Abstract
The movement protein of tobacco mosaic virus, MP30, mediates viral cell-to-cell transport via plasmodesmata. The complex MP30 intra- and intercellular distribution pattern includes localization to the endoplasmic reticulum, cytoplasmic bodies, microtubules, and plasmodesmata and likely requires interaction with plant endogenous factors. We have identified and analyzed an MP30-interacting protein, MPB2C, from the host plant Nicotiana tabacum. MPB2C constitutes a previously uncharacterized microtubule-associated protein that binds to and colocalizes with MP30 at microtubules. In vivo studies indicate that MPB2C mediates accumulation of MP30 at microtubules and interferes with MP30 cell-to-cell movement. In contrast, intercellular transport of a functionally enhanced MP30 mutant, which does not accumulate and colocalize with MP30 at microtubules, is not impaired by MPB2C. Together, these data support the concept that MPB2C is not required for MP30 cell-to-cell movement but may act as a negative effector of MP30 cell-to-cell transport activity.
Plant viruses produce movement proteins required to mediate cell-to-cell spread of the viral genomic information via plasmodesmata (Lucas, 1995; Lazarowitz and Beachy, 1999; McLean and Zambryski, 2000; Tzfira et al., 2000; Haywood et al., 2002; Heinlein, 2002). Tobacco mosaic virus (TMV), a virus with a (+) single-stranded RNA genome encodes one of the most extensively studied movement proteins, MP30. Several biochemical and cell biological features of MP30 were revealed: MP30 binds cooperatively single-stranded nucleic acids (Citovsky et al., 1990), interacts with the tubulin- and actin-based cytoskeleton (Heinlein et al., 1995; McLean et al., 1995), associates to the endoplasmic reticulum (ER; Heinlein et al., 1998; Reichel and Beachy, 1998; Mas and Beachy, 1999; Gillespie et al., 2002), and increases the size exclusion limit of plasmodesmata (Wolf et al., 1989; Waigmann et al., 1994). In the current model, MP30 is proposed to form complexes with genomic TMV-RNA (vRNA) and to move these vRNA-protein complexes from ER-associated sites of synthesis throughout the cell via the cytoskeletal network toward plasmodesmata. There, MP30 induces an increase in plasmodesmal size exclusion limit and facilitates the transport of the vRNA-MP30 complex through the enlarged plasmodesmal channels into adjacent cells (Lazarowitz and Beachy, 1999; Tzfira et al., 2000; Haywood et al., 2002; Heinlein, 2002). In line with this model, microtubules have been functionally implicated in guiding vRNA-MP30 complexes toward plasmodesmata (Mas and Beachy, 2000; Boyko et al., 2000a, 2000b). However, an alternate, non-movement-promoting role of microtubules in providing a route toward an MP30 degradation site was suggested (Padgett et al., 1996; Mas and Beachy, 1999; Reichel and Beachy, 2000). This model is supported by the observation that TMV was able to spread in tissues with functionally impaired microtubules and by studies performed with a mutant variant of MP30, MPR3 (Toth et al., 2002) with strongly decreased affinity to microtubules but increased cell-to-cell movement capacity (Gillespie et al., 2002).
The complex intra- and intercellular transport processes likely require interaction of MP30 with cellular factors that assist in subcellular distribution and cell-to-cell movement. So far, a cell wall-associated pectin methyl esterase (PME; Dorokhov et al., 1999; Chen et al., 2000; Tzfira et al., 2000) was shown to specifically interact with TMV-MP30. Also, binding studies of MP30 with cell fractions enriched in plasmodesmal components indicate the presence of potential receptor molecules mediating protein movement via plasmodesmata (Kragler et al., 1998). Recently, NtNCAPP1, an ER-localized protein, was found to be involved in the plasmodesmal transport pathway of MP30 (Lee et al., 2003). Furthermore, a cell wall-associated kinase is known to phosphorylate the MP30 (Citovsky et al., 1993) thereby modulating its cell-to-cell movement activity (Waigmann et al., 2000).
For a number of movement proteins from other viruses, interacting cellular factors such as a rab receptor-like protein, Hsp40 (DnaK) family members, a homeodomain protein, and a transcriptional coactivator were recently identified (Soellick et al., 2000; Huang et al., 2001; Lin and Heaton, 2001; Desvoyes et al., 2002; Matsushita et al., 2002). The functions of these interesting movement protein interaction partners with respect to cell-to-cell movement have yet to be elucidated.
Here, we report the identification of a previously uncharacterized MP30-interacting plant protein, MPB2C, from the TMV host plant Nicotiana tabacum. MPB2C associates with microtubules in a punctuate manner and colocalizes with MP30. In functional studies, MPB2C mediates MP30 subcellular redistribution to microtubule-associated sites and selectively inhibits MP30 cell-to-cell movement. A potential role of MPB2C as a negative effector of MP30 transport activity is discussed.
RESULTS
Identification of MP30-Interacting Factor MPB2C via the Yeast Sos Recruitment System (SRS)
The partially hydrophobic nature of MP30 (Citovsky et al., 1990; Brill et al., 2000) and its association to cell compartments such as the ER membrane, cytoskeleton, and plasmodesmal receptors (for review, see Haywood et al., 2002; Heinlein, 2002) indicate that a membrane-based interaction screening system may be more suitable to identify MP30-interacting proteins than the conventional two-hybrid system. The yeast SRS (Aronheim et al., 1997) allows detection of interaction of proteins at the plasma membrane in Brewer's yeast (Saccharomyces cerevisiae; Aronheim et al., 1997; Yamanaka et al., 2000). Because the membranous environment provided by the SRS might be more compatible with the observed cellular distribution of MP30, this yeast-based interaction screening system was chosen to identify plant-produced MP30-binding proteins.
In the SRS, the bait protein is fused to the human Ras guanyl nucleotide exchange factor (hSos), which complements the temperature-sensitive (ts) cdc25-2 yeast growth phenotype upon recruitment to the plasma membrane. Prey library proteins are N-terminally linked to a myristoylation signal providing plasma membrane localization. Bait-prey interaction results in recruitment of hSos to the plasma membrane and leads to growth of yeast colonies at the restrictive temperature (Aronheim et al., 1997). Library protein production is regulated by a Gal-dependent promoter, which allows controlled expression of library protein and verification of specific interactions by observing Gal-dependent growth of the yeast colonies.
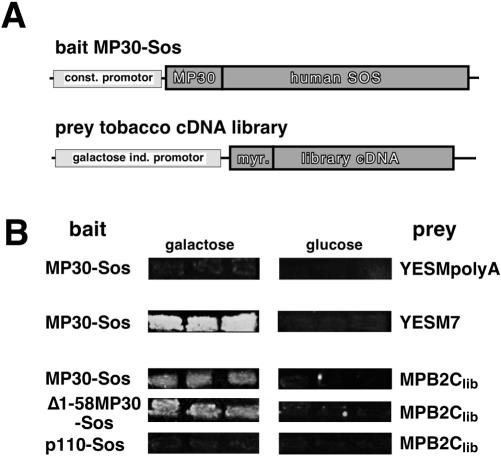
First, a bait MP30-Sos fusion protein expressed by the yeast shuttle vector ADNS-MP (Fig. 1A, top scheme) was constructed. To ensure that MP30-Sos by itself was not able to complement, or to interfere with complementation of, the cdc25-2 ts phenotype, cells were cotransformed with ADNS-MP30 and empty library vector YESMpolyA. No yeast colonies were formed at the restrictive temperature (Fig. 1B, top panels). Thus, MP30 did not mediate hSos localization to the plasma membrane in yeast. Selective Gal-dependent growth was observed when cells were cotransformed with positive control vector YESM7 encoding an hSos-interacting protein and MP30-Sos-expressing bait plasmid (Fig. 1B, panels in second row). This functional test indicates that the MP30-hSos fusion protein is stable, can be translocated to the plasma membrane, and is able to activate the Ras pathway. Thus, MP30 fused to hSos was suitable to be used as bait in a SRS screen.
Figure 1.
SRS screening for MP30 interaction partners. A, Schematic representation of bait and library constructs. B, Complementation of the cdc25 ts phenotype. Bait and prey proteins expressed in transformed cells are indicated to left and right of panels, respectively. Growth of yeast is compared on Gal- and Glc-containing media at the restrictive temperature. Prey proteins are expressed only on Gal-containing medium.
Next, directional cDNA derived from poly(A+)-RNA obtained from leaf, stem, shoot apex, and flower tissue of the natural TMV host plant N. tabacum cv Turkish was used to construct a myristoylation signal-tagged expression library (Fig. 1A, bottom scheme). To isolate cDNAs, which encode interaction partners of MP30, approximately 106 independent cDNA clones were screened. A novel cDNA named movement protein binding 2C, library (MPB2Clib) coding for a 124-amino acid protein (Fig. 3A) was selected for further analysis.
