Abstract
The Pto gene encodes a serine/threonine protein kinase that confers resistance in tomato (Lycopersicon esculentum) to Pseudomonas syringae pv tomato strains that express the type III effector protein AvrPto. Constitutive overexpression of Pto in tomato, in the absence of AvrPto, activates defense responses and confers resistance to several diverse bacterial and fungal plant pathogens. We have used a series of gene discovery and expression profiling methods to examine the effect of Pto overexpression in tomato leaves. Analysis of the tomato expressed sequence tag database and suppression subtractive hybridization identified 600 genes that were potentially differentially expressed in Pto-overexpressing tomato plants compared with a sibling line lacking Pto. By using cDNA microarrays, we verified changes in expression of many of these genes at various time points after inoculation with P. syringae pv tomato (avrPto) of the resistant Pto-overexpressing line and the susceptible sibling line. The combination of these three approaches led to the identification of 223 POR (Pto overexpression responsive) genes. Strikingly, 40% of the genes induced in the Pto-overexpressing plants previously have been shown to be differentially expressed during the human (Homo sapiens) and/or fruitfly (Drosophila melanogaster) immune responses.
Both plants and animals are continually exposed to pathogens, and, as a result, they have evolved intricate defense mechanisms to recognize and defend themselves against a wide array of these disease-causing agents. Recent studies have revealed common mechanisms of pathogen virulence and host resistance underlying plant and animal diseases (Cohn et al., 2001; Staskawicz et al., 2001; Nurnberger and Brunner, 2002). For example, bacterial pathogens of both plants and animals deliver virulence proteins into the host cytoplasm via the type III secretion system (He, 1998; Galan and Collmer, 1999). Pathogen-associated molecular patterns that are similar to those activating immune responses in animals have been shown to mediate activation of plant defenses (Nurnberger and Brunner, 2002). Some plant disease resistance (R) proteins share motifs with signaling components of immune response pathways in mammals and invertebrates (Staskawicz et al., 2001). A striking similarity underlying the molecular mechanisms of pathogen recognition in plants and animals is suggested by the recent isolation of the intracellular receptors NOD1 and NOD2 involved in human (Homo sapiens) Crohn's disease (Ogura et al., 2001a). In common with a large family of plant R proteins, the NOD1/2 proteins contain a putative nucleotide-binding site and a region of Leu-rich repeats (Ogura et al., 2001b).
In tomato (Lycopersicon esculentum), the R gene Pto encodes a Ser/Thr kinase and confers resistance against strains of Pseudomonas syringae pv tomato that express the effector proteins AvrPto or AvrPtoB (Martin et al., 1993; Kim et al., 2002). Overexpression of Pto in tomato under control of the cauliflower mosaic virus (CaMV) 35S promoter has been shown to activate defense responses in the absence of pathogen inoculation. These responses include PR gene expression, an increased level of salicylic acid (SA), and development of spontaneous microscopic lesions similar to the hypersensitive response (HR; Tang et al., 1999; Xiao et al., 2001). Pto-overexpressing plants show resistance not only to P. syringae pv tomato but also to Xanthomonas campestris pv vesicatoria and to the fungal pathogen Cladosporium fulvum. Thus, they exhibit an exaggerated form of the typical plant defense response, making them a useful model to dissect the complicated plant responses that occur during race-nonspecific resistance to several pathogens (Tang et al., 1999). Recently, Xiao et al. (2001) identified a limited number of genes that are expressed constitutively (without inoculation by P. syringae pv tomato expressing avrPto) in the Pto-overexpressing tomato line. Many of the genes encode known PR proteins, confirming that overexpression of Pto activates bona fide defense responses in plants.
We have now used suppression subtractive hybridization (SSH; Diatchenko et al., 1996), an in-depth analysis of the tomato expressed sequence tag (EST) database, and cDNA microarray analysis to characterize more fully the alterations in gene expression caused by the overexpression of Pto. Interestingly, over 40% of the genes that are highly expressed in Pto-overexpressing plants either with or without P. syringae pv tomato infection are also known to be induced in human and/or fruitfly (Drosophila melanogaster) during various defense responses. These findings indicate that many downstream defense responses in plants, humans, and invertebrates, as seen for certain recognition and signaling components, are shared and, thus, are either highly conserved or may have arisen by convergent evolution.
RESULTS
Use of in Silico EST Subtraction to Identify Genes That Are Differentially Expressed upon P. syringae pv tomato Inoculation in Tomato Leaves Overexpressing Pto
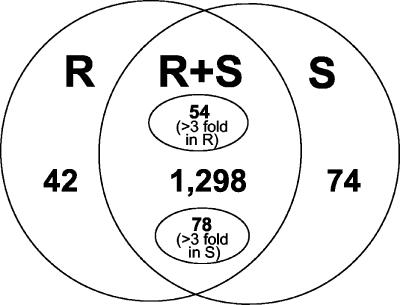
As one method of identifying genes that might be differentially expressed in response to inoculation with P. syringae pv tomato(avrPto), we analyzed the tomato EST database for sequences derived from line R11-12 that expresses the Pto gene from the CaMV 35S promoter and a sibling line R11-13 that has segregated away this transgene. As described in “Materials and Methods,” both lines were inoculated with the avirulent P. syringae pv tomato(avrPto), and leaves were harvested at various time points afterward. We analyzed 10,872 EST sequences from the R11-12 (R) and R11-13 (S) libraries (5,316 for R library and 5,556 for S library). A total of 6,921 of these EST sequences assembled into 1,414 contigs that contained either ESTs all derived from one of the libraries (116 contigs with at least three ESTs; 42 contained ESTs only from the R library, whereas 74 contigs contained ESTs only from the S library; Fig. 1) or a mix of ESTs from both libraries (1,298 contigs). The 3,953 singleton ESTs were excluded from further analysis. Of the 1,298 contigs with mixed R and S sequences, we identified 132 cases where the EST counts within a contig differed by more than 3-fold between R and S libraries (Fig. 1). An additional 51 contigs were identified that encoded known defense genes and that had counts between the libraries differing by more than 2-fold. To test if the genes corresponding to these ESTs were differentially expressed in P. syringae pv tomato-inoculated R11-12 and R11-13 leaves, we selected a representative cDNA clone from each of the 299 contigs (116 cases with at least three ESTs coming only from either R or S library, 132 cases with >3-fold differences, and the 51 cases with >2-fold differences) for microarray analysis (see below).
Figure 1.
Identification of differentially expressed genes by in silico EST subtraction. ESTs derived from the R11-12 (R) and the R11-13 (S) libraries were assembled into contigs. Within each contig, the number of ESTs derived from the R or S libraries was then calculated. Those contigs (with at least three ESTs) with ESTs from only the R library are shown on the left, and those with ESTs from only the S library are shown on the right. Contigs that contained mixtures of ESTs from both libraries are shown in the middle. Fifty-four of these contigs contained 3-fold or more ESTs from the R than S library, whereas 78 contigs contained 3-fold or more ESTs from the S than the R library. The remainder of the contigs (1,298) contained mixtures of ESTs from both libraries but in less than 3-fold ratios.
