Abstract
U1 small nuclear ribonucleoprotein (snRNP)-70K (U1-70K), a U1 snRNP-specific protein, is involved in the early stages of spliceosome formation. In non-plant systems, it is involved in constitutive and alternative splicing. It has been shown that U1snRNP is dispensable for in vitro splicing of some animal pre-mRNAs, and inactivation of U1-70K in yeast (Saccharomyces cerevisiae) is not lethal. As in yeast and humans (Homo sapiens), plant U1-70K is coded by a single gene. In this study, we blocked the expression of Arabidopsis U1-70K in petals and stamens by expressing U1-70K antisense transcript using the AP3 (APETALA3) promoter specific to these floral organs. Flowers of transgenic Arabidopsis plants expressing U1-70K antisense transcript showed partially developed stamens and petals that are arrested at different stages of development. In some transgenic lines, flowers have rudimentary petals and stamens and are male sterile. The severity of the phenotype is correlated with the level of the antisense transcript. Molecular analysis of transgenic plants has confirmed that the observed phenotype is not due to disruption of whorl-specific homeotic genes, AP3 or PISTILLATA, responsible for petal and stamen development. The AP3 transcript was not detected in transgenic flowers with severe phenotype. Flowers of Arabidopsis plants transformed with a reporter gene driven by the same promoter showed no abnormalities. These results show that U1-70K is necessary for the development of sepals and petals and is an essential gene in plants.
Most eukaryotic nuclear genes contain non-coding intervening sequences (introns; Goodall et al., 1991; Sharp, 1994). The introns in the precursor mRNAs are removed, and exons are joined in the nucleus by a process called pre-mRNA splicing to generate functional mRNAs (Sharp, 1994). In the recently completed Arabidopsis genome, it is predicted that 79% of nuclear genes contain one or more introns (Arabidopsis Genome Initiative, 2000; Reddy, 2001). Although plant and animal introns have some common structural features, plant introns differ from non-plant introns in many aspects, suggesting that the early events of spliceosome formation, especially the mechanisms involved in intron recognition in plants, may not be similar to yeast (Saccharomyces cerevisiae) and animals (Luehrsen et al., 1994; Schuler, 1998; Lorkovic et al., 2000; Reddy, 2001). Some pre-mRNAs with multiple introns display complex patterns of alternative splicing, thereby contributing to the proteomic complexity and an additional level of control of gene expression (Smith et al., 1989; Lorkovic et al., 2000; Reddy, 2001). In mammalian genomes, about 30% to 60% of genes produce multiple transcripts by alternative splicing (Hanke et al., 1999; Brett et al., 2000; Lee et al., 2003).
The splicing of nuclear pre-mRNA takes place in a large RNA-protein complex called the spliceosome. In most metazoans, and most likely in plants, there are two (major and recently discovered minor) types of spliceosomes that differ in their composition (for review, see Burge et al., 1999; Reddy, 2001). The major spliceosome contains four small nuclear ribonucleoprotein (snRNP) particles (U1, U2, U4/6, and U5) and a number of non-snRNP proteins (Will and Luhrmann, 1997; Staley and Guthrie, 1998; Burge et al., 1999). The assembly of the spliceosome on the pre-mRNA is an orderly process that involves a series of complex RNA-RNA, RNA-protein, and protein-protein interactions (Fu, 1995; Burge et al., 1999). The U1 snRNP recognizes the 5′ splice site in an ATP-independent manner to form a complex called early complex in mammalian cells or commitment complex in yeast that commits the pre-mRNA to spliceosome assembly. This complex formation involves base pairing between conserved nucleotides at the 5′ end of U1 snRNA and the 5′ splice site of pre-mRNA (Mount et al., 1983; Burge et al., 1999). In metazoans, U1 snRNP contains one U1 snRNA molecule and at least 11 proteins including three U1snRNP-specific proteins (U1-70K, U1-A, and U1-C), whereas the yeast U1snRNP contains an additional six U1snRNP-specific proteins (Fabrizio et al., 1994; Gottschalk et al., 1998). U1 snRNP-specific proteins are required for efficient formation of a complex between U1 snRNA and the 5′ splice site junction (Mount et al., 1983; Heinrichs et al., 1990; Rosbash and Séraphin, 1991).
U1 snRNP-specific protein, U1-70K, is involved in both basic and alternative splicing of pre-mRNAs in animals (Manley and Tacke, 1996; Will and Luhrmann, 1997). U1-70K interacts directly with splicing factors (ASF/SF2 and SC-35) of the SR family that are involved in splice site selection (Wu and Maniatis, 1993; Kohtz et al., 1994; Manley and Tacke, 1996; Will and Luhrmann, 1997). Furthermore, overexpression of U1-70K in cultured animal cells inhibits splicing and nucleocytoplasmic transport (Romac and Keene, 1995). However, Crispino et al. (1994) have shown that nuclear extracts depleted of U1 snRNP could be reconstituted by a high level of Ser/Arg-rich proteins. In another study, SR proteins or purified SC35 protein complemented splicing in extracts where the interaction of U1 snRNP with the 5′ splice site is blocked by antisense RNA (Tarn and Steitz, 1994). The yeast U1-70K has only 30% amino acid identity to human (Homo sapiens) U1-70K and lacks the Arg-rich region (Smith and Barrell, 1991; Kao and Siliciano, 1992). The yeast U1-70K was initially reported to be essential for yeast viability (Smith et al., 1989). Later, it was shown that the lack of U1-70K does not cause lethality; instead, it makes the mutant temperature sensitive and defective in pre-mRNA splicing but still viable (Hilleren et al., 1995). By rescuing the mutant phenotype, the most important functional region of yeast U1-70K was mapped to the amino-terminal part (1–97 amino acids). The other part of U1-70K containing the RNA-binding domain (also called RNA recognition motif [RRM]) and Gly-rich domain was unable to complement the mutant.