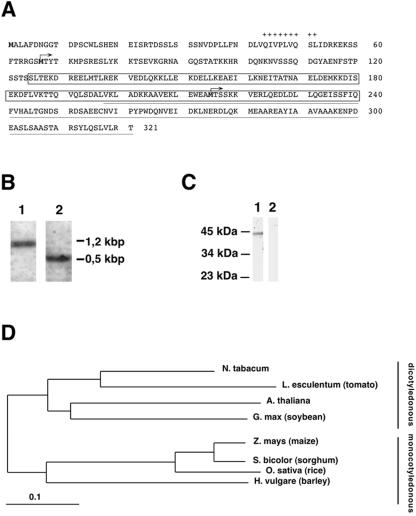
Figure 3.
Expression and sequence analysis of MPB2C. A, Deduced amino acid sequence of full-length MPB2C. N-terminal hydrophobic region (+), the coiled-coil domain (boxed), MPB2Clib sequence (underlined), and the start for N-terminally truncated constructs (arrows) are indicated. B, Northern analysis of MPB2C expression. Tobacco poly(A+)-RNA was probed with an MPB2C-specific probe (lane 1) or a ubiquitin-specific probe as control (lane 2). C, Western-blot analysis of MPB2C expression. Total protein extracted from leaf tissue was incubated with anti-MPB2C-antibody (lane 1) or preimmune antibody (lane 2). D, Phylogenetic tree obtained by MPB2C protein sequence comparison with translated cDNA sequences. The related sequences fall into a dicotyledonous and a monocotyledonous clade, which reflects the evolutionary relation of the plant families. Note that no significantly homologous sequence was observed in available non-flowering plants, fungi, animals, bacteria, and virus sequence databanks. GenBank accession numbers of the cDNA sequences were At5g08120 (Arabidopsis), AI898765 (tomato [Lycopersicon esculentum]), BE347146 (soybean [Glycine max]), BE186113 (maize [Zea mays]), BE356348 (sorghum [Sorghum bicolor]), AU094801 (Oryza sativum), and AL509409 (barley [Hordeum vulgare]). Space bar represents 0.1 substitutions per amino acid.
Specific interaction between MPB2Clib and MP30-Sos was apparent by Gal-dependent growth of cells co-expressing MPB2Clib and MP30-Sos at the restrictive temperature (Fig. 1B, panels in third row). In contrast, upon co-expression of MPB2Clib with a nonrelevant bait protein, p110-Sos, no growth of yeast colonies was observed (Fig. 1B, bottom panels). These complementation tests confirmed the specificity of interaction between MP30 and MPB2Clib. Interestingly, MPB2Clib interacted with Δ1-58MP30-Sos (Fig. 1B, panels in fourth row), a truncated form of MP30 lacking the N-terminal 58 amino acids but retaining the region required for interaction with and movement through plasmodesmata (Waigmann et al., 1994). The cDNA sequence of MPB2Clib was determined, and comparison of the predicted protein against DNA and protein databases using the TBLAST algorithm (Pearson and Lipman, 1988) revealed that MPB2C is an uncharacterized plant protein with structural properties similar to the myosin/kinesin super family (data not shown). Thus, we have isolated the cDNA of a previously uncharacterized tobacco protein, MPB2C, which specifically binds in vivo to MP30 in the SRS interaction assay.
MPB2C and MP30 Interact in Vitro
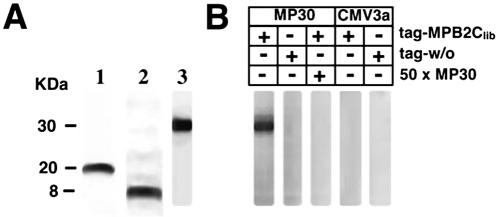
Specificity of the yeast-based interaction studies was confirmed in vitro by overlay assays. First, an Anti-Xpress-tag-MPB2Clib fusion protein of approximately 20 kD, and, as a negative control, tag protein alone with approximately 8 kD were produced in Escherichia coli and detected by western blotting using Anti-Xpress-tag antibody (Fig. 2A, lanes 1 and 2). For overlay assays (Fig. 2B), purified MP30 was blotted onto nitrocellulose, and MP30 presence and size were verified with an antibody directed against MP30 (Fig. 2A, lane 3). In parallel, part of the blot was renatured and incubated with either total E. coli lysate containing tag-MPB2Clib or tag protein alone. Addition of tagged MPB2Clib resulted in strong binding, which could be competed by the addition of an excess of MP30 to the overlay mixture (Fig. 2B). In contrast, no signal was detected in the control reaction with tag protein alone (Fig. 2B). To determine whether MPB2C is capable of interacting with a viral movement protein functionally related to MP30, an immobilized cucumber mosaic virus movement protein (CMV3a; Li and Palukaitis, 1996) was used in a parallel overlay experiment. Here, no interaction of MPB2Clib with CMV3a was observed (Fig. 2B). Together, these results underscore that MP30 and MPB2Clib specifically interact in vitro.
Figure 2.
MPB2C binds to MP30 in in vitro overlay assays. A, Verification of protein expression in E. coli: Tag-MPB2Clib (lane 1), tag protein alone (lane 2), and MP30 (lane 3) were detected by western blotting and labeling with Anti-Xpress-tag antibody (lanes 1 and 2) or anti-MP30 antibody (lane 3). B, Detection of interaction between MPB2Clib and viral MPs by renatured blot overlay assay. Blotted MP30 and CMV3a were incubated with E. coli lysate containing tag-MPB2Clib, tag protein alone, and soluble MP30 as indicated in scheme. Binding was detected with Anti-Xpress-tag antibody.
Sequence and Expression Analysis of MPB2C
By RACE techniques, full-length MPB2C cDNA with a size of 1.2 kb was isolated. Upon northern-blot analysis of poly(A+)-RNA from green tissue of N. tabacum with an MPB2C-specific probe (Fig. 3B, lane 1), a band correlating to the calculated size of MPB2C cDNA was observed. As a positive control for RNA quality, a ubiquitin-specific probe was used (Fig. 3B, lane 2). Together, RACE and northern results indicate that the full-length MPB2C cDNA was isolated.
Full-length MPB2C cDNA translates into a 321-amino acid protein (Fig. 3A) with a calculated molecular mass of 36 kD. MPB2C protein expression was confirmed by western blotting with an anti-MPB2C antibody raised against purified E. coli-generated MPB2C. When total protein extract derived from leaf tissue was analyzed with the anti-MPB2C antibody, a single MPB2C-specific band with a molecular mass of approximately 40 kD was detected (Fig. 3C, lane 1) that was not present upon incubation of the blot with preimmune serum (Fig. 3C, lane 2), indicating that the MPB2C protein is constitutively expressed in leaf tissue. Note that the aberrantly high molecular mass of the MPB2C protein in denaturing polyacrylamide gels was also observed for the recombinant E. coli MPB2C protein (data not shown).
Structural analysis of full-length MPB2C protein revealed a large coiled-coil region typical for cytoskeletal-associated factors (Lupas et al., 1991) stretching from amino acids 125 to 240 (Fig. 3A). In addition, a short hydrophobic region encompassing amino acids 44 to 52 was predicted (Kyte and Doolittle, 1982). When the predicted MPB2C protein sequence was compared with the Arabidopsis database The Arabidopsis Information Resource by FASTA algorithm (Pearson and Lipman, 1988), one homologous protein of similar size of 326 amino acids (approximately 40% identity; gi:8346549), was identified. The carboxy-terminal part of the Arabidopsis MPB2C homolog was previously isolated in a screen for plant cytoskeletal, cell cycle-related, and polarity-related proteins (Xia et al., 1996). TBLASTN searches (http://arabidopsis.org/Blast/) in nonredundant plant databases and subsequent analysis with the ClustalW software (http://www.ebi.ac.uk/clustalw/; Thompson et al., 1994) highlighted a number of uncharacterized cDNAs encoding significantly similar proteins from mono-and dicotyledonous plants with identity ranging from 75% to 37%. The evolutionary relations between MPB2C from N. tabacum and its various homologs from other plant species were compiled (Page, 1996) and depicted as a tree (Fig. 3D). The related proteins fall into a dicotyledonous and a monocotyledonous clade, which reflects the evolutionary relation of the plant families. Interestingly, proteins with significant structural homology to MPB2C seem to be limited to the plant kingdom because no related proteins with overall similarity above 25% were found in databases of non-plant organisms such as viruses, bacteria, yeasts, and animals.