Use of cDNA Subtraction to Identify Genes That Are Differentially Expressed upon P. syringae pv tomato(avrPto) Inoculation in Tomato Leaves Overexpressing Pto
In a second approach to identify differentially expressed genes, we performed cDNA subtraction, an open-architecture gene expression profiling technique. By using SSH (Diatchenko et al., 1996), we “forward” subtracted cDNAs derived from R11-12 plants from cDNAs derived from R11-13 plants and vice versa (“reverse” subtraction) after inoculation with P. syringae pv tomato(avrPto). In the forward subtraction experiments, we used RNA isolated from leaves collected at 4 and 8 h after inoculation to identify genes expressed or overexpressed in R11-12 (tester) as compared with R11-13 (driver). Similarly, we used reverse subtraction to identify genes whose expression was suppressed in R11-12 (driver) as compared with R11-13 (tester). We followed up the subtraction procedure in two ways. First, we directly cloned the resulting PCR products and sequenced about 65 clones from each of the forward and reverse subtractions. Second, we radioactively labeled the subtracted PCR products and used them as a pooled probe on colony blots containing the DNA from a nonredundant EST set developed from the EST sequences of R and S libraries. Based on the relative hybridization intensities, approximately 250 of the spotted cDNA clones were observed to correspond to abundant sequences in the subtraction libraries. Sequences from both of these methods were searched against National Center for Biotechnology Information (NCBI) databases (see below). We further examined the differential expression of genes identified by SSH by using cDNA microarrays and RNA blots (see supplemental data Fig. S1 at http://www.plantphysiol.org).
cDNA Microarray Analysis of Genes Identified by in Silico EST Subtraction and SSH
A total of 600 cDNA clones (some redundancy was observed) obtained from SSH and in silico EST subtraction were selected for microarray analysis. cDNA clones corresponding to known plant defense related genes were also included. Both R11-12 and R11-13 plants were inoculated with P. syringae pv tomato(avrPto), and RNA was isolated at different time points (0, 1, 2, 4, and 8 h) afterward. These RNA samples were used for microarray hybridization. From 299 genes identified by in silico EST subtraction, 159 (53%) of them were differentially expressed (>2-fold) in at least one of the five time points analyzed. Of the 301 clones that were identified by SSH, 158 clones (52%) were shown to be differentially expressed (>2-fold) in at least one of the five time points. Overall, by combining the approaches of in silico EST subtraction, SSH, and cDNA microarrays, we identified 223 nonredundant genes that were significantly differentially expressed at one or more time points between the R11-12 and R11-13 plants (see supplemental data Table S1 at http://www.plantphysiol.org). We designate these genes as POR (Pto overexpression responsive).
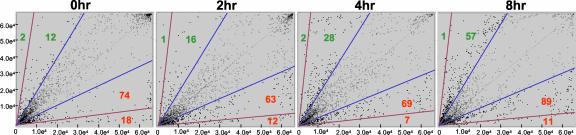
Scatter plots were generated to display the differential expression of the POR genes at different time points after inoculation (Fig. 2). Interestingly, 106 genes were differentially expressed more than 2-fold at the 0-h time point (without any P. syringae pv tomato inoculation), and the transcripts of most of these genes were more abundant in Pto-overexpressing line R11-12 than in the susceptible line R11-13. Remarkably, the transcript abundance of some of the genes (osmotin like, class I chitinase, and basic 30-kD endochitinase precursor) at 0-h time point was greater than 100-fold in R11-12 as compared with R11-13. The number of genes with lower transcript abundance in R11-12 than in R11-13 increased markedly at 8 h after inoculation when compared with the 0-h time point (Fig. 2). The number of genes that were differentially expressed by more than 10-fold in R11-12 compared with R11-13 was reduced at 8 h after inoculation when compared with the 0-h time point. This is not unexpected because many defense-related genes are known to be expressed early in incompatible interactions and to then be very highly expressed later in the compatible interaction. Expression data of the 223 POR genes (those genes with >2-fold expression differences) were used for cluster analysis as described below.
Figure 2.
Scatter plots showing distribution of POR gene expression ratios at different time points after inoculation. Each cDNA insert was replicated three times on the microarray slides (see “Materials and Methods”). The dots (>2-fold induction/repression) and white circles (<2-fold induction/repression) correspond to the signal intensity ratios (R11-12:R11-13) of the individual cDNA spots. The x axis represents the spot intensity of Cy3 (R11-12 RNA; experiment), and the y axis represents the spot intensity of Cy5 (R11-13 RNA; control). Diagonal blue and brown lines represent >2-fold and >10-fold induction:repression ratio cutoffs, respectively, relative to the best fit line through the normalized data (middle black line). Number of cDNA clones that are >2-fold or >10-fold induced (red)/repressed (green) is shown.
Coordinate Regulation of Genes That Are Constitutively Differentially Expressed in Line R11-12 and That Are Differentially Expressed by P. syringae pv tomato(avrPto) Inoculation
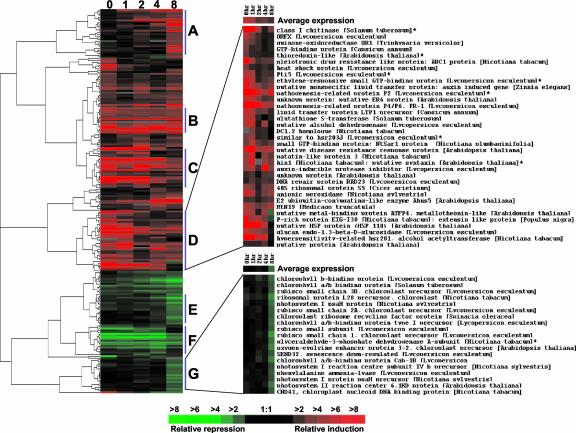
The ratios of spot intensities of the 223 POR genes obtained at different time points after inoculation of R11-12 and R11-13 with P. syringae pv tomato(avrPto) were used to do a hierarchical cluster analysis (Eisen et al., 1998; Fig. 3). From this analysis, we focused on seven clusters (Fig. 3, A–G; see supplemental data Fig. S2 at http://www.plantphysiol.org). Cluster A contains a variety of genes whose expression is induced in R11-12 leaves after inoculation with P. syringae pv tomato(avrPto). Maximum induction of these genes was observed at 8 h after inoculation. Some of the genes that belong to this cluster encode proteins involved in phenylpropanoid metabolism, water transport, cell protection, or stress responses (Fig. S2a). Cluster B genes were highly expressed without P. syringae pv tomato(avrPto) inoculation in R11-12 plants when compared with R11-13 plants, and their transcript levels remained almost constant at time points after inoculation. Cluster B genes encode a variety of proteins, including a glutathione S-transferase (GST), a ferritin, an AP2 domain transcription factor, and some PR genes (Fig. S2a). This cluster also contains Pto whose high expression is driven by the CaMV 35S promoter. Cluster C includes mainly PR genes. It also includes cell protectant genes, an ethylene response factor (ERF) transcription factor Pti4, and a WRKY transcription factor (Fig. S2b). These genes were highly expressed in R11-12 plants before pathogen inoculation, and the ratio of expression between R11-12 and R11-13 plants gradually decreased beginning 2 h after inoculation. This expression pattern is due to the induction of these genes in R11-13 plants after inoculation. Cluster D includes a variety of genes including PR genes, HR-associated genes, general stress-related genes, and another ERF transcription factor Pti5 (Fig. 3). These genes were highly expressed in R11-12 plants before pathogen inoculation, and the ratio of expression between R11-12 and R11-13 decreased beginning 1 h after inoculation. Again, this is due to the induction of these genes in R11-13 plants at this time point.
Figure 3.