In plants, U1-70K has been characterized only from Arabidopsis. It is coded by a single gene, which produces two (short and long) transcripts by inclusion or exclusion of a 910-bp intron (Golovkin and Reddy, 1996). Both U1-70K transcripts are expressed in all tissues, and the level of the transcripts varied in different organs. The deduced amino acid sequence from the short transcript is similar to the animal U1-70K protein and contains an RRM, a Gly hinge, and an Arg-rich region characteristic of the animal U1-70K protein (Golovkin and Reddy, 1996). The long transcript has an in-frame translational termination codon within the 910-bp included intron and produces a truncated protein containing 204 amino acids with part of the RRM containing only RNP2. Plant U1-70K shares some characteristic features with animal U1-70K but differs in others and interacts with novel plant Ser/Arg-rich proteins (Golovkin and Reddy, 1998; 1999). Because the U1snRNP is dispensable in in vitro splicing of some pre-mRNAs and inactivation of U1-70K in yeast does not cause lethality (Crispino et al., 1994; Tarn and Steitz, 1994; Hilleren et al., 1995), we tested to see if it is dispensable in plants. To this end, we blocked the expression of U1-70K using an antisense construct driven by a strong floral organ-specific promoter (AP3 promoter) that is active only in two whorls (petals and stamens) of flowers. Here, we present the evidence that expression of U1-70K antisense transcript in sepals and petals aborts their development, suggesting that U1-70K is essential for the development of these organs.
RESULTS
Expression of U1-70K Antisense with AP3 Promoter Suppressed the Development of Petals and Stamens
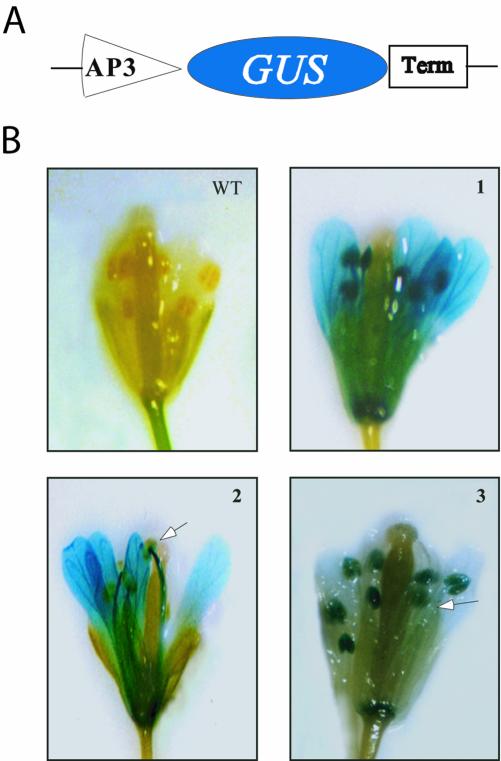
To determine if the U1-70K gene is necessary for development, we chose to block the expression of U1-70K in two floral organs (petals and stamens) using the AP3 promoter that is specific to these organs. The expression of AP3, a floral homeotic gene that is required for specifying petal and stamen identities, begins in young flowers in the precursor cells for the petals and stamens and continues in these organs throughout most of the flower development (Bowman et al., 1989; Jack et al., 1992). It has been shown that the fusion of the 3.7 kb of 5′ AP3 promoter sequence to green fluorescent protein, together with 1.5 kb of 3′ AP3 sequence downstream of GUS, results in expression of the reporter gene specifically in stamens and petals (Jack et al., 1994). In this work, we have used a shorter promoter sequence (approximately 700 bp) of AP3 gene without any 3′ sequence. To determine the activity of this 700-bp region of the AP3 promoter, we transformed Arabidopsis plants with β-glucuronidase (GUS) driven by this promoter (Fig. 1A). As shown in Figure 1B, the expression of reporter gene driven by the 700-bp promoter of AP3 is confined to only petals and stamens. However, the level of reporter gene expression in these organs varied among independent transgenic lines (see Fig. 1B). For example, in one plant GUS expression was very strong in petals and anther filaments with little expression in anther (see Fig. 1B, 2), whereas another line showed strong GUS expression in anthers and low expression in petals and anther filaments (Fig. 1B, 3). The strength of the AP3 promoter in these organs is comparable with the constitutive cauliflower mosaic virus 35 promoter (data not shown). Recently, Tilly et al. (1998) have shown that cis-elements necessary for petal and stamen expression reside within the 700-bp promoter region used in this study. It is worth mentioning that in over 200 transgenic plants containing the GUS reporter driven by the AP3 promoter, we did not observe any flower abnormalities.
Figure 1.
Analysis of 700-bp AP3 promoter activity in transgenic plants. A, Schematic diagram of the promoter-reporter fusion construct. AP3, 700-bp promoter region of AP3 gene; Term, NOS3 terminator. B, Flowers from wild type and three independent transgenic lines were stained for GUS activity. The arrows indicate the parts of the stamens with little or no GUS expression.
