Abstract
Exportin-t was first identified in humans as a protein that mediates the export of tRNAs from the nucleus to the cytoplasm. Mutations in Los1p, the Saccharomyces cerevisiae exportin-t homolog, result in nuclear accumulation of tRNAs. Because no exportin-t mutants have been reported in multicellular organisms, the developmental functions of exportin-t have not been determined. Here, we report the isolation and characterization of two Arabidopsis exportin-t mutants, paused-5 and paused-6. The mutant phenotypes indicate that exportin-t acts pleiotropically in plant development. In particular, paused-5 and paused-6 result in delayed leaf formation during vegetative development. The two paused mutations also cause the transformation of reproductive organs into perianth organs in the hua1-1 hua2-1 background, which is partially defective in reproductive organ identity specification. The floral phenotypes of hua1-1 hua2-1 paused mutants resemble those of mutations in the floral homeotic gene AGAMOUS. Moreover, paused-5 enhances the mutant phenotypes of two floral meristem identity genes, LEAFY and APETALA1. The developmental defects caused by paused mutations confirm the important roles of exportin-t in gene expression in multicellular organisms. In addition, a paused null allele, paused-6, is still viable, suggesting the presence of redundant tRNA export pathway(s) in Arabidopsis.
In plants, postembryonic development results from the activities of the apical meristems. The shoot apical meristem (SAM) generates leaves during the vegetative stage of plant development and floral meristems during reproductive development. In Arabidopsis, four types of floral organs, sepal, petal, stamen, and carpel, are produced by a floral meristem in four concentric rings, or whorls. The identities of the four floral organ types are specified by the combinatorial activities of four classes of floral homeotic genes known as A, B, C, and SEPALLATA genes (for review, see Theissen and Saedler, 2001; Lohmann and Weigel, 2002). AGAMOUS (AG), a class C gene, is a master regulator of reproductive organ identities. Severe loss-of-function mutations in AG result in the transformation of reproductive organs into perianth organs (Bowman et al., 1989). In addition, AG acts in the proper termination of floral meristem activity such that flowers are determinate structures. This AG activity results from its ability to repress the stem cell maintenance gene WUSCHEL (WUS) in the center of flowers (Lenhard et al., 2001; Lohmann et al., 2001).
Several new genes that also act in reproductive organ identity and floral determinacy specification were identified from two genetic screens in sensitized backgrounds. In a genetic screen in the ag-4 background, recessive mutations in HUA1 and HUA2 were found to enhance the phenotypes of the weak ag-4 allele such that ag-4 hua1-1 hua2-1 flowers resemble severe ag mutants, such as ag-1, ag-2, or ag-3 (Chen and Meyerowitz, 1999). The hua1-1 hua2-1 double-mutant flowers have occasional petaloid stamens and gynoecia with partial sepal character (Chen and Meyerowitz, 1999; Chen et al., 2002; Western et al., 2002), phenotypes that indicate partial loss of class C activity. In a genetic screen in the hua1-1 hua2-1 background, extragenic, recessive mutations in three HUA ENHANCER (HEN) loci, HEN1, HEN2, and HEN4, were found to enhance the weak hua1-1 hua2-1 phenotypes to severe ag-like phenotypes, suggesting that the HEN genes also act in reproductive organ identity and floral determinacy specification (Chen et al., 2002, Cheng et al., 2003; Western et al., 2002).
Among the newly identified HUA and HEN genes, HUA1, HUA2, HEN2, and HEN4 appear to promote AG pre-mRNA processing. Mutations in these genes result in the accumulation of AG transcripts containing part of the large AG second intron (Cheng et al., 2003). HUA1 is a nuclear RNA-binding protein (Li et al., 2001) and interacts with HEN4, a protein with KH-type RNA-binding motifs (Cheng et al., 2003). HUA2 encodes a protein with motifs found in transcription coactivators and nuclear RNA-processing proteins (Chen and Meyerowitz, 1999; Cheng et al., 2003). HEN2 encodes a protein with similarity to Saccharomyces cerevisiae Dob1p (Mtr4p), a putative RNA helicase (Western et al., 2002). HEN1 differs from other HUA and HEN genes in that it does not appear to be required for AG pre-mRNA processing. Two recessive mutations in HEN1 result in no or reduced accumulation of microRNAs (Park et al., 2002; W. Park and X. Chen, unpublished data), suggesting that HEN1 acts in microRNA metabolism. The fact that hua1-1 hua2-1 hen1-1 flowers show ag-like phenotypes (Chen et al., 2002) suggests that a microRNA(s) acts in reproductive organ identity and floral determinacy specification.