A hierarchical cluster of gene expression of the 223 POR genes over a time course after inoculation by P. syringae pv tomato expressing avrPto. Each gene is represented by a single row of colored bars, and the columns represent the time points. Colored bars (red, increased transcript abundance; green, decreased transcript abundance; gray, missing or incomplete data; and black, no difference in transcript abundance) represent the ratio of hybridization measurements between corresponding time points in the R11-12 and R11-13 (treated with avirulent P. syringae pv tomato) samples. The cluster tree (left) illustrates the nodes of coregulation of gene expression. The clusters A through G are discussed in the text. Average differential expression and the list of genes in clusters D and G are shown. Genes with asterisks are previously known plant defense-related genes that were deliberately included during microarray experiments.
Cluster E includes a set of genes whose expression in R11-12 is suppressed during the resistance response, in comparison with R11-13 plants, with the suppression starting at 4 h after inoculation and reaching a maximum at 8 h after inoculation. Cluster E contains mostly genes encoding chloroplast- and photosynthesis-related proteins (Fig. S2c). Cluster F contains genes whose expression was suppressed, without pathogen inoculation, in the R11-12 plants when compared with R11-13 plants. Cluster F comprises a variety of genes that encode proteins like translation initiation and elongation factors, ACC oxidase, prohibitin, proteasome alpha-subunit, and chromatin-associated proteins (Fig. S2c). Expression of genes in cluster G is very similar to Cluster E except that the suppression of these genes was observed only at 8 h after inoculation. Cluster G is also enriched with chloroplast- and photosynthesis-related genes (Fig. 3).
Functional Classification of POR Genes
POR genes were classified based on their potential cellular functions into 21 different groups (Table I). Of the 21 classes, the majority of the genes in 16 of the classes were “up-regulated” in R11-12 plants. Viewed broadly, they include genes involved in cell protection (from oxidative stress), cell wall fortification, hormone responses, HR, general metabolism, known plant defense functions, transport, signaling, ubiquitination, and water transport, and genes encoding pathogenesis-related proteins, ribosomal proteins, stress-related proteins, transcription factors, proteins with no homology to an annotated gene in the database, and genes encoding proteins of unknown function. Three classes of genes had both up- and down-regulated genes, and these include genes required for various other miscellaneous functions, phenylpropanoid metabolism, and senescence-associated proteins. It is interesting to note that almost 19% of the POR genes were photosynthesis- and chloroplast-related genes (see supplemental data Table S1 at http://www.plantphysiol.org). Without any exceptions, all the photosynthesis- and chloroplast-related genes were “down-regulated” during the resistance response. All photosynthesis- and chloroplast-related genes were suppressed only after pathogen inoculation and had maximum suppression at 8 h after inoculation.
Table I.
Functional classification of POR genes
| Functional Classification | No. of Up-regulated Genesa | No. of Down-regulated Genesa |
|---|---|---|
| Cell protectant | 18 | 1 |
| Cell wall associated | 8 | — |
| Chloroplast associated | — | 12 |
| Hormone responsive | 3 | — |
| HR related | 3 | — |
| Metabolism | 13 | 3 |
| No matchb | 2 | — |
| Otherc | 10 | 8 |
| Pathogenesis related | 15 | — |
| Phenylpropanoid pathway | 5 | 4 |
| Photosynthesis related | — | 30 |
| Known plant defense related | 14 | 1 |
| Ribosomal protein | 4 | — |
| Senescence associated | 2 | 1 |
| Signaling | 8 | 1 |
| Stress related | 14 | 3 |
| Transcription factor | 9 | 1 |
| Transport | 5 | 1 |
| Ubiquitination pathway | 4 | 1 |
| Unknownd | 12 | 3 |
| Water transport | 4 | — |
| Total | 153 | 70 |
Up- or down-regulated in R11-12 compared with R11-13. b Genes that have no homology to an annotated gene sequence in the database. c Genes that encode proteins of other miscellaneous functions. d Genes that encode proteins of unknown function.
Overexpression of Pto Causes Differential Expression of Genes That Belong to the Same Functional Classes Identified in Tomato Leaves with Pto Expressed from Its Native Promoter
We compared the POR genes with a recently identified set of genes that is differentially expressed 4 h after inoculation with avirulent P. syringae pv tomato(avrPto) in a tomato line expressing Pto from its native promoter (RG-PtoR; Mysore et al., 2002). Approximately 22% of POR genes (48 of 223) were represented in the previously identified set of genes (see supplemental data Table S1 at http://www.plantphysiol.org; Mysore et al., 2002). The differential expression of 16 of these 48 genes was much higher in Pto-overexpressing plants when compared with RG-PtoR plants. Although the other 78% of the genes were distinct from the previously identified set of genes, they fall into functional classes that are similar to the classes of genes identified previously in RG-PtoR plants. Thus, Pto-overexpressing plants express a subset of genes shared with the normal Pto-mediated disease resistance response but also express a distinct set of largely defense-related genes. For example, the majority of genes involved in cell protection and defense were “up-regulated,” and chloroplast- and photosynthesis-related genes were “down-regulated” during the resistance response in both RG-PtoR and R11-12 plants after inoculation with P. syringae pv tomato(avrPto).
Similarities between the Plant Defense Response and the Immune Response of Humans and Fruitfly
Because striking similarities exist among both R proteins and downstream signaling components of plants and certain animal and fruitfly proteins with roles in immunity (Cohn et al., 2001; Staskawicz et al., 2001), we decided to compare the set of POR genes with several expression profiling studies of the mammalian/invertebrate immune response (Gregorio et al., 2001; Huang et al., 2001; Irving et al., 2001). We found that for 62 of 153 POR genes (approximately 40%) that were abundantly expressed in R11-12 plants when compared with R11-13 plants, closely related genes (based on putative function and not by sequence similarity) were induced during the human and/or fruitfly immune responses (Table II). Transcripts of most of these genes were abundant in R11-12 plants without any pathogen inoculation. In addition to these 62 genes, other genes encoding transcription factors, kinases, transporters, and proteins involved in general metabolism were commonly differentially expressed between plants, humans, and fruitfly defense responses. We did not compare the 70 genes that were less abundant in R11-12 (when compared with R11-13) with human and fruitfly defense responses because the majority of them encode photosynthesis- and chloroplast-associated proteins. We also compared our previously identified differentially expressed genes (Mysore et al., 2002) at a very early time (4 h) after infection from RG-PtoR plants inoculated with P. syringae pv tomato(avrPto) with immune response genes from human and fruitfly. Interestingly, very early after infection, only 23 genes of 227 induced genes (approximately 10%) in RG-PtoR were similar to human and/or fruitfly immune response genes (data not shown).
Table II.
Genes that are induced during the Pto-mediated plant defense response
and also during human and/or fruitfly immune response  , Differential
expression; ^, mouse immune response.