To block the expression of U1-70K efficiently, we fused the AP3 promoter (approximately 700-bp region) with the longest cDNA of Arabidopsis U1-70K comprising the entire coding region, 910-bp alternative intron, and extended untranslated region (Golovkin and Reddy, 1996) and used this construct to transform Arabidopsis (Fig. 2A). More than 100 kanamycin-resistant transgenic plants were obtained. Among these, 11% of the plants revealed abnormal flowers that had either rudimentary or partially developed petals and stamens arrested at different stages of their development (Figs. 2 and 3). The number and position of the sepals and carpel remained normal in all transgenic flowers. In seven independent transgenic plants with severe flower phenotype, only sepals and carpel were visible in mature flowers. The inflorescence and a single flower from wild type and one such transgenic plant are shown in Figure 2B. The lines with severe phenotype were completely male sterile. As shown in Figure 3A, in some of the transgenic lines, underdeveloped petal and stamens were present but hidden under sepals. However, flowers of transgenic plants rarely had carpel-like features in place of stamens (Fig. 3B). Transgenic lines with severe phenotype (male sterile) produced viable seeds when pollinated with pollen from wild-type plants, and the kanamycin-resistant plants from such crosses also produced abnormal flowers with rudimentary petals and stamens.
Figure 2.
Targeted expression of U1-70K antisense transcript using the AP3 promoter. A, Schematic diagram of U1-70K antisense cassette used to generate transgenic plants. AP3, Promoter (700 bp) from AP3 gene; U1-70K antisense, U1-70K long cDNA in antisense orientation; Term, NOS3 terminator. B, Expression of U1-70K antisense transcript results in flowers with rudimentary petals and stamens. Left, Inflorescence of a wild-type plant (top) and wild-type flower. Right, Inflorescence of a transgenic plant expressing U1-70K antisense transcript (top) and flower from a transgenic plant with rudimentary petals and stamens (bottom).
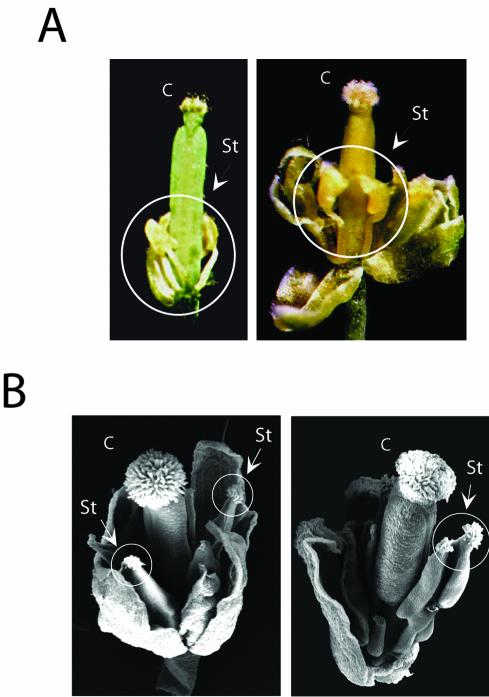
Figure 3.
Variations in the phenotype of flowers expressing U1-70K antisense transcript. A, Opened flowers from two different U1-70K antisense transgenic plants showing variations in stamen development. B, Scanning electron micrograph showing a rarely observed flower phenotype from a transgenic plant with carpel-like structures in place of stamens. Stigmatic papillae on stamens are circled. C, Carpel; St, stamen.
The Severity of Flower Phenotype Is Correlated with the Level of U1-70K Antisense Transcript
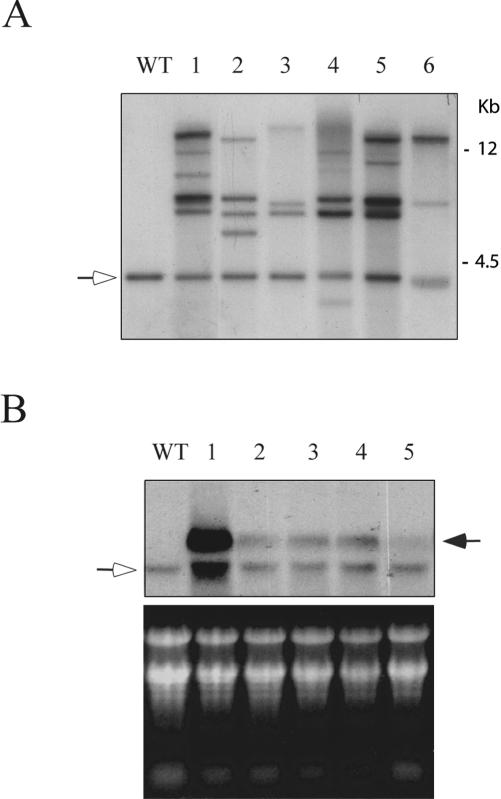
To determine the number of copies of the introduced gene, we performed Southern analysis with independent transgenic lines. The genomic DNA was digested with BamHI, which does not cut the endogenous gene and the introduced AP3/U1-70K antisense cassette. As shown in Figure 4A, the wild-type and transgenic lines yielded the expected size endogenous band. In addition, each transgenic line carried multiple copies of the introduced gene and the number of copies varied significantly from plant to plant (from two in line 6 to several in line 1). To test if the observed flower phenotype correlated with the level of the antisense transcript, we monitored the level of the expression of the introduced gene in five lines. Of these, transgenic line 1 showed severe flower phenotype; lines 2, 3, and 4 showed partially developed stamens; and line 5 showed visibly normal flowers. RNA from flowers and flower buds from wild-type and transgenic plants was probed with a 910-bp fragment that is specific to the long transcript and, therefore, is expected to detect the endogenous long transcript and AP3-driven antisense transcript. The level of the antisense transcript varied considerably between different transgenic plants (Fig. 4B). Plant 1 with severe phenotype (very rudimentary petals and stamens in mature flowers) has a very high level of antisense transcript, whereas the flowers with less severe phenotype (lanes 2–4) showed lower levels of antisense transcript. Flowers from line 5 with visibly normal flowers showed barely detectable levels of antisense transcript. These results show a strong correlation between the level of U1-70K antisense overexpression with the phenotype severity. The flowers of transgenic progeny in the T1 and T2 generations that showed abnormal flower phenotype also showed high levels of antisense transcript (data not shown). Transgenic lines expressing a very high level of U1-70K antisense transcript showed additional smaller size transcripts, including a transcript corresponding to the endogenous transcript (see Fig. 4B, lane 1), which are likely due to the degradation of overexpressed transcript.