MPB2C Localizes to Microtubules in a Punctuate Manner
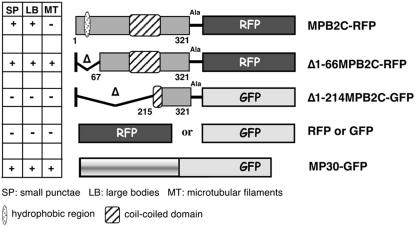
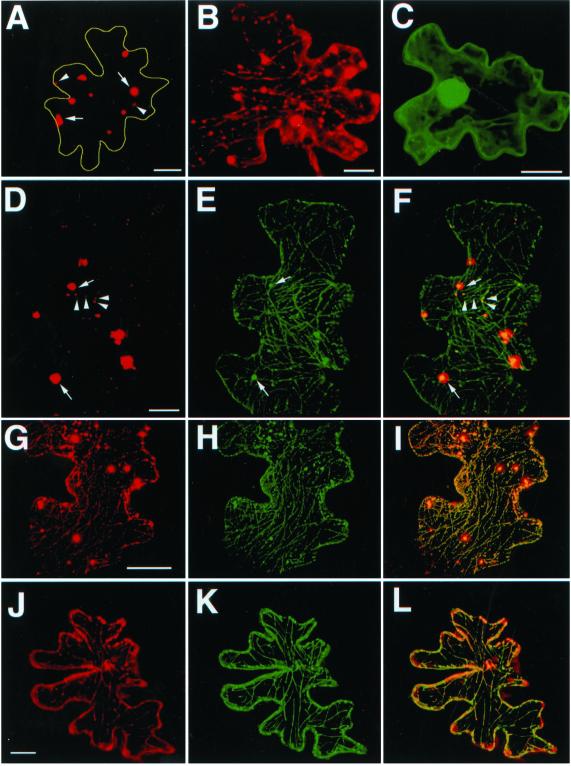
To evaluate intracellular localization, full-length and N-terminally truncated variants of MPB2C lacking either the hydrophobic region (Δ1-66MPB2C), or in addition to the hydrophobic region, a large part of the coiled-coil domain (Δ1-214MPB2C), were fused to red fluorescent protein (RFP; Matz et al., 1999) or green fluorescent protein (GFP; Chalfie et al., 1994; Table I), respectively. Fusion proteins were transiently expressed in epidermal cells of N. tabacum source leaves and were analyzed for subcellular localization patterns by confocal laser scanning microscopy. The full-length MPB2C-RFP appears as small punctae and larger bodies (Fig. 4, A and D, arrows point to bodies, arrowheads to punctae; Table I). Variable relative amounts of MPB2C-RFP bodies and punctae can be observed in individual cells ranging from primarily body-like phenotypes (Fig. 4A) over mixed phenotypes (Figs. 4D and 6A) to predominantly punctuate phenotypes (Figs. 4G and 6D). The truncated Δ1-66MPB2C-RFP lacking the hydrophobic N-terminal domain localizes to filamentous structures as well as to larger bodies and small punctae (Fig. 4B; Table I). Frequently, the bodies and punctae decorated by the MPB2C-RFP fusion protein are in close contact and appear to align along filamentous structures (Figs. 4, B and G, and 6D). Similar localization patterns were observed for MPB2C-GFP and Δ1-66MPB2C-GFP (data not shown). Δ1-214MPB2C-GFP lacks most of the coiled-coil structure in addition to the hydrophobic domain and shows cytoplasmic distribution equivalent to that observed for free soluble GFP in epidermal cells (Fig. 4C; Table I).
Table I.
MPB2C constructs and intracellular localization pattern
Figure 4.
Analysis of intracellular expression patterns of MPB2C. RFP and GFP fusion proteins were transiently expressed in epidermal cells of N. tabacum leaves by biolistic transformation and were analyzed by confocal microscopy after 48 h in the appropriate channels. Red color indicates RFP fluorescence; green color indicates GFP fluorescence. A, MPB2C-RFP; for better orientation, the cellular outline has been redrawn. B, Δ1-66MPB2C-RFP. C, Δ1-214MPB2C-GFP. D to I, Co-expression of MPB2C-RFP (D and G) with MAP4-GFP (E and H); F and I, merged image. J to L, Co-expression of Δ1-66MPB2C-RFP (J) with MAP4-GFP (K); L, merged image. Arrows point to bodies, arrowheads to punctae. Bars in A, G, and J = 20 μm; bars in B through D = 10 μm.
Figure 6.
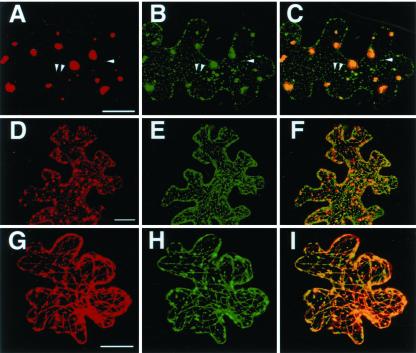
MPB2C proteins colocalize with MP30-GFP. Analysis of expression and presentation of micrographs as described in Figure 4. A to F, Co-expression of MPB2C-RFP (A and D) with MP30-GFP (B and E); C and F, merged image. G to I, Co-expression of Δ1-66MPB2C-RFP (G) and MP30-GFP (H); I, merged image. Arrowheads point to small punctae decorated by MPB2C-RFP and MP30-GFP. Bars in A, D, and G = 20 μm.
The filamentous appearance of the Δ1-66MPB2C-RFP suggests association to microtubules. Therefore, colocalization experiments between MPB2C-RFP fusion proteins and microtubule-associated protein MAP4-GFP (Marc et al., 1998) were performed. The large bodies decorated by MPB2C-RFP (Fig. 4D, arrows) were also decorated by MAP4-GFP (Fig. 4E, arrows) and were in close contact with MAP4-GFP-decorated microtubules (Fig. 4F, arrows). Also, MPB2C-RFP appearing as small punctae (Fig. 4, D [arrowheads] and G) aligned at MAP4-GFP-decorated microtubules (Fig. 4, E and H) in the merged image (Fig. 4, F [arrowheads] and I). Similarly, the truncated Δ1-66MPB2C-RFP fusion protein predominantly decorated punctae and filaments that colocalized to filaments decorated by MAP4-GFP (Fig. 4, J–L).
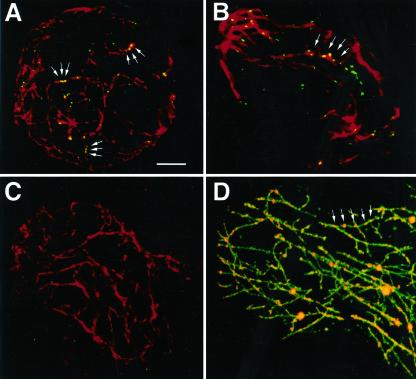
Furthermore, to determine the subcellular localization of endogenous MPB2C, protoplasts derived from leaf tissue were analyzed by immunofluorescence. Fixed protoplasts were incubated with anti-tubulin antibody and anti-MPB2C antibody (Fig. 5, A and B) or, as a negative control, with anti-tubulin antibody and preimmune serum (Fig. 5C). Intracellular localization patterns were evaluated by confocal microscopy. The MPB2C-specific signal was detected in the green channel and manifested as punctae that generally aligned to or were in close contact with microtubular filaments detected in the red channel (Fig. 5, A and B, arrows). In contrast, in protoplasts incubated with preimmune serum, microtubule-associated punctae were not detected (Fig. 5C). For comparison, a high-resolution image of transiently co-expressed MPB2C-RFP and MAP4-GFP is provided. Note that MPB2C-RFP punctae (Fig. 5D, arrows) align to MAP4-GFP-decorated microtubules highly similar to endogenous MPB2C.
Figure 5.
Endogenous MPB2C localizes in punctae to microtubules. Leaf tissue-derived protoplasts were analyzed by immunofluorescence and were observed with a confocal microscope. A and B, Protoplasts incubated with anti-tubulin-antibody (red) and anti-MPB2C-antibody (green). Arrows point to MPB2C punctae. C, Protoplasts incubated with anti-tubulin-antibody (red) and preimmune serum (green). D, High-resolution image of MPB2C-RFP co-expressed with MAP4-GFP in an epidermal leaf cell. Compare the punctuate MPB2C-RFP pattern (arrows) with that of endogenous MPB2C. Bar for A to D in A = 5 μm.
Collectively, these studies indicate that endogenous MPB2C localizes in discrete punctae to microtubular filaments and that this punctuate localization pattern is correctly reflected by transiently expressed MPB2C-RFP. However, in transient expression experiments, we additionally observe large MPB2C-RFP-decorated bodies that do not correctly reflect the localization of endogenous MPB2C and are likely a consequence of the high expression level achieved in the transient assay.
MPB2C Colocalizes to MP30 in Plant Cells
As a necessary prerequisite for interaction between MPB2C and MP30 in plant cells in vivo, both proteins should be present in the same subcellular compartment(s). In line, the filament and body-like appearance of MPB2C resembles described intracellular localization patterns of MP30 (Heinlein et al., 1995, 1998; McLean et al., 1995; Kahn et al., 1998; Mas and Beachy, 1999; Crawford and Zambryski, 2000; Boyko et al., 2000a, 2000b; Kotlizky et al., 2001).