, Differential
expression; ^, mouse immune response.
| GenBank Accession No. | No. of Similar Genes Up-Regulated during Plant Defense | Gene (Best Blast Hit) | Functional Classification | Human Immune Responsea | Fruitfly Immune Responsea | References |
|---|---|---|---|---|---|---|
| AI779773 | 1 | Phospholipid hydroperoxide glutathione peroxidase (tomato, Lycopersicon esculentum) | Cell protectant | Beck (2001) | ||
| AI489449 | 1 | Catalase (tomato) | Cell protectant | Zancope-Oliver et al. (1999) | ||
| AI772349 | 1 | Ferritin tobacco (Nicotiana tabacum) | Cell protectant | Huang et al. (2001) | ||
| AI774583 | 6 | GST (Solanum commersonii) | Cell protectant | Huang et al. (2001); Irving et al. (2001) | ||
| AI775811 | 2 | MT-like protein (tomato) | Cell protectant | Huang et al. (2001); Irving et al. (2001) | ||
| CB751576 | 1 | NADP-dependent isocitrate dehydrogenase-like protein (tomato) | Cell protectant | Jo et al. (2001) | ||
| CB751574 | 1 | Quinone-oxidoreductase QR1 (Triphysaria versicolor) | Cell protectant | Ross et al. (2000) | ||
| AI488458 | 1 | Superoxide dismutase (Cu-Zn SOD; tomato) | Cell protectant | Huang et al. (2001) | ||
| AI776377 | 2 | Thioredoxin, protein disulfide-isomerase precursor (tobacco) | Cell protectant | Huang et al. (2001); Irving et al. (2001) | ||
| AI777808 | 3 | Lipid transfer protein LTP1 precursor (Capsicum annuum) | Plant defense/cell wall associated | Irving et al. (2001) | ||
| AI485587 | 1 | Arg decarboxylase 1 (Datura stramonium) | Metabolism | Efron and Barbul (1999) | ||
| CB751579 | 1 | Fructose-1,6-bisphosphatase precursor (potato, Solanum tuberosum) | Metabolism | Mabondzo et al. (1991) | ||
| AI778001 | 1 | Glu synthetase/glutamate-ammonia ligase (Nicotiana plumbaginifolia) | Metabolism | Karinch et al. (2001) | ||
| AI781779 | 1 | S-adenosylmethionine decarboxylase proenzyme (potato) | Metabolism | Di Leo et al. (1999) | ||
| AI772723 | 1 | Thiamin biosynthetic enzyme (soybean, Glycine max) | Metabolism | Axelrod (1981) | ||
| AI488639 | 1 | UDP-galactose/Glc 4-epimerase-like protein (Arabidopsis) | Metabolism | Gregorio et al. (2001) | ||
| CB751578 | 1 | Uricase/urate oxidase (Lotus japonicus) | Metabolism | Gregorio et al. (2001); Irving et al. (2001) | ||
| CB751580 | 1 | Actin (Malva pusilla) | Other | Irving et al. (2001) | ||
| CB751550 | 1 | DNA repair protein RAD23 (tomato and fruitfly) | Other | Brocksted et al. (1998) | ||
| AI775854 | 1 | Ferredoxin-nitrite reductase (tobacco) | Other | Gregorio et al. (2001) | ||
| AI488991 | 1 | Histone H2A (Euphorbia esula) | Other | Huang et al. (2001) | ||
| CB751552 | 1 | Vacuolar proton-inorganic pyrophosphatase (tobacco) | Other | Komissarenko et al. (1986) | ||
| AI484482 | 5 | Chitinase (tomato) | Pathogenesis-related | Irving et al. (2001) | ||
| CB751582 | 1 | Cytochrome P450 monooxygenase (tobacco) | Phenylpropanoid pathway | Huang et al. (2001); Irving et al. (2001) | ||
| AW032166 | 1 | Hin1 (tobacco), putative syntaxin (Arabidopsis) | Known plant defense related | Shukla et al. (2000) | ||
| CB751584 | 1 | Leu aminopeptidase (tomato) | Known plant defense related | Gregorio et al. (2001); Irving et al. (2001) | ||
| CB751558 | 1 | Lipoxygenase (tomato) | Known plant defense related | Malaviya and Abraham (2001) | ||
| AI774536 | 4 | Ribosomal protein (N. plumbaginifolia) | Ribosomal protein | Huang et al. (2001); Irving et al. (2001) | ||
| CB751586 | 1 | Aconitate hydratase or aconitase (potato) | Signaling |
|
Pitarch et al. (2001) | |
| AI782784 | 2 | Heat shock protein (tomato) | Stress related | Huang et al. (2001); Gregorio et al. (2001) | ||
| AI777200 | 1 | Putative trypsin inhibitor (Arabidopsis); Lemir (tomato) | Stress related | Gregorio et al. (2001) | ||
| BG625896 | 3 | Putative alcohol dehydrogenase (tomato) | Stress/plant defense related |
|
Irving et al. (2001); Pitarch et al. (2001) | |
| CB751569 | 1 | ABC1 protein (N. plumbaginifolia); PDR5-like ABC transporter | Transport | Irving et al. (2001) | ||
| CB751568 | 1 | ADP-ribosylation factor (Arabidopsis) | Transport | Gregorio et al. (2001) | ||
| AI485102 | 1 | Synaptobrevin-like V snare protein (yeast, Saccharomyces cerevisiae, and human, Homo sapiens) | Transport | Irving et al. (2001) | ||
| AI775530 | 4 | Water channel protein (Nicotiana excelsior); aquaporin-like protein (Petunia × hybrida) | Transport | Irving et al. (2001) | ||
| AI488816 | 3 | E2 ubiquitin-conjugating-like enzyme Ahus5 (Arabidopsis) | Ubiquitination pathway | Huang et al. (2001); Irving et al. (2001) | ||
| CB751590 | 1 | Polyubiquitin (maize, Zea mays) | Ubiquitination pathway | Ishii et al. (1999) |
Similarity is inferred by putative function and not by sequence relationships.
DISCUSSION
Signaling pathways leading to activation of defense response genes in mammals, insects, and plants share many similar components (Cohn et al., 2001; Staskawicz et al., 2001). For example, the resistance protein Pto shares sequence similarity with the IRAK proteins in human and Pelle in fruitfly, both of which play central roles in controlling the innate immune response (Cohn et al., 2001). To investigate whether “downstream” defense responses are similar among mammals, insects, and plants, we compared the POR genes with previously published human and fruitfly immune response genes (Gregorio et al., 2001; Huang et al., 2001; Irving et al., 2001). Strikingly, 40% of the POR genes that were abundantly expressed in Pto-overexpressing plants are functionally similar to genes previously shown to be induced during human and/or fruitfly immune responses (Table II). Some of these could be general stress-responsive genes and may not be specific to pathogen-induced immune response. However, they may also still play a role in conferring immunity against pathogens. These results suggest that defense responses in these diverse organisms are either conserved or may have arisen by convergent evolution. For example, genes encoding metallothionein (MT), GST, thioredoxin (TRX), cytochrome P450 (P450), heat shock proteins (HSPs), ribosomal proteins, and ubiquitin-conjugating enzymes are all induced during defense responses in Pto-overexpressing plants, humans, and fruitfly (Table II). Although these genes are reasonably well characterized in mammalian systems, their actual role in immunity is not clear. Because of the availability of high-throughput techniques like virus-induced gene silencing and reverse genetics in plants, it might be easier to investigate the function of these in immunity by using a plant model system.
The oxidative burst that precedes HR, a common phenomenon triggered by an incompatible plant pathogen interaction, has been suggested to be a primary event responsible for triggering the cascade of defense responses in various plant species against infection with avirulent pathogens or pathogen-derived elicitors. The oxidative burst may be followed by activation of genes encoding antioxidant enzymes in tissue surrounding the initial infection site (Lamb and Dixon, 1997) to protect the surrounding cells against oxidative damage. We found that several of these cell protectant genes (POR1–POR19), like glutathione peroxidase, separation anxiety protein, catalase, superoxide dismutase, ferritin, GST, MT, NADP-dependent isocitrate dehydrogenase, ascorbate oxidase, quinone-oxidoreductase, and TRX, were up-regulated during the Pto-mediated disease resistance response. A majority of these was constitutively up-regulated in R11-12 plants without any pathogen infection. Interestingly, all these genes, with the exception of separation anxiety protein and ascorbate oxidase, were also shown to be induced during human immune response (Table II). This suggests that the cell protectants that are induced during an immune response are highly conserved among animals and plants.
