functionally related genes such as the ribosomal protein group and photosynthetic genes. With these candidate genes and processes identified, future studies can be directed toward particular UV-B responses in maize and other plants and toward the regulation of specific pathways. Because of the diversity of physiological responses to UV-B and of possible signal transduction pathways responding to this environmental parameter, dissecting the regulatory circuitry will be challenging.
MATERIALS AND METHODS
Plant Material
Three near-isogenic maize (Zea mays) lines that differ in flavonoid phenotype were constructed in the W23 inbred: (a) Bz2, B, Pl; (b) Bz2, b, pl; and (c) bz2, weak B, pl. The weak B allele is a paramutant form of the strong B-I allele recovered after spontaneous paramutation (Walbot, 2001). The Mutator line is in a mainly (>80%) W23 background that is weak B, pl; it contains >10 copies of the transposase-encoding MuDR element, numerous Mu elements, and the reporter allele bz2-mu2.
Radiation Treatments and Measurements
UV treatments were done either in the field (exclusion of UV), using different plastic filters to screen either UV-B or both UV-B and UV-A, or in the greenhouse using UV-B lamps. In field experiments, solar UV-B radiation was removed to produce the minus UV-B treatment using PE filters (100-μm clear PE plastic, Tap Plastics, Mountain View, CA). This PE filter absorbs UV-B without significantly affecting UV-A or visible radiation (Fig. 1B). For the no UV treatment, both solar UV-B and UV-A were removed using LE sheets (3-mm LE plastic, Tap Plastics). To control for differences in wind or humidity under plastic sheeting, CA sheeting was used (100-μm extra clear CA plastic, Tap Plastics); the CA sheeting transmits most radiation from sunlight and is designated as the full UV treatment. Seeds from the four maize lines were planted during the summer of 2001 outdoors at the Stanford Plant Growth Facility. Approximately 14 d after sowing and 7 d after germination, 1.5 × 3.2 m of each plastic was draped over 1- × 2.5-m wooden frames that were erected in the field; the excess plastic was stapled to the sides of the frames to reduce light exposure. The N and S sides were left open to allow air to circulate. However, 50-cm-long curtains with the same plastic were made on the east and west sides to avoid early morning and late afternoon UV exposure. The frames were maintained about 30 cm above the plant canopy during the course of the experiments. Temperature and humidity in the soil and in the leaves were recorded using an infrared thermometer (model 210ALCS microcomputer based agri-term infrared thermometer, Everest Interscience Inc., Fullerton, CA) and a relative humidity hygrometer (Thermo-Hygro 800016, Sper Scientific, Scottsdale, AZ); there were no differences among the plots. Average canopy temperatures under the filters were always within ±0.5°C of each other, and in no case were consistent differences in temperature detected between filter treatments. The same was observed when humidity was recorded, showing differences lower than 25% between treatments. Measurements of incoming solar radiation were obtained using a spectroradiometer (model 752, Optronics Laboratories, Orlando, FL) that was calibrated against a National Bureau of Standards certified radiation source before each use. The spectrum under each filter was recorded periodically with 1-nm resolution across the entire sunlight spectrum (290–800 nm) to check for changes in the transmittance of the filters. Under our conditions, there were no significant reductions of the transmittance of the filters after 3 weeks of exposure to solar UV, equivalent to the duration of our experiments. Consequently, the filters were not replaced, but because they accumulate dust and could develop small tears, we cleaned them at least every 4th d, and we replaced areas with small rips. Plants were grown under the specified conditions for 21 d; adult leaf samples from leaf 9 or 10 were collected for RNA extractions at 4 pm in all treatments. The experiment was repeated twice during the summer of 2001 using different replicate plots (with at least 10 plants of each genotype per plot; Supplementary Fig. 1) to control the variables that might influence the expression profiles. At the end of the treatments, plant height was measured, and significant differences were analyzed using two-factor ANOVA using Microsoft Excel 2002 (Microsoft, Redmond, WA), with P < 0.05.