Upon transient expression in epidermal cells of N. tabacum source leaves, MPB2C-RFP-decorated bodies and punctae (Fig. 6, A and D) colocalize to MP30-GFP (Fig. 6, B and E) in the merged image (Fig. 6, C and F; arrowheads in A, B, and C point to some of the colocalizing punctae). In addition, an MP30-GFP signal is also observed in small regular cortical punctae (Fig. 6B) reflecting ER-associated MP30-GFP (Gillespie et al., 2002), but these punctae are not tagged by MPB2C-RFP proteins (Fig. 6C). Upon coexpression with Δ1-66MPB2C-RFP, MP30-GFP is predominantly detected at filaments that overlap with those decorated by Δ1-66MPB2C-RFP (Fig. 6, G–I). In summary, colocalization between MP30 and MPB2C is observed at the level of microtubules but not when MP30 is associated to the ER. This observation indicates that in vivo interaction between MP30 and MPB2C takes place at microtubules, in line with the endogenous localization of MPB2C.
Interestingly, in the presence of transiently expressed MPB2C-RFP fusion proteins, MP30-GFP was nearly exclusively localized to microtubule-associated sites and to the ER (Fig. 6, B, E, and H) and was localized in only 4% of co-expressing cells to the characteristic cell wall-associated punctae, reflecting plasmodesmal association of MP30 (compare Fig. 7A; Crawford and Zambryski, 2000, 2001; Boyko et al., 2000a, 2000b; Kotlizky et al., 2001). In contrast, when MP30-GFP was expressed in the absence of MPB2C, 24% of expressing cells show the cell wall-associated punctuate expression pattern (compare Fig. 7A; K. Trutnyeva, R. Bachmaier, and E. Waigmann, unpublished data). A similar effect on MP30-GFP intracellular distribution was observed in Nicotiana benthamiana: when MP30-GFP was expressed by itself, it localized to cell wall-associated punctae in 61% of expressing cells (pattern similar to Fig. 7A), whereas upon co-expression with MPB2C-RFP, the cell wall-associated pattern of MP30-GFP was detected in only 26% of expressing cells. The reduction in cell wall-associated pattern was accompanied by a corresponding increase in the ER and microtubule-associated expression pattern of MP30-GFP in the presence of MPB2C-RFP (pattern similar to Fig. 6, B and E). Thus, co-expression of MPB2C-RFP results in redistribution of MP30-GFP from the cell periphery toward microtubule-associated sites, thereby further supporting the concept that MPB2C and MP30 interact at microtubules in vivo.
Figure 7.
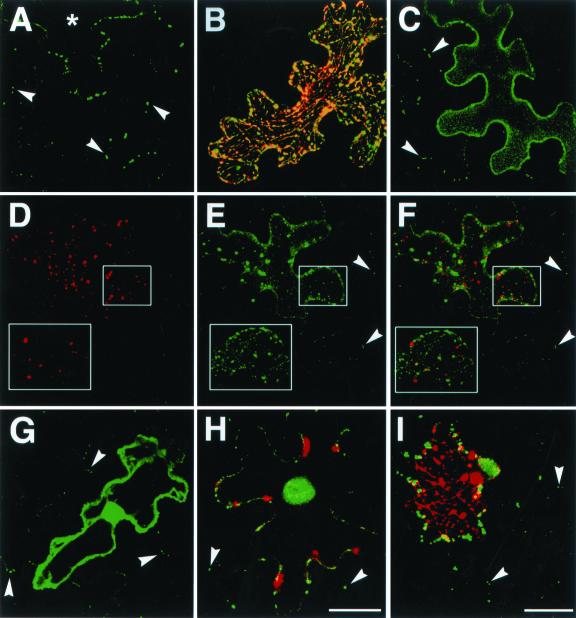
Cell-to-cell movement of viral MPs in epidermal leaf cells expressing MPB2C proteins. Analysis of expression and presentation of micrographs as described in Figure 4, except that experiments depicted in C to F were done in epidermal leaf cells of N. benthamiana. A, Cell-to-cell movement of MP30-GFP in the absence of MPB2C-RFP. *, Expressing cell. B, Lack of cell-to-cell movement in cell co-expressing MP30-GFP and Δ1-66MPB2C-RFP. C, Cell-to-cell movement of MPR3-GFP. D to F, Cell-to-cell movement of MPR3-GFP in cell co-expressing Δ1-66MPB2C-RFP. D, Δ1-66MPB2C-RFP; E, MPR3-GFP; F, merged image. G, Cell-to-cell movement of CMV3a-GFP. H and I, Cell-to-cell movement of CMV3a-GFP in cells co-expressing MPB2C-RFP (H) or Δ1-66MPB2C-RFP (I). In B, H, and I, merged images are shown. Arrowheads point to MP30-GFP, MPR3-GFP, or CMV3a-GFP that has moved into neighboring cell. Bar for A to C and H in H = 20 μm; bar for D to G and I in I = 40 μm.
MPB2C Interferes with MP30 Cell-to-Cell Movement
Following biolistic bombardment of epidermal cells, MP30-GFP fusion proteins are known to move from cell to cell via plasmodesmata (Fig. 7A; Crawford and Zambryski, 2000, 2001; Kotlizky et al., 2001). Interestingly, upon co-expression of MP30-GFP and MPB2C-RFP or Δ1-66MPB2C-RFP constructs, no MP30-GFP cell-to-cell movement to adjacent cells could be detected in more than 50 cells evaluated on 10 independent leaves from different N. tabacum plants (Fig. 7B; compare also Fig. 6, B, E, and H).
Inhibition of intercellular transport of MP30-GFP in cells expressing MPB2C-RFP proteins could be due to specific interaction between MPB2C and MP30 or could result from general down-regulation of plasmodesmal transport potentially induced by overexpression of MPB2C proteins. To discriminate between these possibilities, the viral movement protein CMV3a, which is functionally related to MP30 but does not interact with MPB2C in vitro (compare Fig. 2B), was employed. Like MP30-GFP, a GFP-tagged CMV3a fusion protein moves between cells (Fig. 7G; Itaya et al., 1997). Moreover, even in the presence of the MPB2C-RFP protein variants, CMV3a-GFP cell-to-cell movement was clearly detectable (Fig. 7, H and I). Note that in co-expressing cells, CMV3a-GFP localization did not overlap with that of MPB2C-RFP or Δ1-66MPB2C-RFP (Fig. 7, H and I, respectively). Together, these results imply that MPB2C selectively interferes with intercellular transport of MP30.
The negative effect on cell-to-cell transport of MP30-GFP might be due to improper amounts of fluorescently tagged MPB2C proteins and/or the RFP tag which might mask a potential positive involvement of MPB2C in MP30 cell-to-cell transport. To further clarify the function of MPB2C in intercellular transport of MP30, we employed a L72V point mutant of MP30 (MPR3; Gillespie et al., 2002; Toth et al., 2002) with enhanced cell-to-cell movement capacity in N. benthamiana. Because the MPR3 is primarily localized to ER-associated sites (Gillespie et al., 2002), we predicted that the MPR3 should not interact with the microtubule-associated MPB2C protein. Consequently, transient expression of MPB2C-RFP proteins should not lead to accumulation of MPR3-GFP at microtubules and should not interfere with MPR3 cell-to-cell movement. Because the MPR3 mutant has been characterized in N. benthamiana, we used this host for quantitative, comparative movement studies.
When MPR3-GFP was co-expressed with full-length MPB2C-RFP or Δ1-66MPB2C-RFP, the percentage of cells with movement was only slightly reduced compared with the percentage observed in the control experiment where no MPB2C variants were present (Table II). In contrast, wild-type MP30-GFP movement was reduced to less than one-half in the presence of MPB2C-RFP proteins (Table II). Furthermore, in the presence of co-expressed MPB2C-RFP proteins, the MPR3-GFP was not observed to accumulate at microtubules but retained its ER-associated localization pattern (Fig. 7, D–F). Results indicate that MPB2C and MPR3 are present in different subcellular compartments, and thus do not interact in vivo. Considering that the MPR3 protein is movement competent, these data suggest that interaction with MPB2C is not required for movement of MPR3.
Table II.