Figure 4.
Molecular characterization of transgenic plants. A, DNA gel-blot analysis. Genomic DNA from the wild type (Wt) and six independent transgenic plants was digested with BamHI and probed with a U1-70K. Arrow indicates the endogenous U1-70K gene. B, Expression of antisense U1-70K transcript in flowers of transgenic plants. RNA from wild type (Wt) and five transgenic plants was hybridized with the 910-bp “alternative intron” fragment that is present only in the large transcript of plant U1-70K (top). A hollow arrow indicates expression of endogenous U1-70K. Solid arrow indicates the level of antisense transcript. Stained gel with ribosomal RNA is shown in the bottom panel.
The Observed Flower Phenotypes Are Not Due to the Disruption of Homeotic Genes, AP3 or PI (PISTILLATA), That Control the Development of Petals and Stamens
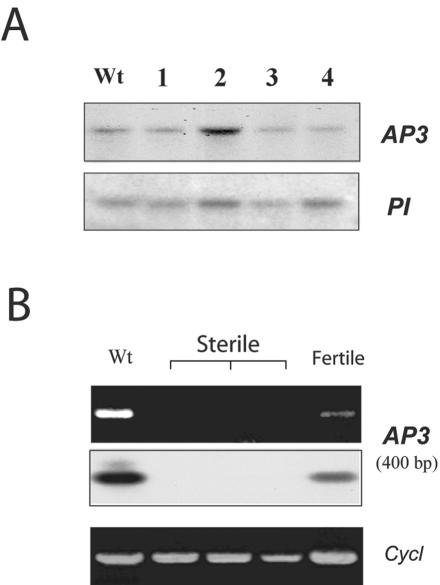
Floral organ identity is based on the combinatorial activities of three classes (ABC) of homeotic genes (Coen and Meyerowitz, 1991; Clark and Meyerowitz, 1994). Each class of genes acts in two adjacent whorls in a combinatorial fashion to specify organ identity. Class A genes are active in sepals and petals, Class B genes control petals and stamens development, and Class C genes are active in stamens and carpels. Expression of A and B leads to petal development, whereas the expression of B and C leads to stamen development. There are two class B genes (AP3 and PI) in Arabidopsis (Bowman et al., 1991; Coen and Meyerowitz, 1991). The expression of Class B genes, AP3 and PI, is restricted to petals and stamens (Hill and Lord, 1989; Jack et al., 1992). Mutations in Class B genes result in flowers that contain sepals in place of petals and carpels in place of stamens (Bowman et al., 1991). Thus, the disruption of the normal function of the B class genes (AP3 or PI) leads to the development of abnormal flowers. It is possible but very unlikely that the integration of introduced gene into either AP3 or PI could have caused the phenotype that we observed. To rule out this possibility, we verified the integrity of AP3 and PI genes. We hybridized BamHI-digested genomic DNA of the individual transgenic plants with the 7.5-kb BamHI DNA fragment of the AP3 gene, which contains the entire gene and its promoter or PI cDNA. As shown in Figure 5A, a single expected size fragment hybridized in both wild-type and transgenic plants with both probes, suggesting that AP3 and PI genes are intact in the transgenic plants.
Figure 5.
Analysis of AP3 and PI genes in plants expressing U1-70K antisense transcript. A, DNA-blot analysis of transgenic plants with AP3 and PI genes. Genomic DNA from wild type (Wt) and four transgenic plants (lanes 1–4) was digested by BamHI and hybridized with AP3 and PI genes. The size of the BamHI fragment is about 7.5 kb with the APETALA3 probe and 12 kb with the PISTILLATA probe. Based on the bacterial artificial chromosome sequence in this region, the hybridizing bands correspond to the expected sizes and contain the whole gene and the promoter. B, Expression of AP3 in transgenic plants. Total RNA from buds and flowers of wild type (Wt) and transgenic plants with severe phenotype (sterile) and fertile (fertile) were used for reverse transcriptase (RT)-PCR analysis using AP3-specific forward and reverse primers corresponding to exons one and seven. Top, Stained gel of amplified products. Middle, Hybridization of a blot prepared from the top gel with 32P-labeled AP3-specific probe. Bottom, Amplification of cyclophilin (Cycl) by RT-PCR showing the presence of template in all reactions.