For experiments with supplemental UV-B radiation, plants from the four lines were grown in the greenhouse with supplemental visible lighting to 10% of summer noon radiation and no UV-B for 3 weeks (Fig. 1C). After that, plants were illuminated using UV-B lamps using fixtures mounted 30 cm above the plants (F40UVB 40 W and TL 20 W/12, Phillips, Eindhoven, The Netherlands) for 8 h, and leaf samples were collected at 4 pm immediately after the end of the light treatment; this simulates a 10-fold increase in UV-B at 305 nm compared with the Stanford field in August at noon (Fig. 1C). The bulbs were covered using CA filters to exclude wavelengths lower than 280 nm. As a control, plants were exposed for the same period of time under the same lamps covered with PE (no UV-B treatment; Fig. 1C). The output of the UV-B source in the greenhouse was recorded using the Optronics spectroradiometer.
Flavonoid Extraction
One-half gram of fresh leaf tissue was frozen in liquid nitrogen and was ground to a powder with a mortar and pestle. The powder was extracted for 8 h with 3 mL of acidic methanol (1% [v/v] HCl in methanol), followed by a second extraction with 6 mL of chloroform and 3 mL of distilled water. The extracts were vortexed and then centrifuged for 2 min at 3,000g. The extract from b, pl leaves was used as a reference when spectra were recorded using Spectra MAX 250 spectrophotometer (Molecular Devices, Sunnyvale, CA).
RNA Isolation, mRNA Purification, and Probe Synthesis
Multiple adult leaves from each radiation treatment were collected for RNA extraction. Because of the sensitivity of microarrays, plant-to-plant variation was reduced by bulk harvesting at least six leaves of different plants collected from each experimental treatment. RNA was extracted from samples from different experiments to use as biological replicates. RNA was extracted using either RNA wiz (Ambion, Austin, TX) or TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturers' recommendations. Poly(A+) RNA was isolated using Oligotex (Qiagen USA, Valencia, CA), and 4 μg of poly(A+) RNA was used for each cDNA synthesis using Superscript II reverse transcriptase (Invitrogen). cDNA was labeled using 100 μm Cy5-dUTP or Cy3-dUTP (Amersham Biosciences, Piscataway, NJ). Excess nucleotides and primers were removed using QIAquick PCR Purification Kit (Qiagen USA).
RNA Gel-Blot Hybridization
RNA (10 μg of total RNA) was analyzed by electrophoresis on a 2% (v/v) formaldehyde/1.5% (w/v) agarose gels and blotted onto Hybond-N+ nylon membrane (Amersham Biosciences). DNA probes were labeled with [α-32P]dCTP by the random primer method and purified from unincorporated nucleotides using probe purification columns. Hybridizations were done at 42°C using formamide overnight. Quantification of hybridization signals was achieved using Kodak ds 1D Digital Science (Scientific Imaging System, New Haven, CT).
Microarray Experiments
Microarray analysis was done following the Minimum Information About a Microarry Experiment (MIAME) guidelines (Brazma et al., 2001). Maize microarrays were type 606 arrays fabricated by the Maize Gene Discovery Project (for more information, see http://gremlin3.zool.iastate.edu/zmdb/microarray/). About 90% of the spots showed significant hybridization when leaf mRNA was used for the experiments. Within these arrays, the cDNA samples are printed three times next to each other, and many of the 2,500 element types were printed several times on each slide. Consequently, average signal intensities and the ratio between cohybridized samples could be assessed multiple times within each microarray and within experiments. These 606 arrays also contain numerous internal control cDNAs such as genes of the anthocyanin pathway, photosynthetic genes, and MRP genes (http://gremlin3.zool.iastate.edu/zmdb/microarray/Data-for-606-Image1.txt). The experimental and reference samples were labeled with either Cy5-dUTP or Cy3-dUTP fluorescent dye (Amersham Biosciences). Two samples, one labeled with each dye, were mixed and then hybridized to a microarray for 15 h at 60°C. The slides were washed and then scanned with a GenePix 4000B Scanner (Axon Instruments, Union City, CA). Normalization between the Cy3 and Cy5 fluorescent dye emission channels was achieved by adjusting the levels of both image intensities. Pair wise and loop designs were used for comparisons to minimize potential biases (Churchill, 2002; Yang and Speed, 2002). For pair wise comparisons, dye-swapping experiments were done. In these experiments, the RNA samples were labeled reciprocally, both as a biological and technical repetition for comparing the reproducibility of the experiments.