Effect of MPB2C on movement of MP30-GFP and MPR3-GFP in N. benthamiana
| Bombarded Plasmid DNA | Cells with Movement | No. of Analyzed Cells |
|---|---|---|
| % | ||
| MP30-GFP | 62.7 | 43 |
| MP30-GFP/MPB2C-RFP | 28.6 | 42 |
| MP30-GFP/Δ1-66MPB2C-RFP | 16.3 | 43 |
| MPR3-GFP | 60.3 | 53 |
| MPR3-GFP/MPB2C-RFP | 55.1 | 49 |
| MPR3-GFP/Δ1-66MPB2C-RFP | 58.0 | 50 |
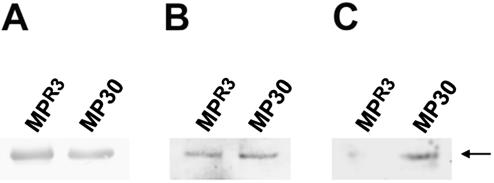
We further tested whether in vitro binding between MPB2C and MPR3 would be reduced as compared with binding between MPB2C and MP30. For this purpose, MPR3 and MP30 were overexpressed and purified from E. coli (Fig. 8A). Equal amounts of purified MPR3 and MP30 protein were blotted onto nitrocellulose, renatured, and overlayed with purified MPB2C protein. Binding of MPB2C to MP30 or MPR3 was detected with an antibody directed against the MPB2C protein. Several molar ratios of MPB2C to MPR3 and MP30 were tested. When the molar amount of MPB2C exceeded that of MPR3 or MP30 2-fold, MPR3 showed slightly reduced binding to MPB2C as compared with MP30 (Fig. 8B, compare lanes MPR3 and MP30). When MPB2C was present at a 2.5-fold lower molar amount than MPR3 or MP30, binding of MPB2C to MPR3 was clearly reduced as compared with MP30 (Fig. 8C, compare lanes MPR3 and MP30). Together, these results indicate that in vitro MPB2C binds with lower affinity to the MPR3 protein than to the MP30 protein.
Figure 8.
Comparison of MP30 and MPR3 binding to MPB2C. A, Western blot of purified MPR3 and MP30 detected by anti-MP30 antibody. B and C, Renatured blot overlay assay. Twenty-three picomoles of MP30 and MPR3 was overlayed with 45 pmol of MPB2C (B) or 9 pmol of MPB2C (C) in the overlay mixture. Binding was detected by anti-MPB2C antibody. Arrow points to MP30 or MPR3, respectively.
DISCUSSION
The frequently observed localization patterns of MP30 comprising accumulation at ER-associated punctae, microtubules, large bodies, and cell wall-associated punctae have been combined in a model for TMV cell-to-cell transport that involves transfer of MP30 and viral RNA from the ER via a microtubular delivery mechanism toward and through plasmodesmata (Lazarowitz and Beachy, 1999; Tzfira et al., 2000; Haywood et al., 2002; Heinlein, 2002), thereby indicating a movement promoting role of microtubules in transport of the MP30-vRNA complex (Heinlein et al., 1995, 1998; McLean et al., 1995; Mas and Beachy, 2000; Boyko et al., 2000a, 2000b; Kotlizky et al., 2001). On the other hand, microtubules have also been implicated in MP30 degradation (Padgett et al., 1996; Mas and Beachy, 1999; Reichel and Beachy, 2000). In line, an MP30 mutant that escapes accumulation at microtubules but displays enhanced cell-to-cell movement was recently described (MPR3; Gillespie et al., 2002), indicating that accumulation at microtubules is detrimental for MP30 movement.
The complex MP30 localization pattern likely requires interaction with several plant endogenous factors. Here, we describe the isolation and characterization of a previously uncharacterized, plant-specific MP30-binding protein, MPB2C, that localizes in discrete punctae to microtubules. The C-terminal one-third of the 321-amino acid-long MPB2C is sufficient to bind MP30. The N-terminal part of MPB2C harbors a hydrophobic region and a coiled-coil domain involved in association to microtubules. As shown by northern and western analysis, MPB2C is constitutively expressed in N. tabacum leaf tissue. Massively Parallel Signature Sequencing data on the Arabidopsis homolog of MPB2C also demonstrate constitutive expression at low levels throughout plant tissues (http://allometra.com/mpss.shtml), indicating a general housekeeping function of MPB2C.
The localization of MPB2C to discrete punctae along microtubules points to the presence of specific target sites on microtubules and is reminiscent of patterns observed for other microtubule-associated proteins such as a kinesin-related motor protein (Cai et al., 2000) or microtubule-plus end-associated proteins, for example CLIP-170 (Perez et al., 1999). Interestingly, the localization of MPB2C to discrete punctae shifts toward a more continuous decoration of microtubules when its 66 N-terminal amino acids are deleted (compare Fig. 4, A and B). Similar observations with CLIP-170 deletion mutants were interpreted as evidence for regions in the CLIP-170 protein that control and regulate its binding to microtubules (Pierre et al., 1994). In line, the N-terminal region of MPB2C might also exert a modulating function on microtubule binding of MPB2C.
What may be the nature of large bodies that are detected in addition to microtubule-associated punctae upon transient expression of MPB2C? In co-expression experiments, MPB2C bodies are also tagged by MP30-GFP and MAP4-GFP and form in close proximity to the microtubular network (compare Fig. 4, D–F), suggesting that these bodies may contain a mixture of microtubule-associated proteins. Interestingly, microtubule-connected bodies are also observed upon overexpression of CLIP-170-GFP (Perez et al., 1999) and tubulin-GFP (F. Kragler, unpublished data) and upon expression of MP30-GFP either by itself (Kotlizky et al., 2001; K. Trutnyeva and E. Waigmann, unpublished data) or in the context of the virus (Gillespie et al., 2002), suggesting that their formation might represent a general response to high-level expression of microtubule-associated proteins.
In transient expression studies, both full-length MPB2C and Δ1-66MPB2C colocalize to MP30 at the level of microtubules and effectively interfere with MP30 localization to the characteristic punctuate pattern, indicating plasmodesmal localization (compare Fig. 7A). In contrast, MPB2C neither colocalizes to ER-associated MP30 nor was it observed to interfere with MP30 ER localization. This is in line with the view that MP30 is located at the ER before its transfer onto microtubules or transport toward plasmodesmata (Gillespie et al., 2002; K. Trutnyeva, R. Bachmaier, and E. Waigmann, unpublished data).
Functional analysis by transient expression studies revealed a specific, MPB2C-mediated decrease in MP30 cell-to-cell movement, suggesting a negative impact of MPB2C on MP30 transport activity. The negative effect of MPB2C on MP30 transport activity was analyzed in two different host plants and varied from a complete block in N. tabacum to a more than 50% reduction in N. benthamiana. This host-dependent difference may be correlated to the well-known high susceptibility of N. benthamiana for viral infection (Gibbs et al., 1997) and the less tightly controlled plasmodesmal transport activity of N. benthamiana as compared with N. tabacum (Waigmann et al., 2000).
Further support for a non-movement-promoting role of MPB2C was derived from analyzing the impact of MPB2C proteins on intercellular transport activity of MPR3, an L72V point mutant of MP30 with improved cell-to-cell movement capacity and decreased affinity for microtubules (Gillespie et al., 2002; Toth et al., 2002). Cell-to-cell transport of MPR3-GFP was not impaired by the presence of MPB2C proteins (Table II). Moreover, even in the presence of MPB2C proteins, MPR3-GFP was located at the ER (Fig. 7, D–F; Gillespie et al., 2002) and was not redistributed to microtubules. These results suggest that MPR3 and MPB2C do not encounter each other in living cells and because the MPR3 protein is highly movement competent (Table II; Gillespie et al., 2002), that interaction with MPB2C is not required for MPR3 cell-to-cell movement. In support, in vitro binding of MPR3 to MPB2C is reduced when compared with binding between MP30 and MPB2C (Fig. 8). This reduction in binding affinity, likely together with changes in binding affinity to other cellular factors, may contribute to the difference in subcellular localization of MPR3 as compared with MP30 and to the lack of inhibition of MPR3 cell-to-cell movement in the presence of transiently expressed MPB2C proteins.
Considering that the MPR3 protein and wild-type MP30 are distinguished only by a point mutation, it is likely that the two proteins move cell-to-cell via a similar mechanism, and that interaction with MPB2C might also not be required for cell-to-cell movement of wild-type MP30. Because the MPR3 protein is characterized by enhanced transport activity and because we observed an MPB2C-mediated decrease of MP30 transport activity in transient expression studies, we presently favor the hypothesis that endogenous MPB2C serves a negative role in MP30 cell-to-cell transport. However, to further elucidate and define the function of MPB2C in cell-to-cell transport of TMV-MP and in intercellular spread of TMV, it will be necessary to generate plants with reduced endogenous MPB2C levels such as anti-sense or silenced plants.
In line with our hypothesis, we propose a model wherein endogenous MPB2C encounters and binds to MP30 at microtubules and acts in a pathway that promotes MP30 accumulation at microtubules. This pathway is not involved in MP30 cell-to-cell transport but might be assigned for degradation as previously suggested (Padgett et al., 1996; Mas and Beachy, 1999; Reichel and Beachy, 2000; Gillespie et al., 2002). When MPB2C is present at endogenous levels, a number of MP30 molecules produced in infected cells escape the MPB2C involving microtubular accumulation pathway and engage in cell-to-cell transport. When MPB2C is transiently overexpressed, the equilibrium is shifted toward the MPB2C pathway, and directs MP30 away from the cell-to-cell transport pathway toward accumulation at microtubules. Hence we observed a negative effect of MPB2C on MP30 movement in the transient overexpression assay.
In summary, we report the identification and characterization of MPB2C, a plant endogenous, microtubule-localized MP30 interaction partner. Future studies will be directed toward a detailed evaluation of the impact of MPB2C on local and systemic spread of TMV and to reveal the as yet unknown endogenous function of the MPB2C protein.