MTs and GSTs are a group of stress and immune response proteins that contribute to cellular survival due to oxidative damage (Borghesi and Lynes, 1996). TRX plays multiple roles in intracellular signaling and resistance against oxidative stress (Hirota et al., 2002). In humans, the TRX gene contains a series of stress-responsive elements; in turn, TRX promotes activation of transcription factors such as NF-kappa B, AP-1, and p53 that are components of the human immune system (Hirota et al., 2002). In vertebrates, the wide variety of P450s is a key to many of the known defense mechanisms of vertebrates against all xenobiotic forms (Morfin, 2002). Taken together, overexpression of MTs, GSTs, TRX, and P450s during immune responses in humans, insects, and plants seems to participate in the same basic function of cell protection. HSPs act as chaperones, are widely distributed in nature, and are among the most highly conserved molecules of the biosphere (Zugel and Kaufmann, 1999). HSP synthesis is increased to protect prokaryotic or eukaryotic cells from various insults during periods of stress caused by infection, inflammation, or similar events (Zugel and Kaufmann, 1999). Reversal of polypeptide unfolding and preventing protein aggregation are major functions of HSPs, especially under stress, and they seem to perform the same functions in animals, plants, and insects (Zugel and Kaufmann, 1999).
Protein translation plays an important role during innate immunity in plants, animals, and insects. For example, the fruitfly Thor gene has been shown to be required for innate immunity in fruitfly by preferential translation of immune transcripts (Bernal and Kimbrell, 2000). During the immune and defense responses in mammals, insects, and plants, massive production of antimicrobial proteins is required, and translation of mRNAs encoding proteins with counteractive effects must be suppressed. Hence, modulation of ribosomal gene expression during immune responses in humans, fruitfly and plants is probably essential.
Protein ubiquitination is not only essential for the normal physiological turnover of proteins but appears to have been adapted as part of an intracellular surveillance system that can be activated by altered, damaged, or foreign proteins and organelles and has evolved to be an “intracellular immune system” (Ben-Neriah, 2002). Ubiquitination pathway genes have been shown to be induced during both human and fruitfly immune response (Huang et al., 2001; Irving et al., 2001; Table II). Recently, it has been shown that mutations in a plant homolog of animal SGT1, a component of ubiquitin ligase, disable early defenses conferred by multiple R genes (Austin et al., 2002; Azevedo et al., 2002). SGT1 is also shown to play an important role in N gene-mediated resistance response to tobacco mosaic virus (Liu et al., 2002b). In this study, we have shown that four ubiquitination pathway-related genes (POR200 and POR202-204) were induced during Pto-mediated disease resistance response. These data suggest that the ubiquitination-mediated protein degradation pathway plays an important role in both plant and animal defense.
Pto-overexpressing plants (R11-12) are slightly stunted, and they develop small white veins in the leaves as they age (Li et al., 2002). It is possible that these phenotypes are due to pleiotropic effects of Pto overexpression that are unrelated to the defense response. However, it is unlikely that differential expression of most of the POR genes is due to pleiotropic effects because we found that the majority of the genes are differentially expressed upon inoculation by P. syringae pv tomato(avrPto), and they fall into the same functional classes that were shown to be differentially expressed during a disease resistance response in RG-PtoR plants (Mysore et al., 2002; see supplemental data Table S1 at http://www.plantphysiol.org).
Requirement of Prf for the constitutive plant defense response mediated by Pto overexpression (Xiao et al., 2003) suggests that both R11-12 and RG-PtoR plants initiate similar defense responses in response to P. syringae pv tomato expressing avrPto. Even though the POR genes belong to the same functional classes identified in this earlier study, approximately 80% represent previously undiscovered genes. This lack of significant overlap could be due to several reasons. First, the expression profiling we present in this paper is likely not as comprehensive as that reported earlier; further overlap of the genes might be revealed with a more in-depth profiling approach such as GeneCalling (Mysore et al., 2002). Second, it seems likely that Pto overexpression “amplifies” the gene expression differences that normally occur in plants expressing Pto from its native promoter. This exaggerated response might have allowed the identification of genes in the Pto-overexpressing line that are normally expressed at much lower levels during the “natural” Pto-mediated resistance response. Third, the differentially expressed genes identified in this study were from tissue collected at 4 and 8 h after infection, whereas in the previous study it was only from tissue collected at 4 h after infection. Fourth, a recent report suggests that the AvrPto-independent defense activation in R11-12 and gene-for-gene resistance mediated by Pto are functionally separable. Even though Pto-mediated gene-for-gene resistance and AvrPto-independent resistance require both Pto and Prf, it was recently shown that AvrPto-independent resistance appears to be distinct from gene-for-gene resistance in terms of the requirement of downstream components, sensitivity to mutations in Pto, and defense activation (Xiao et al., 2003).
A recent report from Xiao et al. (2001) provided some insights about differential gene expression in the Pto-overexpressing line, and we have now significantly expanded on that study. Our study differs from that of Xiao et al., first, by investigating not only genes that are differentially expressed without pathogen inoculation in the Pto-overexpressing line but also those genes that are induced in this line upon inoculation by P. syringae pv tomato(avrPto). Second, we also identified a set of genes that are suppressed in the Pto-overexpressing plants. Third, because results are now available from a comprehensive transcript profiling experiment of a tomato line expressing Pto from its native promoter, we were able to compare the POR genes with those differentially expressed during the “normal” resistance response. Fourth, we examined the expression of the POR genes over a time course, and this allowed us to cluster them based on their expression patterns. We did observe some overlap of the POR genes with those identified by Xiao et al. Approximately 52% of the genes identified by Xiao et al. are identical to our POR genes. Together, these two studies have identified a total of 255 genes that are differentially expressed in the Pto-overexpressing lines.
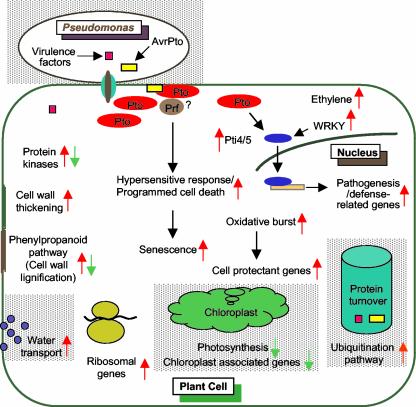
We identified 21 different classes of genes that were differentially expressed in R11-12 after inoculation with P. syringae pv tomato expressing avrPto (Table I). Interestingly, genes belonging to these classes, with the exception of chloroplast- or photosynthesis-related genes, water transport-related genes, and ubiquitination pathway genes, were constitutively expressed in the R11-12 plants in the absence of P. syringae pv tomato expressing avrPto. Figure 4 depicts a model for constitutive plant defense responses due to Pto overexpression.
Figure 4.
A summary of the plant responses to overexpression of Pto and P. syringae pv tomato infection. Red arrows indicate genes that were induced and green arrows indicate genes that were suppressed due to Pto overexpression. Black arrows indicate either possible steps in signal transduction pathways or host responses that might directly impact other responses. Classes of genes shaded in gray are also responsive to P. syringae pv tomato infection.
Phenylpropanoids have been proposed to serve as flower pigments (anthocyanin), UV protectants, defense chemicals (phytoalexins and insect repellents), allelopathic agents, and signal molecules in plant-microbe interactions. We identified 10 different genes (POR97-105) involved in the phenylpropanoid pathway that are differentially expressed in the Pto-overexpressing plants, R11-12. Five of these 10 genes were constitutively differentially expressed in R11-12 plants. Cell wall reinforcement and thickening are associated with plant defense during resistance response. In this study, we have shown that majority of the cell wall-associated genes (POR20-21, POR23-24, and POR26-27) were constitutively up-regulated in R11-12 plants.