Data Analysis
The hybridization intensities of each microarray element were measured using ScanAlyze 4.24 (available at http://genome-www4.stanford.edu/MicroArray/SMD/restech.html). The two channels were normalized in log space using the z-score normalization on a 95% trimmed data set. We removed unreliable spots according to the following criteria: Spots flagged as having false intensity caused by dust or background on the array were removed, and spots for which intensity was less than 3-fold above background were also eliminated. Signals from triplicate spots were averaged. Multiple experiments were analyzed using Cluster and Treeview software (Eisen et al., 1998; http://genome-www4.stanford.edu/MicroArray/SMD/restech.html). We used the default options of hierchical clustering using the uncentered correlation similarity metric. We performed the analysis using both normalized and non-normalized data; the outcomes were essentially the same.
Supplementary Material
Acknowledgments
We thank Yangrae Cho for his generous help with microarray hybridization and interpretation, Tom DeHoog for his assistance with the field experiments, Nikki Reyes for performing many of the spectrophotometric recordings, and Dean Goodman for providing MRP probes for RNA-blot analysis. Yangrae Cho, George Rudenko, Matt Fitzgerald, Eduardo Rodrí-guez, Valeria Lara, and Diego Gómez Casati provided helpful comments on a draft of the manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.022871.
This study was supported in part by the National Science Foundation (grant no. IBN 98–72657). P.C. is a postdoctoral fellow of Fundación Antorchas and a member of the Research Career of the Consejo Nacional de Investigaciones Científicas y Técnicas.
The online version of this article contains Web-only data. The supplemental material is available at http://www.plantphysiol.org.
References
- Alfenito MR, Souer E, Goodman CD, Buell R, Mol J, Koes R, Walbot V (1998) Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases. Plant Cell 10: 1135–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnots T, Murphy TM (1991) A comparison of the effects of a fungal elicitor and ultraviolet radiation on ion transport and hydrogen peroxide in rose cells. Environ Exp Bot 31: 209–216 [Google Scholar]
- Ballaré CL, Rousseaux MC, Searles PS, Zaller JG, Giordano CV, Robson TM, Caldwell MM, Sala OE, Scopel AL (2001) Impacts of solar ultraviolet-B radiation on terrestrial ecosystems of Tierra del Fuego (southern Argentina): an overview of recent progress. J Photochem Photobiol B Biol 62: 67–77 [DOI] [PubMed] [Google Scholar]
- Bieza K, Lois R (2001) An Arabidopsis mutant tolerant to lethal ultraviolet-B levels shows constitutively elevated accumulation of flavonoids and other phenolics. Plant Physiol 126: 1105–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof K, Krabs G, Wiencke C, Hanelt D (2002) Solar ultraviolet radiation affects the activity of ribulose-1,5-bisphosphate carboxylase-oxygenase and the composition of photosynthetic and xanthophyll cycle pigments in the intertidal green alga Ulva lactuca L. Planta 215: 502–509 [DOI] [PubMed] [Google Scholar]
- Bjorn LO (1994) Introduction. In RE Kendrick, GHM Kronenberg, eds, Photomorphogenesis in Plants, Ed 2. Kluwer Academic Publishers, Boston, pp 3–25
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC et al. (2001) Minimum information about a microarray experiment (MIAME): toward standards for microarray data. Nat Genet 29: 365–371 [DOI] [PubMed] [Google Scholar]
- Britt AB (1996) DNA damage and repair in plants. Annu Rev Plant Physiol Plant Mol Biol 4: 75–100 [DOI] [PubMed] [Google Scholar]