MATERIALS AND METHODS
Plant Growth Conditions
Six- to 8-week-old Nicotiana tabacum cv Turkish or Nicotiana benthamiana plants were used. Plants were grown in a controlled environment with a cycle of 16 h of light at 22°C and 8 h of darkness at 20°C.
Bait Plasmid and SRS cDNA Library Construction
Conventional PCR cloning techniques were employed to construct the MP30-Sos expressing bait plasmids ADNS-MP30 and ADNS Δ1-58MP30. pET-MP30 DNA was amplified with primers FK63 (5′-GATCCCAAGCTTAGGATCCAGAAGATGGCTCTAGTTGTTAAAGG-3′) and FK70 (5′-CGGAGCCCGGGAAACGAATCCGATTCGGCG-3′) and was ligated as HindIII-XbaI fragments (partial HindIII digest for full-length MP30 open reading frame) into HindIII-XbaI digested ADNS 5′SOS vector (Aronheim et al., 1997). After sequencing, cdc25-2 yeast was transformed with ADNS-MP30 and ADNS Δ1-58MP30, and MP30 expression was verified with polyclonal rabbit MP30 antibody (data not shown). Poly(A+)-RNA was isolated from the following tissues of N. tabacum cv Turkish plants according to information supplied by the manufacturer (RNeasy plant mini kit and Oligotex mRNA kit, Qiagen USA, Valencia, CA): leaves (sink and source), inflorescence and shoot apex including young leaves (< 1 cm), stem, and whole young plants (4 cm high). A directional cDNA synthesis was carried out (cDNA synthesis kit, Stratagene, La Jolla, CA), and the resulting EcoRI-XhoI cDNA fragments with sizes >200 bp were ligated with EcoRI-XhoI-digested YESΔpolyA plasmid (Aronheim et al., 1997) and transformed into Epicurian Coli XL1-Blue MRF′ supercompetent Escherichia coli cells following the manufacturer's instructions (Stratagene). Transformed E. coli cells were grown in LB-Amp media (Ausubel et al., 1987), and cDNA plasmids were isolated (Plasmid Midi kit, Qiagen USA).
SRS Screen
Compositions of yeast (Saccharomyces cerevisiae) Gal and Glc containing growth media and transformation reaction techniques and growth conditions were employed as described (Ausubel et al., 1987; Aronheim et al., 1997) with the following exception: The thermosensitive yeast cdc25-2 (Matα, ura3-5, his3-200, ade2-101, lys2-801, trp1-109, leu2-3, cdc25-2, and Gal-) strains transformed with library plasmid and various bait plasmids were first incubated in a 21°C incubation chamber on selectable Glc plates (non-restrictive conditions). Subsequently, colonies of transformed cells were replica plated in parallel onto plates containing Gal- or Glc-selective medium. Plates were incubated at 36C° (restrictive conditions) for >5 d to evaluate bait-prey interaction-mediated growth complementation of the cdc25-2 ts phenotype (Aronheim et al., 1997). Standard control plasmids ADNS-5′p110-SOS, YESM7, YESMpolyA, and YESMΔpolyA are described (Aronheim et al., 1997; SRS protocol provided by A. Aronheim to F.K.).
RNA Expression Analysis and RACE
Poly(A+)-RNA was isolated as described above, and northern analysis was carried out using the NorthernMax-Gly Kit according to the manufacturer's protocol (Ambion, Austin, TX). An MPB2C-specific probe was produced by PCR of the library plasmid YES-MPB2Clib with primers FK76 and FK94. The ubiquitin-specific probe was prepared as described (Sablowski and Meyerowitz, 1998). Probes were radioactively labeled with Random primed DNA labeling kit according to the manufacturer's protocol (Roche Diagnostics, Mannheim, Germany). Note that the hybridized blot was exposed five times longer compared with the ubiquitin probed membrane to observe the MPB2C signal. To obtain full-length MPB2C cDNA, 5′-RACE reactions were carried out following the instructions supplied by the manufacturer (Marathona cDNA amplification kit; BD Biosciences Clontech, Palo Alto, CA) using primers FK100 (5′-TTATGTTCTCAGAACAAGTGATTTTGCAGG-3′) and FK110 (5′-ATCTCATGAGGATCCCCTGTTCTCAGAAC-3′).
Production of MPB2C Protein and Anti-MPB2C Antibodies
A glutathionine S-transferase (GST)-MPB2C expression construct was obtained by PCR amplification of MPB2C from 35S::MPB2C-RFP using primers 5′-GCAGGATCCGATGACGATGACAAGATGGCGCTTGCATTTG-3′ and 5′-GCAGAATTCTTATGTTCTCAGAACAAG-3′. The resulting fragment was digested with BamHI/EcoRI and was cloned into BamHI/EcoRI-digested vector pGEX-6P-1 encoding a GST moiety followed by a PreScission Protease cleavage site (Amersham Biosciences, Uppsala) to yield construct pGEX-MPB2C. To obtain soluble GST-MPB2C, BL21-CodonPlus bacteria transformed with pGEX-MPB2C were grown at 30°C until optical density = 0.6. Induction was performed with 0.1 mm isopropyl-β-d-thiogalactopyranoside for 1 h at 15°C. Bacteria were collected by centrifugation and were lysed by sonication in phosphate-buffered saline (PBS; Ausubel et al., 1987) containing 1% (v/v) Triton X-100 and Protease Inhibitor Cocktail Tablets (Roche Diagnostics, Germany). To obtain purified MPB2C protein, cleared supernatant was incubated with Glutathione Sepharose 4B (Amersham Biosciences) and was subsequently released from the matrix with PreScission Protease according to the manufacturer's instructions (Amersham Biosciences). Because purified MPB2C protein migrated aberrantly high on denaturing polyacrylamide gels (observed molecular mass, approximately 40 kD; calculated molecular mass, 36 kD) the identity of MPB2C was confirmed by N-terminal sequencing and mass spectroscopy before antibody production. Polyclonal antibodies against MPB2C were obtained by immunization of rabbit with 200 μg of purified MPB2C and a boost with 150 μg of purified MPB2C after 8 weeks.
Western-Blot Analysis of MPB2C Protein
N. tabacum leaf tissue was frozen and ground to powder in liquid N2. Proteins were extracted by shaking the powder in the same volume of 2× Laemmli sample buffer (Ausubel et al., 1987) for 30 min and boiling for 10 min. The extract was cleared from insoluble material by ultracentrifugation at 35,000g for 1 h. Proteins were separated on denaturing polyacrylamide gels and were blotted onto nitrocellulose membrane. The blot was blocked in 5% (w/v) milk powder, 10 mm Tris-HCl, pH 7.5, and 150 mm NaCl (M-TBS) for 30 min, incubated for 2 h with anti-MPB2C antibodies or preimmune serum at a dilution of 1:1,000 in M-TBS, washed three times with M-TBS, and then incubated for 1 h with alkaline phosphatase-coupled goat anti-rabbit antibody (Pierce, Rockford, IL) at a dilution of 1:2,000 in M-TBS. After washing three times with M-TBS and Tris-buffered saline, the blot was developed with nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl-phosphate.
In Vitro Interaction of MPB2C and MP30
The MPB2C cDNA fragment isolated in the SRS-screen was PCR amplified with specific primers FK76 (5′-CCGGCTCGAGAGTTAAGTTAGCTGACAAGAAAGCAGC-3′) and FK94 (5′-CGGAATTCCGTTAAGTTAGCTGACAAGAAAGCAGCGGTAG-3′) and was cloned as an XhoI/EcoRI fragment into XhoI/EcoRI-digested pRSETA to obtain the expression plasmid pRSETA-Δ1-197MPB2C giving rise to protein tag-MPB2Clib. The MPR3 coding sequence was amplified by PCR from plasmid MPR3:GFP (Gillespie et al., 2002) using primers R3start (5′-GCACATATGGCTCTAGTTGTTAAAGG) and R3back (5′-GCAGGATCCTTAAAAGGAATCCGATTCGGC) and was cloned as an NdeI/BamHI fragment into NdeI/BamHI digested pET3A vector giving rise to expression plasmid pET-MPR3. MP30, MPR3, and CMV3a proteins were overexpressed in E. coli from plasmids pET-MP30 (Citovsky et al., 1990), pET-MPR3, and pET-CMV3a (Li and Palukaitis, 1996), respectively. Inclusion body isolation, PAGE, and western blotting were done as described (Citovsky et al., 1990). Tag-MPB2Clib was overexpressed from plasmid pRSETA-Δ1-197MPB2C, and western-blot analysis of tag-MPB2Clib was done according to the manufacturer's protocol (Invitrogen, Carlsbad, CA). Conditions for renatured blot overlay assay are based on previously described dot-blot assays (Kragler et al., 1998). In brief, MP30 and CMV3a were blotted onto nitrocellulose membranes, treated twice for 15 min at room temperature with incubation buffer (40 mm HEPES, pH 6.8, and 10% [v/v] glycerol) containing 0.2% (v/v) Triton X-100 and Complete Protease Inhibitor Cocktail Tablets (Roche Diagnostics). After washing three times and blocking the membrane with overlay buffer (incubation buffer containing 0.1% [v/v] Tween 20 and 2% [w/v] bovine serum albumin), nitrocellulose stripes were overlaid with overlay buffer containing total E. coli lysate from cells expressing tag-MPB2Clib protein or the Anti-Xpress-tag protein alone. For competition experiments, an approximately 50× molar access of purified MP30 over MPB2Clib was added to the overlay buffer. After 48 h of incubation at 10°C, washing five times with overlay buffer, and treatment with incubation buffer containing 2% (v/v) diglutaraldehyde for 1 min at room temperature, bound MPB2Clib was detected with commercially available Anti-Xpress-tag antibody following the manufacturer's protocol (Invitrogen). In experiments designed to compare the affinity of MP30 and that of MPR3 with MPB2C, 0.7 μg corresponding to 23 pmol of MP30 and MPR3 protein, respectively, was blotted onto nitrocellulose. A small strip encompassing the MP30 and MPR3 bands was cut out and incubated with either 45 or 9 pmol of MPB2C protein in overlay buffer. The experimental procedure for renatured blot overlay assay was as described above. Binding of MPB2C was detected with anti-MPB2C antibody at a dilution of 1:2,000.