Overlap between the leaf senescence and pathogen defense programs has been reported earlier (Quirino et al., 1999). Obvious visual symptoms of leaf senescence are loss of chlorophyll pigments due to decline in photosynthesis and chloroplast disintegration (Buchanan-Wollaston, 1997). It has been suggested that in cells undergoing HR, a decline in photosynthesis might act as an inducer of senescence (Quirino et al., 2000). In this study, we show 30 photosynthesis-related genes (POR106–135) that were suppressed during the Pto-mediated disease resistance response. We also identified 12 genes encoding chloroplast associated proteins (POR28–39) that were also suppressed during the resistance response. Interestingly, pathogen inoculation was required for the suppression of these genes. These observations suggest that, in the face of pathogen attack, the plant is rapidly redirecting its resources from growth processes to defense responses.
Water is essential for pathogen multiplication in the plant tissue. One of the P. syringae pv tomato disease symptoms is water-soaked lesions on tomato leaves. In this study, we have shown that four aquaporin/tonoplast intrinsic proteins (POR220-223) were induced during the resistance response. Interestingly, aquaporin genes were suppressed during a previously studied Pto (native promoter driven)-mediated disease resistance response (Mysore et al., 2002). It seems like the Pto overexpression is slightly up-regulating the aquaporin genes, and its expression is enhanced after pathogen infection. The actual role of water transport during plant defense response is yet to be determined.
Several genes encoding known components of the Pto signal transduction pathway were deliberately included in the microarray experiments to study their expression in Pto-overexpressing plants (see supplemental data Table S1 at http://www.plantphysiol.org). As expected, the Pto gene (POR161) was abundantly expressed in the R11-12 plants when compared with the azygous plants at the 0-h time point. Pto was slightly induced due to P. syringae pv tomato expressing avrPto inoculation. This was probably due to the CaMV 35S promoter used to overexpress the Pto gene. Tomato plants infected by the avirulent P. syringae pv tomato have been shown to accumulate SA (Oldroyd and Staskawicz, 1998), and the CaMV 35S promoter is responsive to SA (Qin et al., 1994). Pti4, Pti5, and Pti6 are ERF transcription factors that interact with Pto; expression of the Pti4/5/6 genes is induced in both the compatible and incompatible interactions, whereas the induction of Pti5 is further enhanced during incompatible interaction when compared with compatible interaction (Zhou et al., 1997; Thara et al., 1999; Gu et al., 2000). Here, we have shown that genes encoding Pti4 (POR186) and Pti5 (POR185) are constitutively and abundantly expressed in R11-12 plants without any pathogen inoculation. Previously, using RNA-blot hybridizations, Pti4 was shown to be equally expressed in both Pto-overexpressing and azygous lines (Tang et al., 1999; Gu et al., 2000). This discrepancy is most likely due to the differences in the sensitivity of RNA-blot hybridization and microarray hybridization. It is interesting to note that Pti4 was also induced in the RG-PtoR plants, in response to P. syringae pv tomato expressing avrPto, when compared with Rio-Grande tomato plants without Pto (Mysore et al., 2002). Pti6 was not differentially expressed due to Pto overexpression (data not shown).
Api1 and Api2 encode tomato proteins that interact with AvrPto in a yeast two-hybrid system (Bogdanove and Martin, 2000). Using microarrays, we have shown that Api1 (POR139) was suppressed in R11-12 plants at 4 and 8 h after inoculation with P. syringae pv tomato expressing avrPto. Api1 encodes a stress-related protein that is induced by heavy metal, wounding, and virus inoculation. It is possible that Api1 is a virulence target for AvrPto, and, hence, the plant is suppressing the gene expression as a defense mechanism. Api2 (POR42), which encodes a Ras-related Rab8-like protein, was constitutively and abundantly expressed in R11-12 plants without any pathogen inoculation. Api2 is involved in vesicular protein trafficking, and its role in disease formation is yet to be determined. Adi1 encodes an AvrPto-dependent Pto-interacting tomato protein (Bogdanove and Martin, 2000). Adi1 is a catalase that is involved in scavenging hydrogen peroxide (Bogdanove and Martin, 2000). In this study, we have identified a tomato catalase (POR7) that is constitutively abundantly expressed in the R11-12 plants without any pathogen inoculation. This catalase is most likely involved in protecting the cells from excessive damage due to oxidative burst that results from Pto overexpression.
The possible role of Pto in a basal defense response is suggested by our recent report that the RG-PtoR plants constitutively induce several defense-related genes, including PR genes without any pathogen infection (Mysore et al., 2002). When Pto is overexpressed, such basal defense response is exaggerated to up-regulate an array of defense-related genes (POR genes). Constitutive overexpression of ERF transcription factors Pti4 and Pti5 (Zhou et al., 1997), without any pathogen infection, in Pto-overexpressing plants further corroborates a role for Pto in basal defense and supports the notion that Pto can activate Pti4 and Pti5 in the absence of AvrPto (Zhou et al., 1997). Interestingly, many of the POR genes are also known to be induced during immune responses in human and/or fruitfly. This observation further supports the notion that defense responses among plants, mammals, and insects have been conserved during evolution. Future characterization of the POR genes by virus-induced gene silencing (Liu et al., 2002a) and other loss- and gain-of-function methods should help to clarify the potential role of these genes in disease resistance.
MATERIALS AND METHODS
Plant Materials, Bacterial Strains, Plant Inoculation, and RNA Isolation
Tomato (Lycopersicon esculentum) Pto-transgenic line R11-12 (35S::Pto/35S::Pto) and a sibling line R11-13 that has segregated away the Pto transgene were described previously (Tang et al., 1999). Seeds of these two genotypes were germinated on moist Whatman paper (Whatman, Clifton, NJ) and grown in soil in the greenhouse at 26°C to 28°C and 16 h of light per day. Bacterial inoculations were performed as described previously (Gu et al., 2000) and below under cDNA library construction. For SSH, cDNA microarray, and RNA-blot experiments, plants were inoculated with bacteria by vacuum infiltration at a concentration of 2.2 × 107 colony-forming units (cfu) mL–1. RNA was isolated using the hot phenol method as described previously (Gu et al., 2000).
Construction of cDNA Libraries and EST Sequencing
To create cDNA libraries, 4-week-old plants of tomato lines R11-12 and R11-13 were vacuum infiltrated with Pseudomonas syringae pv tomato strain T1 expressing avrPto at concentrations of either 105 or 108 cfu mL–1. At the 108 cfu mL–1 (high titer) inoculation level, leaves were harvested at 0, 2, 4, 6, and 8 h after inoculation. Effectiveness of the high-titer inoculations was verified by assessing defense-related gene expression (in R11-13) or by observation of the HR (in R11-12). At the 105 cfu mL–1 (low titer) inoculation level, leaves were harvested at 0, 12, 24, 36, and 48 h after inoculation. Effectiveness of the low-titer inoculations was verified by assessing defense-related gene expression (in both R11-12 and R11-13). Equal amounts of leaf tissues from the R11-12 and R11-13 plants from the different time points of high- and low-titer inoculations for each line were pooled and used to extract total RNA.