- Britt AB, Chen JJ, Wykoff D, Mitchell D (1993) A UV-sensitive mutant of Arabidopsis defective in the repair of pyrimidine-pyrimidinone (6–4) dimmers. Science 261: 1571–1574 [DOI] [PubMed] [Google Scholar]
- Brosche M, Schuler MA, Kalbina I, Connor L, Strid A (2002) Gene regulation by low level UV-B radiation: identification by DNA array analysis. Photochem Photobiol Sci 1: 656–664 [DOI] [PubMed] [Google Scholar]
- Caldwell MM, Robberecht R, Flint SD (1983) Internal filters: prospects for UV-acclimation in higher plants. Physiol Plant 58: 445–450 [Google Scholar]
- Catterou M, Dubois F, Schaller H, Aubanelle L, Vilcot B, Sangwan-Norreel BS, Sangwan RS (2001) Brassinosteroids, microtubules and cell elongation in Arabidopsis thaliana: II. Effects of brassinosteroids on microtubules and cell elongation in the bul1 mutant. Planta 212: 673–683 [DOI] [PubMed] [Google Scholar]
- Chiang PK, Gordon RK, Tal J, Zeng GC, Doctor BP, Pardhasaradhi K, McCann PP (1996) S-Adenosylmethionine and methylation. FASEB J 10: 471–480 [PubMed] [Google Scholar]
- Churchill GA (2002) Fundamentals of experimental design for cDNA microarrays. Nat Genet 32: 490–495 [DOI] [PubMed] [Google Scholar]
- Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR (2000) Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci USA 97: 3260–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q, Yan B, Huang S, Liu X, Peng S, Miranda MLM, Chavez AQ, Vegara BS, Olszyk D (1997) Response to oxidative stress defense systems in rice (Oryza sativa) leaves with supplemental UV-B radiation. Physiol Plant 101: 301–308 [Google Scholar]
- Desikan R, Mackerness SAH, Hancock JT, Neill SJ (2001) Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol 127: 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner HK (1983) Coordinate genetic regulation of flavonoid biosynthetic enzymes in maize. Mol Gen Genet 189: 136–141 [Google Scholar]
- Edwards R, Dixon DP, Walbot V (2000) Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health. Trends Plant Sci 5: 193–198 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt KE, Wilson MI, Greenberg BM (1999) Tryptophan photolysis leads to a UVB-induced 66 kDa photoproduct of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) in vitro and in vivo. Photochem Photobiol 70: 49–56 [Google Scholar]
- Gould KS, McKelvie J, Markham KR (2002) Do anthocyanins function as antioxidants in leaves? Imaging of H2O2 in red and green leaves after mechanical injury. Plant Cell Environ 25: 1261–1269 [Google Scholar]
- Green R, Fluhr R (1995) UV-B induced PR-1 accumulation is mediated by active oxygen species. Plant Cell 2: 203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S, Knebel A, Tenev T, Neininger A, Gaestel M, Herrlich P, Bohmer FD (1999) Inactivation of protein-tyrosine phosphatases as mechanism of UV-induced signal transduction. J Biol Chem 274: 26378–26386 [DOI] [PubMed] [Google Scholar]
- Harborne JB, Grayer RJ (1988) The anthocyanins. In JB Harborne, ed, The Flavonoids. Chapman and Hall, London, pp 1–20
- Ingvardsen C, Veierskov B (2001) Ubiquitin- and proteasome-dependent proteolysis in plants. Physiol Plant 112: 451–459 [DOI] [PubMed] [Google Scholar]
- Jansen MAK, Gaba V, Greenberg BM (1998) Higher plants and UV-B radiation: balancing damage, repair and acclimation. Trends Plant Sci 3: 131–135 [Google Scholar]
- Jordan BR (1996) The effects of UV-B radiation on plants: a molecular perspective. In JA Callow, ed, Advances in Botanical Research. Academic Press, Boca Raton, FL, pp 97–162
- Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K, Galbraith D, Bohnert HJ (2001) Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 13: 889–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klessig DF, Durner J, Noad R, Navarre DA, Wendehenne D, Kumar D, Zhou JM, Shah J, Zhang SQ, Kachroo P et al. (2000) Nitric oxide and salicylic acid signaling in plant defense. Proc Natl Acad Sci USA 97: 8849–8855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer ER, Scheuringer N, Podtelejnikov V, Mann M, Peters JM (2000) Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell 11: 1555–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48: 251–275 [DOI] [PubMed] [Google Scholar]
- Landry LG, Chapple CCS, Last RL (1995) Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol 109: 1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry LG, Stapleton AE, Lim J, Hoffman P, Hays JB, Walbot V, Last RL (1997) An Arabidopsis photolyase mutant is hypersensitive to ultraviolet-B radiation. Proc Natl Acad Sci USA 94: 328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann E, Tavernaro A, Schulz WG, Somssich IE, Hahlbrock K (2000) UV light selectively coinduces supply pathways from primary metabolism and flavonoid secondary product formation in parsley. Proc Natl Acad Sci USA 97: 1903–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann E, Wu SC, Schroder J, Schmelzer E, Somssich IE, Hahlbrock K (1995) Gene activation by UV light, fungal elicitor or fungal infection in Petroselinum crispum is correlated with repression of cell cycle-related genes. Plant J 8: 865–876 [DOI] [PubMed] [Google Scholar]
- Mackerness SAH, John CF, Jordan B, Thomas B (2001) Early signaling components in ultraviolet-B responses: distinct roles for different reactive oxygen species and nitric oxide. FEBS Lett 489: 237–242 [DOI] [PubMed] [Google Scholar]
- Mackerness SAH, Jordan BR (1999) Changes in gene expression in response to ultraviolet B-induced Stress. In M Pessarakli, eds, Handbook of Plant and Crop Stress, Ed 2. Marcel Dekker, New York, pp 749–768
- Mackerness SAH, Thomas B, Jordan BR (1997) The effect of supplementary ultraviolet-B radiation on transcripts, translation and stability of chloroplast proteins and pigment formation in Pisum sativum L. J Exp Bot 48: 729–738 [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA (2000) The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nature Gen 26: 403–410 [DOI] [PubMed] [Google Scholar]
- Marty I, Brugidou C, Chartier Y, Meyer Y (1993) Growth related gene expression in Nicotiana tabacum mesophyll protoplasts. Plant J 4: 265–278 [DOI] [PubMed] [Google Scholar]
- Mazza CA, Boccalandro HE, Giordano CV, Battista D, Scopel AL, Ballaré CL (2000) Functional significance and induction by solar radiation of ultraviolet-absorbing sunscreens in field-grown soybean crops. Plant Physiol 122: 117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles GP, Samuel MA, Ellis BE (2002) Suramin inhibits oxidant signalling in tobacco suspension-cultured cells. Plant Cell Environ 25: 521–527 [Google Scholar]
- Mironov V, DeVeylder L, VanMontagu M, Inzé D (1999) Cyclin-dependent kinases and cell division in plants: the nexus. Plant Cell 11: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noah JW, Shapkina T, Wollenzien P (2000) UV-induced crosslinks in the 16S rRNAs of Escherichia coli, Bacillus subtilis and Thermus aquaticus and their implications for ribosome structure and photochemistry. Nucleic Acids Res 28: 3785–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos YA, Gordon RJ, McRae KB, Bush RS, Belanger G, Butler EA, Fillmore SAE, Morrison M (1999) Current and elevated levels of UV-B radiation have few impacts on yields of perennial forage crops. Global Change Biol 5: 847–856 [Google Scholar]