Protoplast Preparation and Immunofluorescence
Protoplasts were prepared as described previously (Koop et al., 1996) and were fixed in 4% (w/v) paraformaldehyde (Science Services, Munich, Germany) and 5% (v/v) dimethyl sulfoxide in PBS for 1 h at room temperature. Fixed protoplasts were washed five times with PBS and were incubated with anti-MPB2C antibody or preimmune serum at a dilution of 1:20 and mouse anti-α-tubulin antibody (Molecular Probes, Eugene, OR) at a dilution of 1:1,000 in PBS for 1 h. After washing five times with PBS, protoplasts were incubated for 1 h with Alexa Fluor 488-coupled goat anti-rabbit and Alexa Fluor 568-coupled goat anti-mouse secondary antibodies (Molecular Probes) diluted 1:1,000 in PBS. After washing three times in PBS, protoplasts were mounted on slides and analyzed by confocal microscopy.
Construction and Transient Expression of GFP and RFP Fusion Proteins
The pUC18-based vectors pZY226 and pZY260 containing a 10× Ala linker at the C or N terminus of a 35S driven mGFP6, respectively, were a generous gift to F.K. by D. Jackson (Cold Spring Harbor Laboratories, Cold Spring Harbor, NY). GFP expression vectors producing soluble smGFP (CD3–326; Arabidopsis Biological Resource Center, Ohio State University, Columbus), ER-GFP (Haseloff et al., 1997), MAP4-GFP (Marc et al., 1998), CMV3a-GFP (Itaya et al., 1997), MP30-GFP (McLean et al., 1995), and MPR3-GFP (Gillespie et al., 2002) were described previously. Plasmid 35S::RFP was a generous gift from V. Citovsky (State University of New York, Stony Brook).
To obtain MPB2C-RFP fusion proteins with a separating Ala linker between the two protein moieties, a 35S::GFP-RFP plasmid was constructed first by cloning a SalI/XbaI-digested RFP PCR fragment amplified from pDsRed1-N1 (BD Biosciences Clontech) using primers FK111 (5′-GCCGTCGACATGGTGCGCTCCTCC-3′) and FK112 (5′-CGCCTCTAGACTACAGGAACAGGTGG-3′) into SalI/XbaI-digested pZY260. A BamHI/XbaI fragment of the 35S::GFP-RFP plasmid containing the Ala linker and RFP was ligated with BsaI(NcoI)/BamHI-digested MPB2C PCR fragments (primer combinations: FK131 (5′-GCCGGTCTCCCATGGCGCTTGCATTTGAC-3′) or FK 114 (5′-GCCGGTCTCCCATGACCTATACCAAA-3′) with FK110) into NcoI/XbaI-digested pZY226 resulting in expression plasmids 35S::MPB2C-RFP or 35S::Δ1-66MPB2C-RFP. Construct 35S::Δ1-214MBP2C-GFP was generated by PCR amplification of MPB2C with primers FK132 (5′-AGGTCATGACTTCCAGCAAG-3′) and FK110 (5′-GATCTCATGAGGATCCCCTGTTCTCAGAAC-3′) and subsequent cloning of the resulting fragment into vector pZY260.
Transient Expression and Confocal Microscopy
DNA was coated onto 1-μm gold particles, and N. tabacum or N. benthamiana source leaves (>20 cm) were bombarded using a Helios Gene Gun following the manufacturer's protocol (Bio-Rad Laboratories, Hercules, CA). Each bombardment experiment was repeated at least five times with the total number of analyzed cells ranging from 40 to 100. Images were obtained with a TCS-SP confocal microscope (Leica Microsystems, Heidelberg). GFP and RFP were excited with a ArKr laser at 476 and 568 nm, respectively. GFP was detected at 500 to 520 nm, and RFP was detected at 600 to 620 nm. Micrographs were assembled from serial optical sections and were prepared for presentation by Photoshop 5.1 software (Adobe Systems, San Jose, CA).
Accession Numbers
The cDNA sequence of MPB2C has been deposited at GenBank under accession number AF326729.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes with the exception of the anti-MPB2C antibodies, which are available only in limited amounts.
Acknowledgments
We thank A. Aronheim for providing plasmids and the yeast strain required for the SRS screen. We are also indebted to Dave Jackson for providing vectors pZY260 and pZY226, to Peter Palukaitis for providing vector pET-CMV3a, to Vitaly Citovsky for providing vector 35S::RFP, to Biao Ding for providing vector pRTL-CMV3a-GFP, to Richard Cyr for providing a MAP4-GFP expression construct, and to Karl Oparka for providing MPR3-GFP expression construct. E.W. is thankful to Andrea Barta for her invaluable support throughout the whole project.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.022269.
This work was supported by the Austrian Science Foundation (grant nos. P12614–MOB and Sfb17, project part 08 to E.W.). E.W. was supported by an Austrian Programme for Advanced Research and Technology fellowship (APART 441) from the Austrian Academy of Science.