Petal- and Stamen-Specific Expression of U1-70K Antisense Transcript Affected the Level of AP3 mRNA in These Organs
Because the AP3 gene is intact in the transgenic lines, we expect it to be expressed in flowers with partially developed petals and stamens. It has been shown previously that the AP3 promoter is active throughout the development of petals and stamens (Jack et al., 1994; Tilly et al., 1998). Furthermore, the maintenance of AP3 expression is dependent on its own transcription (autoregulation; Goto and Meyerowitz, 1994; Jack et al., 1994). In rudimentary petals and stamens of transgenic plants with high levels of antisense transcript, we expect severe impairment of splicing due to low or no U1-70K, resulting in little or no processed mRNA. Hence, we tested the level of AP3 mRNA in these transgenic floral tissues using RT-PCR analysis. AP3 pre-mRNA contains six introns that are spliced to produce functional mRNA in petals and stamens. Using primers corresponding to the first and seventh exons of the AP3 gene, we detected the expected size (about 400 bp) PCR-amplified product in the RT sample from the RNA of the wild type and the transgenic flowers with less severe (fertile) phenotype (Fig. 5B). However, in RT samples obtained from the floral RNA of severe phenotype (sterile), no PCR products were detected (Fig. 5B). Hybridization with the same AP3-specific probe (7.5-kb BamHI fragment) confirmed these results (Fig. 5B, middle). These data suggest that there is no mature mRNA from AP3 gene present in transgenic flowers with severe phenotype, whereas there is a high level of AP3-driven U1-70K antisense product. The presence of first strand cDNA in all samples was verified by amplifying cyclophilin (Fig. 5B, bottom; Golovkin and Reddy, 1999). These results show a strong correlation between the disappearance of mature AP3 transcripts in flowers with a severe phenotype and a high level of antisense U1-70K transcript. The probable reason for the lack of AP3 transcripts in flowers with severe phenotype is that the unspliced AP3 mRNA is unstable and undergoes degradation. Alternatively, it is possible that the AP3 promoter of the transgenic construct may compete with the endogenous AP3 promoter for the trans-acting factors such as AP1, AP3, PI, and AG that are known to interact with the AP3 promoter (Hill et al., 1998), resulting in no expression of AP3 mRNA. The fact that we did not observe ap3 mutant phenotype in about 200 transgenic plants transformed with GUS reporter driven by AP3 promoter indicates that the second possibility is very unlikely.
DISCUSSION
In metazoans, U1-70K has been shown to interact with Ser/Arg-rich splicing factors of the SR family (SC35, ASF/SF2, and Sip1) and control both basic and alternative splicing (Wu and Maniatis, 1993; Kohtz et al., 1994; Manley and Tacke, 1996; Zhang and Wu, 1998). The yeast U1-70K also interacts with a large number of proteins (Fromont-Racine et al., 1997). The Arabidopsis U1-70K shares significant similarity with human and metazoan U1-70K and interacts with a novel set of proteins including SR45, a plant-specific SR protein, two 9G8-like splicing factors, and an SC35-like protein (Golovkin and Reddy, 1998, 1999; Lopato et al., 1999), suggesting the plant U1-70K is also likely to regulate basic and alternative splicing of pre-mRNAs. Because U1-70K interacts with several other proteins involved in splicing, it was suggested that U1-70K serves as an assembly focus for the functional components of the splicing/transport machinery (such as SR proteins) and plays a key regulatory role in early events of spliceosome assembly (Romac and Keene, 1995; Golovkin and Reddy, 1999; Lopato et al., 1999). Hence, it is reasonable to expect that expression of U1-70K antisense transcript would inhibit splicing of many pre-mRNAs and the production of corresponding proteins resulting in adverse affects on cellular processes. In fact, transient overexpression of full-length and the truncated versions of U1-70K in cultured animal cells affected the nuclear distribution of splicing factor SC35 and led to the inhibition of both splicing and nucleocytoplasmic transport of mRNA (Romac and Keene, 1995). Quantitative analysis of nuclear and cytoplasmic RNA revealed an increase of the amount of both intronless and unspliced nuclear messages due to U1-70K overexpression. Interestingly, overexpression of the carboxy-terminal part of U1-70K alone showed a similar effect. These results suggest that excess of a splicing component can cause an imbalance in splicing factors and disruption at the very early stages of spliceosome formation. A yeast strain lacking the U1-70K can grow at low temperature, suggesting that U1-70K is dispensable in yeast (Hilleren et al., 1995). However, it is not known if U1-70K is dispensable in multicellular eukaryotes in which both basic and alternative splicing play an important role in growth and development.
In this study, we addressed this question by expressing the U1-70K antisense transcript in a tissue-specific manner in petals and stamens to determine if it is essential for the development of these organs. Because U1-70K is coded by a single gene in Arabidopsis (Golovkin and Reddy, 1996; Arabidopsis Genome Initiative, 2000) it is an ideal candidate to block the expression using an antisense approach. Several reports have shown that inhibition of expression of endogenous genes in plants can be achieved at a reasonable frequency by overexpressing the antisense transcript corresponding to the endogenous gene (Kuipers et al., 1997). Our antisense approach to block U1-70K in specific organs (petal and stamens) resulted in suppression of development of these organs, suggesting that in plants, and most likely in other multicellular organisms, U1-70K is essential. Several lines of evidence presented here indicate that the observed phenotype of flowers is due to expression of U1-70K antisense transcript and not due to disruption of the AP3 or PI gene. First, Southern analysis with these genes verified that they are intact in transgenic plants (Fig. 5A). Second, the severity of the phenotype was correlated with the extent of expression of U1-70K antisense transcript in petals and stamens (Fig. 4B). Third, unlike in mutants of AP3 or PI where petals and stamens are transformed to sepals and carpels, respectively, flowers of transgenic plants expressing U1-70K antisence RNA had either rudimentary or partially developed petals and stamens (Fig. 3A). Fourth, several independent transgenic lines showed similar phenotype. Finally, we did not observe this phenotype in hundreds of transgenic plants that are transformed with the same promoter fused to a GUS reporter gene. Taken together, our results clearly show that the targeted expression of U1-70K antisense transcript in petals and stamens effectively suppresses their development. Based on the role of U1-70K, it is likely that low or no expression of U1-70K adversely affects the splicing of pre-mRNAs, thereby altering the levels of several proteins.