- Paul ND, Gwynn-Jones D (2003) Ecological roles of solar UV radiation: towards an integrated approach. Trends Ecol Evol 18: 48–55 [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozema J, vandeStaaij J, Bjorn LO, Caldwell M (1997) UV-B as an environmental factor in plant life: stress and regulation. Trends Ecol Evol 12: 22–28 [DOI] [PubMed] [Google Scholar]
- Schaffer R, Landgraf J, PerezAmador M, Wisman E (2000) Monitoring genome-wide expression in plants. Curr Opin Biotechnol 11: 162–167 [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA 97: 11655–11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurmann P, Jacquot JP (2000) Plant thioredoxin systems revisited. Annu Rev Plant Physiol Plant Mol Biol 51: 371–400 [DOI] [PubMed] [Google Scholar]
- Searles PS, Flint SD, Caldwell MM (2001) A meta analysis of plant field studies simulating stratospheric ozone depletion. Oecologia 127: 1–10 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K (2001) Monitoring the expression pattern of 1,300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan JJ (1996) Sinapate esters provide greater UV-B attenuation than flavonoids in Arabidopsis thaliana (Brassicaceae). Am J Bot 83: 679–686 [Google Scholar]
- Stapleton AE, Thornber CS, Walbot V (1997) UV-B component of sunlight causes measurable damage in field-grown maize (Zea mays L): Developmental and cellular heterogeneity of damage and repair. Plant Cell Environ 20: 279–290 [Google Scholar]
- Stapleton AE, Walbot V (1994) Flavonoids can protect maize DNA from the induction of ultraviolet-radiation damage. Plant Physiol 105: 881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann JW, Stelmach BA, Weller EW, Ryan CA (2000) UVB/UVA radiation activates a 48 kDa myelin basic protein kinase and potentiates wound signaling in tomato leaves. Photochem Photobiol 71: 116–123 [DOI] [PubMed] [Google Scholar]
- Thimm O, Essigmann B, Kloska S, Altmann T, Buckhout TJ (2001) Response of Arabidopsis to iron deficiency stress as revealed by microarray analysis. Plant Physiol 127: 1030–1043 [PMC free article] [PubMed] [Google Scholar]
- Turunen M, Heller W, Stich S, Sandermann H, Sutinen ML, Norokorpi Y (1999) UV screening in lodgepole pine (Pinus contorta ssp. latifolia) cotyledons and needles. Environ Pollut 106: 219–228 [DOI] [PubMed] [Google Scholar]
- Valéry C, Bon B, Barge J, Grob JJ, Verrando P (2001) Gene expression profiling with cDNA microarray technology of ultraviolet-B irradiated normal human melanocytes: relation to melanocarcinogenesis [Abstract]. J Inv Dermatol 117: 799. [DOI] [PubMed] [Google Scholar]
- Walbot V (1999) UV-B damage amplified by transposons in maize. Nature 397: 398–399 [DOI] [PubMed] [Google Scholar]
- Walbot V (2001) Imprinting of R-r, paramutation of B-I and Pl, and epigenetic silencing of MuDR/Mu transposons in Zea mays L. are coordinately affected by inbred background. Genet Res 77: 219–226 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B (2001) Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtaszek P (1997) Oxidative burst: an early plant response to pathogen infection. Biochem J 322: 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Speed T (2002) Design issues for cDNA microarray experiments. Nat Rev 3: 579–588 [DOI] [PubMed] [Google Scholar]
- Zavala JA, Scopel AL, Ballaré CL (2001) Effects of ambient UV-B radiation on soybean crops: impact on leaf herbivory by Anticarsia gemmatalis. Plant Ecol 156: 121–130 [Google Scholar]
- Yalamanchili RD, Stratmann JW (2002) Ultraviolet-B activates components of the systemin signaling pathway in Lycopersicon peruvianum suspension-cultured cells. J Biol Chem 277: 28424–28430 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.