References
- Aronheim A, Zandi E, Hennemann H, Elledge SJ, Karin M (1997) Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol Cell Biol 17: 3094–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Smith JA, Seidman JG, Struhl K (1987) Current Protocols in Molecular Biology. Greene Publishing-Wiley Interscience, New York
- Boyko V, Ferralli J, Ashby J, Schellenbaum P, Heinlein M (2000a) Function of microtubules in intercellular transport of plant virus RNA. Nat Cell Biol 2: 826–832 [DOI] [PubMed] [Google Scholar]
- Boyko V, Ferralli J, Heinlein M (2000b) Cell-to-cell movement of TMV RNA is temperature-dependent and corresponds to the association of movement protein with microtubuli. Plant J 22: 315–325 [DOI] [PubMed] [Google Scholar]
- Brill LM, Nunn SR, Kahn TW, Yeager M, Beachy RN (2000) Recombinant tobacco mosaic virus movement protein is an RNA-binding, α-helical membrane protein. Proc Natl Acad Sci USA 97: 7112–7117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G, Romagnoli S, Moscatelli A, Ovidi E, Gambellini G, Tiezzi A, Cresti M (2000) Identification and characterization of a novel microtubule-based motor associated with membranous organelles in tobacco pollen tubes. Plant Cell 12: 1719–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC (1994) Green fluorescent protein as a marker for gene expression. Science 263: 802–805 [DOI] [PubMed] [Google Scholar]
- Chen M-H, Sheng J, Hind G, Handa AK, Citovsky V (2000) Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J 19: 913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V, Knorr D, Schuster G, Zambryski P (1990) The P30 movement protein of tobacco mosaic virus is a single strand nucleic acid binding protein. Cell 60: 637–647 [DOI] [PubMed] [Google Scholar]
- Citovsky V, McLean BG, Zupan J, Zambryski P (1993) Phosphorylation of tobacco mosaic virus cell-to-cell movement protein by a developmentally-regulated plant cell wall-associated protein kinase. Genes Dev 7: 904–910 [DOI] [PubMed] [Google Scholar]
- Crawford KM, Zambryski PC (2000) Subcellular localization determines the availability of non-targeted proteins to plasmodesmatal transport. Curr Biol 10: 1032–1040 [DOI] [PubMed] [Google Scholar]
- Crawford KM, Zambryski PC (2001) Non-targeted and targeted protein movement through plasmodesmata in leaves in different developmental and physiological states. Plant Physiol 125: 1802–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvoyes B, Faure-Rabasse S, Chen M-H, Park J-W, Scholthof HB (2002) A novel plant homeodomain protein interacts in a functionally relevant manner with a virus movement protein. Plant Physiol 129: 1521–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorokhov YL, Makinen K, Frolova OY, Merits A, Saarinen J, Kalkkinen N, Atabekov JG, Saarma M (1999) A novel function for a ubiquitous plant enzyme pectin methylesterase: the host-cell receptor for the tobacco mosaic virus movement protein. FEBS Lett 461: 223–228 [DOI] [PubMed] [Google Scholar]
- Gibbs MJ, Armstrong J, Weiller GF, Gibbs AJ (1997) Virus evolution: the past, a window on the future? In M Tepfer, E Balazs, eds, Virus-Resistant Transgenic Plants: Potential Ecological Impact. Springer-Verlag, Berlin, pp 1–17
- Gillespie T, Boevink P, Haupt S, Roberts AG, Toth R, Valentine T, Chapman S, Oparka KJ (2002) Functional analysis of a DNA-shuffled movement protein reveals that microtubules are dispensable for the cell-to-cell movement of tobacco mosaic virus. Plant Cell 14: 1207–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge SSO (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94: 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood V, Kragler F, Lucas W (2002) Plasmodesmata: pathways for protein and ribonucleoprotein signaling. Plant Cell Suppl 14: S303–S325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinlein M (2002) The spread of tobacco mosaic virus infection: insights into the cellular mechanism of RNA transport. Cell Mol Life Sci 59: 58–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinlein M, Epel BL, Padgett SH, Beachy RN (1995) Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science 270: 1983–1985 [DOI] [PubMed] [Google Scholar]
- Heinlein M, Padgett HS, Gens JS, Pickard BG, Caspar SJ, Epel BL, Beachy RN (1998) Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 10: 1107–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Andrianov VM, Han Y, Howell SH (2001) Identification of Arabidopsis proteins that interact with the cauliflower mosaic virus (CaMV) movement protein. Plant Mol Biol 47: 663–675 [DOI] [PubMed] [Google Scholar]
- Itaya A, Hickman H, Bao Y, Nelson R, Ding B (1997) Cell-to-cell trafficking of cucumber mosaic virus movement protein:green fluorescent protein fusion produced by biolistic bombardment in tobacco. Plant J 12: 1223–1230 [Google Scholar]
- Kahn T, Lapidot M, Heinlein M, Reichel C, Cooper B, Gafny R, Beachy RN (1998) Domains of the TMV movement protein involved in subcellular localization. Plant J 15: 15–25 [DOI] [PubMed] [Google Scholar]
- Koop H-U, Steinmüller K, Wagner H, Rössler C, Eibl C, Sacher L (1996) Integration of foreign sequences into the tobacco plastome via polyethylene glycol-mediated protoplast transformation. Planta 199: 193–201 [DOI] [PubMed] [Google Scholar]
- Kotlizky G, Katz A, van der Laak J, Boyko V, Lapidot M, Beachy RN, Heinlein M, Epel BL (2001) A dysfunctional movement protein of tobacco mosaic virus interferes with targeting of wild-type movement protein to microtubules. Mol Plant-Microbe Interact 14: 895–904 [DOI] [PubMed] [Google Scholar]
- Kragler F, Monzer J, Shash K, Xonocostle-Cázares B, Lucas WJ (1998) Cell-to-cell transport of proteins: requirement for unfolding and characterization of binding to a putative plasmodesmal receptor. Plant J 15: 367–381 [Google Scholar]
- Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–142 [DOI] [PubMed] [Google Scholar]
- Lazarowitz SG, Beachy RN (1999) Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell 11: 535–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-Y, Yoo B-C, Rojas MR, Gomez-Ospina N, Staehelin LA, Lucas WJ (2003) Selective trafficking of non-cell-autonomous proteins mediated by NtNCAPP1. Science 299: 392–396 [DOI] [PubMed] [Google Scholar]
- Li Q, Palukaitis P (1996) Comparison of the nucleic acid- and NTP-binding properties of the movement protein of cucumber mosaic cucumovirus and tobacco mosaic tobamovirus. Virology 216: 71–79 [DOI] [PubMed] [Google Scholar]
- Lin B, Heaton LA (2001) An Arabidopsis thaliana protein interacts with a movement protein of turnip crinkle virus in yeast cells and in vitro. J Gen Virol 82: 1245–1251 [DOI] [PubMed] [Google Scholar]
- Lucas WJ (1995) Plasmodesmata: intercellular channels for macromolecular transport in plants. Curr Opin Cell Biol 7: 673–680 [DOI] [PubMed] [Google Scholar]
- Lupas A, van Dyke M, Stock J (1991) Predicting coiled coils from protein sequences. Science 252: 1162–1164 [DOI] [PubMed] [Google Scholar]
- Marc J, Granger CL, Brincat J, Fisher DD, Kao T, McCubbin AG, Cyr RJ (1998) A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10: 1927–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P, Beachy R (1999) Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J Cell Biol 147: 945–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P, Beachy R (2000) Role of microtubules in the intracellular distribution of tobacco mosaic virus movement protein. Proc Natl Acad Sci USA 97: 12345–12349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita Y, Miyakawa M, Nisiguchi M, Nyunoya H (2002) Cloning of a tobacco cDNA coding for a putative transcriptional coactivator MBF1 that interacts with the tomato mosaic virus movement protein. J Exp Bot 53: 1531–1532 [PubMed] [Google Scholar]
- Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA (1999) Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol 17: 969–973 [DOI] [PubMed] [Google Scholar]
- McLean BG, Zambryski P (2000) Interactions between viral movement proteins and the cytoskeleton. In CJ Staiger, F Baluska, D Volkmann, PW Barlow, eds, Actin: A Dynamic Framework for Multiple Plant Cell Functions. Kluwer Academic Publishers, Dordrecht, The Netherlands
- McLean BG, Zupan J, Zambryski PC (1995) TMV P30 movement protein associates with the cytoskeleton in tobacco cells. Plant Cell 7: 2101–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett HS, Epel BL, Kahn TW, Heinlein M, Watanabe Y, Beachy RN (1996) Distribution of tobamovirus movement protein in infected cells and implications for cell-to-cell spread of infection. Plant J 10: 1079–1088 [DOI] [PubMed] [Google Scholar]
- Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ (1988) Improved tools for biological sequence comparison. Proc Natl Acad Sci USA 85: 2444–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez F, Diamantopoulos GS, Stalder R, Kreis TE (1999) CLIP-170highlights growing microtubule ends in vivo. Cell 96: 517–527 [DOI] [PubMed] [Google Scholar]
- Pierre P, Pepperkok R, Kreis TE (1994) Molecular characterization of two functional domains of CLIP-170 in vivo. J Cell Sci 107: 1909–1920 [DOI] [PubMed] [Google Scholar]
- Reichel C, Beachy RN (1998) Tobacco mosaic virus infection induces severe morphological changes of the endoplasmic reticulum. Proc Natl Acad Sci USA 95: 11169–11174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichel C, Beachy RN (2000) Degradation of tobacco mosaic virus movement protein by the 26S proteasome. J Virol 74: 3330–3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski RWM, Meyerowitz EM (1998) A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92: 93–103 [DOI] [PubMed] [Google Scholar]
- Soellick T-R, Uhrig JF, Bucher GL, Kellmann J-W, Schreier PH (2000) The movement protein NSm of tomato spotted wilt tospovirus (TSWV): RNA binding, interaction with the TSWV N protein, and identification of interacting plant proteins. Proc Natl Acad Sci USA 97: 2373–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth RL, Pogue GP, Chapman S (2002) Improvement of a plant virus based vector through DNA shuffling. Plant J 30: 593–600 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Rhee Y, Chen M-H, Citovsky V (2000) Nucleic acid transport in plant-microbe interactions: the molecules that walk through the walls. Annu Rev Microbiol 54: 187–219 [DOI] [PubMed] [Google Scholar]
- Waigmann E, Chen M-H, Bachmaier R, Goshroy S, Citovsky V (2000) Regulation of plasmodesmal transport by phosphorylation of tobacco mosaic virus cell-to-cell movement protein. EMBO J 19: 4875–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waigmann E, Lucas W, Citovsky V, Zambryski P (1994) Direct functional assay for tobacco mosaic virus cell-to-cell movement protein and identification of a domain involved in increasing plasmodesmal permeability. Proc Natl Acad Sci USA 91: 1433–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S, Deom CM, Beachy RN, Lucas WJ (1989) Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 246: 377–379 [DOI] [PubMed] [Google Scholar]
- Xia G, Ramachandran S, Hong Y, Chan y, Simanis V, Chua N-H (1996) Identification of plant cytoskeletal, cell cycle-related and polarity-related proteins using Schizosaccharomyces pombe. Plant J 10: 761–769 [DOI] [PubMed] [Google Scholar]
- Yamanaka T, Ohta T, Takahashi M, Meshi T, Schmidt R, Dean C, Naito S, Ishikawa M (2000) TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc Natl Acad Sci USA 97: 10107–10112 [DOI] [PMC free article] [PubMed] [Google Scholar]