Poly(A+) RNA was purified using the poly(A+) tract mRNA isolation system (Promega, Madison, WI). An oriented Uni-ZAP XR library was prepared using a ZAP-cDNA synthesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The cDNA (0.4 μg) was ligated into Uni-ZAP XR digested with XhoI and EcoRI. This yielded 1.4 × 106 and 1.2 × 106 primary plaques for the R11-12 (R) library and the R11-13 (S) library. Mass excision was performed on 1 × 107 plaque-forming units from each library. Escherichia coli strain XL1-Blue MRF was used for propagating the library. The bacterial cultures were subsequently arrayed into 384-well plates and used for sequencing. Additional information about the libraries can be found online (http://sgn.cornell.edu or http://www.tigr.org/tdb/tgi/lgi). Approximately 5,500 clones from each of the R and S libraries were sequenced by The Institute for Genomics Research (TIGR; http://www.tigr.org/tdb/tgi/lgi).
In Silico EST Subtraction and cDNA Subtraction by SSH
For in silico EST subtraction (Stekel et al., 2000), 10,872 EST sequences from the R and S libraries were downloaded from the TIGR database and assembled using “TIGR Assembler” running on a Linux operating system. Custom scripts were written in PERL to index each contig resulting from the assembly. The indexing process calculated the number of ESTs within each contig derived from the R or S libraries. Contigs that contained more than 2-fold the number of ESTs from one library than the other were flagged for further analysis (see “Results”).
SSH was done by using the CLONTECH PCR-Select cDNA Subtraction Kit (CLONTECH Laboratories Inc., Palo Alto, CA). Equal amounts of RNA from leaves 4 and 8 h after inoculation with P. syringae pv tomato(avrPto) were pooled for each of the R11-12 and R11-13 plants and used for SSH. Both forward (R11-12 as tester and R11-13 as driver) and reverse (R11-13 as tester and R11-12 as driver) subtractions were performed according to the manufacturer's protocol. The final PCR products (enriched for cDNAs corresponding to differentially expressed transcripts) resulting from the SSH were either digested with NotI and directly cloned into pBluescript vector or labeled with radioactive P32 (DECAprime DNA labeling kit, Ambion Inc., Austin, TX) and used as a probe on colony blots containing a nonredundant EST collection from the R and S libraries. PCR products cloned into pBluescript vector were sequenced (65 each from forward and reverse subtractions), and the sequences were searched against the NCBI databases. EST sequences corresponding to colonies that hybridized to the radiolabeled PCR product were obtained from the TIGR databases and were subsequently used for searches of the NCBI databases.
Microarray Preparation, Hybridization, and Data Analysis
Inserts from approximately 650 cDNA clones corresponding to ESTs (http://www.tigr.org) were PCR amplified using T3 and T7 primers. This number included about 600 ESTs that corresponded to genes obtained by SSH (301) and in silico EST subtraction (299), 38 ESTs that corresponded to known defense response genes, and 12 controls (e.g. alpha-tubulin, beta-tubulin, and genes encoding ribosomal proteins). PCR products were purified using MagBeads (Bangs Lab., Fishers, IN). Purified PCR products were vacuum dried and resuspended in 30 μl of spotting buffer (3× SSC + 0.1% [w/v] Sarkosyl). DNA spotting, labeling, hybridization, and data analyses were performed as described earlier (Mysore et al., 2002). Changes in mRNA expression in excess of 2-fold between the two samples were considered for these experiments. Cluster and Treeview software (Eisen et al., 1998) were used (http://rana.lbl.gov/EisenSoftware.htm) to group and display genes with similar expression profiles. We used the default options of hierarchical clustering.
Several quality control measures were implemented in our analysis of the microarray data to meet the Minimum Information About a Microarray Experiment (Brazma et al., 2001) guidelines (http://www.mged.org/Workgroups/MIAME/miame.html). The variation in abundance (coefficient of variation shown in supplementary Table S1 at http://www.plantphysiol.org) for each transcript was estimated using two biological replications and at least two technical replications for each biological replication. PCR products corresponding to each cDNA were replicated three times on the same slide to control for possible variations in hybridization within a slide. The ratios shown in Table S1 (http://www.plantphysiol.org) are averages of up to 12 independent ratios representing each gene (two biological replications × two technical replications × three spots per replicate). The correlation coefficient between biological replicates was 0.812 and between technical replicates was 0.923. By dye swapping, we showed that there was no significant variation due to possible biases in direct labeling method of Cy3 or Cy5 used in this study. The correlation coefficient for the dye-swap experiment was 0.918. All the identified POR genes were resequenced from both 3′ and 5′ ends to confirm their gene identity. To conform with Minimum Information About a Microarray Experiment guidelines, we have provided the raw data (before normalization) of spot intensities and an example of an overlay image in supplementary Table S2 and Fig. S3 (http://www.plantphysiol.org), respectively.
Supplementary Material
Acknowledgments
We thank Paul Debbie (Boyce Thompson Institute Center for Gene Expression Profiling) for technical help with microarray experiments and David Wendell for deriving nonredundant cDNAs.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.022731.
This work was partly supported by Boyce Thompson Institute (innovation grant to K.S.M.), by the Noble Foundation (grants to K.S.M.), and by the National Science Foundation (Plant Genomics grant nos. IBN–9872617 and IBN–0109633 to G.B.M.).
The online version of this article contains Web-only data. The supplemental material is available at http://www.plantphysiol.org.
References
- Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JDG, Parker JE (2002) Regulatory role of SGT1 in Early R gene-mediated plant defenses. Science 295: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Axelrod AE (1981) Role of the B vitamins in the immune response. Adv Exp Med Biol 135: 93–106 [DOI] [PubMed] [Google Scholar]
- Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P (2002) The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295: 2073–2076 [DOI] [PubMed] [Google Scholar]
- Beck MA (2001) Antioxidants and viral infections: host immune response and viral pathogenicity. J Am Coll Nutr 20: 384S–388S [DOI] [PubMed] [Google Scholar]
- Ben-Neriah Y (2002) Regulatory functions of ubiquitination in the immune system. Nat Immunol 3: 20–26 [DOI] [PubMed] [Google Scholar]
- Bernal A, Kimbrell DA (2000) Drosophila Thor participates in host immune defense and connects a translational regulator with innate immunity. Proc Natl Acad Sci USA 97: 6019–6024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove AJ, Martin GB (2000) AvrPto-dependent Pto-interacting proteins and AvrPto-interacting proteins in tomato. Proc Natl Acad Sci USA 97: 8836–8840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghesi LA, Lynes MA (1996) Stress proteins as agents of immunological change: some lessons from metallothionein. Cell Stress Chaperones 1: 99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC et al. (2001) Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet 29: 365–371 [DOI] [PubMed] [Google Scholar]
- Brocksted E, Rickers A, Kostka S, Laubersheimer A, Dorken B, Wittmann-Liebold B, Bommert K, Otto A (1998) Identification of apoptosis-associated proteins in a human Burkitt lymphoma cell line: cleavage of heterogeneous nuclear ribonucleoprotein A1 by caspase 3. J Biol Chem 273: 28057–28064 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V (1997) The molecular biology of leaf senescence. J Exp Bot 48: 181–199 [Google Scholar]
- Cohn J, Sessa G, Martin GB (2001) Innate immunity in plants. Curr Opin Immunol 13: 55–62 [DOI] [PubMed] [Google Scholar]