Expression of antisense genes in transgenic plants has been used successfully in plants to block expression of the corresponding genes (Chuang and Meyerowitz, 2000). The reductions in endogenous mRNA, protein, and/or enzyme activity level could vary greatly in different transgenic lines (Kuipers et al., 1997; Chuang and Meyerowitz, 2000). Here, we observed the expected phenotype only in 11% of transgenic lines. Furthermore, although the lines with abnormal flowers were all male sterile, the extent to which petals and stamens developed varied. In some lines, flowers showed very rudimentary petals and stamens, whereas in others, these organs were partially developed. The observed variation in phenotype is due to the level of expression of antisense transcript because high levels of antisense transcript correlated with the severe phenotype. The reason for strong level of expression in some plants but not in others could be due to the number of copies of the introduced gene, the influence of adjacent plant genomic DNA sequences, and/or chromosomal structure at the integration site (Peach and Velten, 1991). Variability in expression level of introduced gene and its phenotype is a general feature of antisense lines (Kuipers et al., 1997; Chuang and Meyerowitz, 2000). Similar variations in promoter activity were observed with other promoters also (Peach and Velten, 1991). As shown in Figure 1, with the AP3-GUS construct there is significant difference in the strength and specificity of the expression in petals and stamens. The appearance of carpel-like structures rarely in some flowers could be caused by splicing alterations due to partial suppression of U1-70K, which may have resulted in the unbalanced expression of homeotic flower development genes (Jack et al., 1992; Goto and Meyerowitz, 1994). The presence of very rudimentary petals and stamens suggests that either the expression of U1-70K is not completely blocked by antisense RNA or the U1-70K protein present in the precursor cells of stamens and petals, before activation of the AP3 promoter, may be sufficient for the development of these rudimentary organs. The latter possibility is most likely if the U1-70K protein is stable with a long half-life. Despite the fact that the depletion of U1snRNP in in vitro splicing extracts derived from animal cells can be complemented with an excess amount of a single SR protein or mixture of SR proteins (Crispino et al., 1994; Tarn and Steitz, 1994), our in vivo studies show that U1-70k is essential in plants and likely in other multicellular organisms.
MATERIALS AND METHODS
Preparation of U1-70K Antisense Construct
We used pCGN1547-based pLB215 plasmid (Jack et al., 1994) to prepare U1-70K antisense construct. The bacterial reporter gene uidA (GUS) in pLB215, which is between the APETALA3 (AP3) promoter and NOS3 terminator, was replaced with the long cDNA of U1-70K in antisense orientation. First, a fragment containing both the AP3 promoter and GUS reporter was removed from pLB215 plasmid by partial XbaI/BamHI digestion and placed into SpeI/XbaI of pBluescript (KS) plasmid. The GUS gene was then excised from this construct by BsmI/SpeI digestion. The resulting plasmid contained the AP3 promoter and a small (50-bp) GUS fragment with a unique BamHI site. The 750-bp AP3 promoter from this construct was released by XbaI/BamHI digestion and subcloned back into the same XbaI/BamHI pLB215 vector from the first step. The resulting plasmid contained AP3 promoter and NOS3 terminator with a convenient BamHI site in between. The Arabidopsis U1-70K cDNA from the large transcript (Golovkin and Reddy, 1996) was obtained by ApaI/SmaI digestion. The fragment was blunted with T4 polymerase and inserted into the unique BamHI site (blunted by Klenow) to generate the pAP3-U1-70K antisense construct.
Generation of Transgenic Plants
A vacuum infiltration procedure was used to generate Arabidopsis transgenic plants essentially as described by Bechtold et al. (1993). Plasmid constructs (AP3-GUS or AP3-U1-70K) were introduced into Agrobacterium tumefaciens strain ASE by electroporation using a Cell Porator (Invitrogen, Carlsbad, CA), and the transformants were selected on Luria-Bertani medium plates with antibiotics (kanamycin, chloramphenicol, and gentamycin). Three- to four-week-old Arabidopsis (Columbia) plants were infiltrated and grown to maturity. Seeds were collected, dried for at least 10 d, sterilized, and stratified at 4°C in a 0.1% (w/v) agarose solution for 3 d. Seeds were then spread on selection plates (Murashige and Skoog medium containing 1% [w/v] Suc, 50 μg mL–1 kanamycin, and 500 μg mL–1 timentin) and grown under continuous light. Plants that grew on these plates were transferred to fresh plates and finally to soil. About 100 transgenic plants were obtained and analyzed. Using cross-pollination with pollen from wild-type Arabidopsis (Columbia) flowers, we were able to maintain the mutant phenotype of these plants for several generations. Flowers from transgenic plants carrying the GUS reporter driven by the AP3 promoter were stained for GUS activity.
Scanning Electron Microphotography
Young inflorescences were incubated in 2% (w/v) glutaraldehyde in 0.025 m sodium phosphate (pH 7.0) at 4°C. Samples were transferred to 1% (w/v) osmium tetroxide in 0.025 m sodium phosphate buffer (pH 7.0) for 4 to 6 h at room temperature and in 0.05 m sodium cocodylate buffer (pH 7.0) for 12 h. Dehydration was performed through a standard ethanol battery (50%, 70%, 80%, 90%, and twice at 100% [v/v]) and then dried in liquid CO2. Individual flowers were removed from the inflorescence and mounted on a scanning electron microphotography table. Flower buds were carefully opened using glass needles and then coated with gold (4:1 [w/v]) and photographed at an accelerating voltage of 20 kV.
DNA and RNA Gel-Blot Analyses
DNA gel-blot analysis of individual transgenic plants was performed as described earlier (Golovkin and Reddy, 1996). Genomic DNA was digested with BamHI, which leaves the U1-70K intact. Ten micrograms of genomic DNA was separated in a 0.8% (w/v) Tris-acetate EDTA agarose gel, blotted onto a Hybond N+ (Amersham, Buckinghamshire, UK) nylon filter, and probed with a 1.1-kb fragment of 32P-labeled U1-70K cDNA. Hybridization was carried out for 3 h at 65°C. Total RNA from flowers and flower buds was isolated using “Trizol” (Life Technologies/Gibco-BRL). RNA (50 μg) was resolved in a formaldehyde-containing agarose gel and transferred onto a Hybond N+ (Amersham) nylon filter. The 910-bp PCR product of an “alternative intron” from the Arabidopsis U1-70K gene was used to detect both the U1-70K antisense transcript and the endogenous U1-70K large transcript.
To exclude the possibility that flower developmental genes are disrupted by T-DNA integration, we used the Arabidopsis AP3 gene and the PI cDNA probes to hybridize with the same blots of BamHI-digested genomic DNA.
RT-PCR Analysis for AP3 Gene Expression
One microgram of total RNA from wild type and transgenic plants was treated with RNAse-free DNAse and used to synthesize first strand cDNA. Primers 5′-CGATTATCATGTTCTCTAGCTCC-3′ and 3′-TTGTCTACTAGTCCATAGTGAGG-5′ specific to the first and seventh exon of AP3 gene were used to amplify a approximately 400-bp fragment of AP3 transcript. The PCR reactions were performed using the “Expand Long template” PCR system (Boehringer Mannheim/Roche, Basel) in a final volume of 50 μL. The first six cycles of PCR consisted of 92°C for 1 min, 58°C for 30 s, and 68°C for 3 min, and the next 29 cycles consisted of 92°C for 1 min, 60°C for30 s, and 68°C for 3 min with a 10-s gradient increase in extension time. The amplified products were separated in a 1% (w/v) agarose gel and blotted onto a nylon membrane. A 7.5-kb BamHI fragment of the AP3 gene from pD275 plasmid was used as a probe to detect the amplified 400-bp product. The amount of first strand cDNA in each reaction was verified using the primers (forward, 5′-GTC TGATAGAGATCTCACGT-3′; and reverse, 5′–AATCGGCAACAACCACAG GC-3′) corresponding to a constitutively expressed cyclophilin gene (Golovkin and Reddy, 1999).
Acknowledgments
We thank Dr. Thomas Jack for providing the AP3 gene and A. tumefaciens ASE strain and Dr. Day for her critically reading the manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.023192.
This work was supported by the Department of Energy, Division of Energy Biosciences (grant no. DE–FG03–01ER15199 to A.S.N.R.).
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci 316: 1194–1199 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM (1989) Genes directing flower development in Arabidopsis. Plant Cell 1: 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM (1991) Genetic interactions among floral homeotic genes of Arabidopsis. Development 112: 1–20 [DOI] [PubMed] [Google Scholar]
- Brett D, Hanke J, Lehmann G, Haase S, Delbruck S, Krueger S, Reich J, Borka P (2000) EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett 474: 83–86 [DOI] [PubMed] [Google Scholar]
- Burge CB, Tushl T, Sharp PA (1999) Splicing of precursors to mRNAs by the spliceosomes. In RF Gesteland, TR Cech, JF Atkins, eds, The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 525–560
- Chuang CF, Meyerowitz EM (2000) Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA 97: 4985–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Meyerowitz EM (1994) Arabidopsis flower development. In EM Meyerowitz, CR Somerville, eds, Arabidopsis. Cold Spring Harbor Press, Cold Spring Harbor, NY, pp 435–466
- Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Crispino JD, Blencowe BJ, Sharp PA (1994) Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science 265: 1866–1869 [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Esser S, Kastner B, Lührmann R (1994) Isolation of S. cerevisiae snRNPs: comparison of U1 and U4/U6.U5 to their human counterparts. Science 264: 261–265 [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M, Rain J-C, Legrain P (1997) Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet 16: 277–282 [DOI] [PubMed] [Google Scholar]
- Fu X (1995) The superfamily of arginine/serine-rich splicing factors. RNA 1: 663–680 [PMC free article] [PubMed] [Google Scholar]
- Golovkin M, Reddy AS (1998) The plant U1 small nuclear ribonucleoprotein particle 70K protein interacts with two novel serine/arginine-rich proteins. Plant Cell 10: 1637–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkin M, Reddy AS (1999) An SC35-like protein and a novel serine/arginine-rich protein interact with Arabidopsis U1-70K protein. J Biol Chem 274: 36428–36438 [DOI] [PubMed] [Google Scholar]
- Golovkin M, Reddy ASN (1996) Structure and expression of a plant U1 snRNP 70K gene: Alternative splicing of U1 snRNP 70K pre-mRNAs produces two different transcripts. Plant Cell 8: 1421–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall GJ, Kiss T, Filipowicz W (1991) Nuclear RNA splicing and small nuclear RNAs and their genes in higher plants. Oxford Surv Plant Mol Cell Biol 7: 255–296 [Google Scholar]
- Goto K, Meyerowitz EM (1994) Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev 13: 1548–1560 [DOI] [PubMed] [Google Scholar]