- Di Leo A, Messa C, Russo F, Linsalata M, Amati L, Caradonna L, Pece S, Pellegrino NM, Caccavo D, Antonaci S et al. (1999) Helicobacter pylori infection and host cell responses. Immunopharmacol Immunotoxicol 21: 803–846 [DOI] [PubMed] [Google Scholar]
- Diatchenko L, Lau Y-FC, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED et al. (1996) Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA 93: 6025–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron DT, Barbul A (1999) Arginine and nutrition in renal disease. J Ren Nutr 9: 142–144 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JE, Collmer A (1999) Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284: 1322–1328 [DOI] [PubMed] [Google Scholar]
- Gregorio ED, Spellman PT, Rubin GM, Lemaitre B (2001) Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci USA 98: 12590–12595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YQ, Yang C, Venkatappa TK, Zhou J, Martin GB (2000) Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 12: 771–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He SY (1998) Type III protein secretion systems in plant and animal pathogenic bacteria. Annu Rev Phytopathol 36: 363–392 [DOI] [PubMed] [Google Scholar]
- Hirota K, Nakamura H, Masutani H, Yodoi J (2002) Thioredoxin superfamily and thioredoxin-inducing agents. Ann N Y Acad Sci 957: 189–199 [DOI] [PubMed] [Google Scholar]
- Huang Q, Liu D, Majewski P, Schulte LC, Korn JM, Young RA, Lander ES, Hacohen N (2001) The plasticity of dendritic cell responses to pathogens and their components. Science 294: 870–875 [DOI] [PubMed] [Google Scholar]
- Irving P, Troxler L, Heuer TS, Belvin M, Kopczynski C, Reichhart JM, Hoffman JA, Hetru C (2001) A genome-wide analysis of immune responses in Drosophila. Proc Natl Acad Sci USA 98: 15119–15124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, Itoh K, Sato H, Bannai S (1999) Oxidative stress-inducible proteins in macrophages. Free Radic Res 31: 351–355 [DOI] [PubMed] [Google Scholar]
- Jo SH, Son MK, Koh HJ, Lee SM, Song IH, Kim YO, Lee YS, Jeong KS, Kim WB, Park JW et al. (2001) Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J Biol Chem 276: 16168–16176 [DOI] [PubMed] [Google Scholar]
- Karinch AM, Pan M, Lin CM, Strange R, Souba WW (2001) Glutamine metabolism in sepsis and infection. J Nutr 131: 2535S–2538S [DOI] [PubMed] [Google Scholar]
- Kim YJ, Lin N-C, Martin GB (2002) Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109: 589–598 [DOI] [PubMed] [Google Scholar]
- Komissarenko SV, Gulaia NM, Gaivoronskaia GG, Karlova NP, Tarusova NB (1986) Inorganic pyrophosphatase activity of the mouse spleen in the immune response and after treatment with bis-phosphonates. Ukr Biokhim Zh 58: 22–27 [PubMed] [Google Scholar]
- Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48: 251–275 [DOI] [PubMed] [Google Scholar]
- Li J, Shan L, Zhou J-M, Tang X (2002) Overexpression of Pto induces a salicylate-independent cell death but inhibits necrotic lesions caused by salicylate-deficiency in tomato plants. Mol Plant-Microbe Interact 15: 654–661 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002a) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Serino G, Deng X-W, Dinesh-Kumar SP (2002b) Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene mediated resistance response to Tobacco mosaic virus. Plant Cell 14: 1483–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabondzo A, Le Naour R, Raoul H, Clayette P, Lafuma C, Barre-Sinoussi FC, Cayre Y, Dormont D (1991) In vitro infection of macrophages by HIV: correlation with cellular activation, synthesis of tumour necrosis factor alpha and proteolytic activity. Res Virol 142: 205–212 [DOI] [PubMed] [Google Scholar]
- Malaviya R, Abraham SN (2001) Mast cell modulation of immune responses to bacteria. Immunol Rev 179: 16–24 [DOI] [PubMed] [Google Scholar]
- Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, Wu T, Earle ED, Tanksley SD (1993) Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262: 1432–1436 [DOI] [PubMed] [Google Scholar]
- Morfin R (2002) Involvement of steroids and cytochrome P(450) species in the triggering of immune defenses. J Steroid Biochem Mol Biol 80: 273–290 [DOI] [PubMed] [Google Scholar]
- Mysore KS, Crasta OR, Tuori RP, Folkerts O, Swirsky PB, Martin GB (2002) Comprehensive transcript profiling of Pto- and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J 32: 299–315 [DOI] [PubMed] [Google Scholar]
- Nurnberger T, Brunner F (2002) Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr Opin Plant Biol 5: 318–324 [DOI] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH et al. (2001a) A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 411: 603–606 [DOI] [PubMed] [Google Scholar]
- Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G (2001b) Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kB. J Biol Chem 276: 4812–4818 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Staskawicz BJ (1998) Genetically engineered broadspectrum disease resistance in tomato. Proc Natl Acad Sci USA 95: 10300–10305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitarch A, Diez-Orejas R, Molero G, Pardo M, Sanchez M, Gil C, Nombela C (2001) Analysis of the serological response to systematic Candida albicans infection in a murine model. Proteomics 1: 550–559 [DOI] [PubMed] [Google Scholar]
- Qin XF, Holuigue L, Horvath DM, Chua NH (1994) Immediate early transcription activation by salicylic acid via the cauliflower mosaic virus as-1 element. Plant Cell 6: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirino BF, Noh YS, Himelblau E, Amasino RM (2000) Molecular aspects of leaf senescence. Trends Plant Sci 5: 278–282 [DOI] [PubMed] [Google Scholar]
- Quirino BF, Normanly J, Amasino RM (1999) Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol Biol 40: 267–278 [DOI] [PubMed] [Google Scholar]
- Ross D, Kepa JK, Winski SL, Beall HD, Anwar A, Siegel D (2000) NAD(P) H:quinone oxidoreductase 1 (NQO1): chemoprotection, bioactivation, gene regulation and genetic polymorphisms. Chem Biol Interact 129: 77–97 [DOI] [PubMed] [Google Scholar]
- Shukla A, Berglund L, Nielsen LP, Nielsen S, Hoffman HJ, Dahl R (2000) Regulated exocytosis in immune functions: are SNARE-proteins involved? Respir Med 94: 10–17 [DOI] [PubMed] [Google Scholar]
- Staskawicz BJ, Mudgett MB, Dangl JL, Galan JE (2001) Common and contrasting themes of plant and animal diseases. Science 292: 2285–2289 [DOI] [PubMed] [Google Scholar]
- Stekel DJ, Git Y, Falciani F (2000) The comparison of gene expression from multiple cDNA libraries. Genome Res 10: 2055–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Xie M, Kim YJ, Zhou J, Klessig DF, Martin GB (1999) Overexpression of Pto activates defense responses and confers broad resistance. Plant Cell 11: 15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thara VK, Tang X, Gu YQ, Martin GB, Zhou JM (1999) Pseudomonas syringae pv tomato induces the expression of tomato EREBP-like genes Pti4 and Pti5 independent of ethylene, salicylate and jasmonate. Plant J 20: 475–483 [DOI] [PubMed] [Google Scholar]
- Xiao F, Lu M, Li J, Zhao T, Yi SH, Thara VK, Tang X, Zhou J (2003) Pto mutants differentially activate Prf-dependent, avrPto-independent resistance and gene-for-gene resistance. Plant Physiol 131: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Tang X, Zhou J (2001) Expression of 35S::Pto globally activates defense-related genes in tomato plants. Plant Physiol 126: 1637–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zancope-Oliver RM, Reiss E, Lott TJ, Mayer LW, Deepe GS (1999) Molecular cloning, characterization, and expression of the M antigen of Histoplasma capsulatum. Infect Immunol 67: 1947–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Tang X, Martin GB (1997) The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J 16: 3207–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zugel U, Kaufmann SHE (1999) Role of heat shock proteins in protection from and pathogenesis of infectious diseases. Clin Microbiol Rev 12: 19–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.