- Gottschalk A, Tang J, Puig O, Salgado J, Neubauer G, Colot HV, Mann M, Seraphin B, Rosbash M, Luhrmann R et al. (1998) A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA 4: 374–393 [PMC free article] [PubMed] [Google Scholar]
- Hanke J, Brett D, Zastrow I, Aydin A, Delbruck S, Lehmann G, Luft F, Reich J, Bork P (1999) Alternative splicing of human genes: more the rule than the exception? Trends Genet 15: 389–390 [DOI] [PubMed] [Google Scholar]
- Heinrichs V, Bach M, Winkelmann G, Lührmann R (1990) U1-specific protein C needed for efficient complex formation of U1 snRNP with a 5′ splice site. Science 247: 69–72 [DOI] [PubMed] [Google Scholar]
- Hill JP, Lord EM (1989) Floral development in Arabidopsis thaliana: a comparison of the wild-type and the homeotic pistillata mutant. Can J Bot 67: 2922–2936 [Google Scholar]
- Hill TA, Day CD, Zondlo SC, Thackeray AG, Irish VF (1998) Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development 125: 1711–1721 [DOI] [PubMed] [Google Scholar]
- Hilleren PJ, Kao HY, Siliciano PG (1995) The amino-terminal domain of yeast U1-70K is necessary and sufficient for function. Mol Cell Biol 15: 6341–6350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM (1992) The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68: 683–697 [DOI] [PubMed] [Google Scholar]
- Jack T, Fox GL, Meyerowitz EM (1994) Arabidopsis homeotic gene APETALA3 ectopic expression: transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76: 703–716 [DOI] [PubMed] [Google Scholar]
- Kao H-Y, Siliciano PG (1992) The yeast homolog of the U1 snRNP protein 70K is encoded by the SNP1 gene. Nucleic Acids Res 20: 4009–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohtz JD, Jamison SF, Will CL, Zuo P, Lührmann R, Garcia-Blanco MA, Manley JL (1994) Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 368: 119–124 [DOI] [PubMed] [Google Scholar]
- Kuipers AGJ, Jacobsen E, Visser RGF (1997) Application of antisense technology in plants. In C Lichtenstein, W Nellen, eds, Antisense Technology: A Practical Approach. IRL Press, Oxford, pp 191–219
- Lee C, Atanelov L, Modrek B, Xing Y (2003) ASAP: the alternative splicing annotation project. Nucleic Acids Res 31: 101–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato S, Gattoni R, Fabini G, Stevenin J, Barta A (1999) A novel family of plant splicing factors with a Zn knuckle motif: examination of RNA binding and splicing activities. Plant Mol Biol 39: 761–773 [DOI] [PubMed] [Google Scholar]
- Lorkovic ZJ, Wieczorek Kirk DA, Lambermon MH, Filipowicz W (2000) Pre-mRNA splicing in higher plants. Trends Plant Sci 5: 160–167 [DOI] [PubMed] [Google Scholar]
- Luehrsen KR, Taha S, Walbot V (1994) Nuclear pre-mRNA processing in higher plants. Nucleic Acids Res 47: 149–193 [DOI] [PubMed] [Google Scholar]
- Manley JL, Tacke R (1996) SR proteins and splicing control. Genes Dev 10: 1569–1579 [DOI] [PubMed] [Google Scholar]
- Mount SM, Pettersson I, Hinterberger M, Karmas A, Steitz JA (1983) The U1 small nuclear RNA-protein complex selectively binds a 5′ splice site in vitro. Cell 33: 509–518 [DOI] [PubMed] [Google Scholar]
- Peach C, Velten J (1991) Transgene expression variability (position effect) of CAT and GUS reporter genes driven by linked divergent T-DNA promoters. Plant Mol Biol 17: 49. [DOI] [PubMed] [Google Scholar]
- Reddy ASN (2001) Nuclear pre-mRNA splicing in plants. CRC Crit Rev Plant Sci 20: 523–571 [Google Scholar]
- Romac JM, Keene JD (1995) Overexpression of the arginine-rich carboxy-terminal region of U1 snRNP 70K inhibits both splicing and nucleocytoplasmic transport of mRNA. Genes Dev 9: 1400–1410 [DOI] [PubMed] [Google Scholar]
- Rosbash M, Séraphin B (1991) Who's on first? The U1 snRNP-5′ splice site interaction and splicing. Trends Biochem Sci 16: 187–190 [DOI] [PubMed] [Google Scholar]
- Schuler MA (1998) Plant pre-mRNA splicing. In J Bailey-Serres, DR Gallie, eds, A Look Beyond Transcription: Mechanisms Determining mRNA Stability and Translation in Plants. American Society of Plant Physiologists, Rockville, MD, pp 1–19
- Sharp PA (1994) Split genes and RNA splicing. Cell 77: 805–815 [DOI] [PubMed] [Google Scholar]
- Smith CWJ, Patton JG, Nadal-Ginard B (1989) Alternative splicing in the control of gene expression. Annu Rev Genet 23: 527–577 [DOI] [PubMed] [Google Scholar]
- Smith V, Barrell BG (1991) Cloning of a yeast U1 snRNP 70K protein homologue: Functional conservation of an RNA-binding domain between humans and yeast. EMBO J 10: 2627–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C (1998) Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92: 315–326 [DOI] [PubMed] [Google Scholar]
- Tarn W-Y, Steitz JA (1994) SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes Dev 8: 2704–2717 [DOI] [PubMed] [Google Scholar]
- Tilly JJ, Allen DW, Jack T (1998) The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development 125: 1647–1657 [DOI] [PubMed] [Google Scholar]
- Will CL, Luhrmann R (1997) Protein functions in pre-mRNA splicing. Curr Opin Cell Biol 9: 320–328 [DOI] [PubMed] [Google Scholar]
- Wu JY, Maniatis T (1993) Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75: 1061–1070 [DOI] [PubMed] [Google Scholar]
- Zhang W-J, Wu JY (1998) Sip1, a novel RS domain-containing protein essential for pre-mRNA splicing. Mol Cell Biol 18: 676–684 [DOI] [PMC free article] [PubMed] [Google Scholar]