In this report, we describe two recessive mutations in a locus that we initially named HUA ENHANCER 5 (HEN5), hen5-1 and hen5-2, which cause stamen-topetal and carpel-to-sepal transformation in the hua1-1 hua2-1 background. We show that HEN5 is identical to PAUSED (PSD), a gene previously identified as a regulator of developmental phase transitions (Telfer et al., 1997). We have renamed hen5-1 and hen5-2 as psd-5 and psd-6, respectively. We show that PSD is required for the proper function of not only the SAM, but also the floral meristems and floral organ primordia. We demonstrate that PSD is required for the normal accumulation of AG mRNA and protein.
PSD encodes the only protein in the Arabidopsis genome with significant similarity to human exportin-t and yeast Los1p, which are nuclear export receptors of tRNA (Arts et al., 1998; Hellmuth et al., 1998; Kutay et al., 1998). All three proteins have a conserved Ran-binding domain, a signature of the importin-β protein family (Görlich et al., 1997), at the N terminus of the proteins (see below). Ran is a small GTPase and is a crucial component in many nucleocytoplasmic transport processes (for review, see Görlich and Kutay, 1999; Kuersten et al., 2001). Members of the importin-β family can bind Ran and many act in nucleocytoplasmic transport (for review, see Görlich and Kutay, 1999; Cullen, 2000; Grosshans et al., 2000,Grosshans et al., 2000). Despite the critical function of Los1p in nucleocytoplasmic transport of tRNAs, los1 mutants are viable (Hurt et al., 1987), suggesting the presence of redundant tRNA export pathways in yeast. Exportin-5, another mammalian importin-β protein, has been recently demonstrated to also serve as a tRNA export receptor (Bohnsack et al., 2002; Calado et al., 2002). We demonstrate that psd-6 is a null allele with undetectable PSD mRNA but is still viable and fertile. Therefore, exportin-t, which appears to be dispensable in yeast, is also dispensable in a multicellular organism for viability, suggesting that redundant tRNA export pathway(s) must exist. However, the pleiotropic phenotypes of psd mutants suggest that exportin-t is required for multiple developmental pathways in Arabidopsis.
RESULTS
PSD Is Required for the Proper Activities of the SAM
We isolated the psd-5 mutation from the hua1-1 hua2-1 mutagenesis screen as a recessive mutation that enhanced the weak hua1-1 hua2-1 loss-of-C-function phenotypes. The psd-5 allele was later segregated away from the hua1-1 hua2-1 background and was found to exhibit a vegetative phenotype on its own. While a wild-type SAM continues to generate leaves on its flanks during vegetative development, leaf formation was variably delayed in psd-5. In most psd-5 plants, the leaf-generating activity of the SAM appeared to be compromised such that fewer leaves were formed than wild type in the same period of time (Fig. 1, A and B). In approximately 17% of the plants, no leaf or only one leaf was generated at d 15 after the transfer of the psd-5 seeds to the growth room (Fig. 1, C and D). At the time of bolting, there were fewer rosette leaves produced in psd-5 plants (data not shown). After the cloning of the PSD gene (see below), we identified another mutant, psd-6, which contained a T-DNA insertion in the PSD coding region from the Salk T-DNA collection. psd-6, which was from a different ecotype, exhibited similar phenotypes as psd-5 (Fig. 1, E–H), although the frequency of plants with no or only 1 leaf at d 15 was only 4%. These vegetative phenotypes were similar to those of previously described psd mutants (Telfer et al., 1997). The layered structure of the SAM is abnormal in psd mutants, and psd seedlings appear to contain dead cells at the meristem center (Telfer et al., 1997). The SAM defects may lead to the observed delay in leaf appearance in psd plants (Telfer et al., 1997). Thus, PSD is required for the proper activity of the SAM.
Figure 1.
psd phenotypes in seedlings. A, A 15-d-old Landsberg erecta (Ler) plant. B, A 15-d-old psd-5 plant with fewer true leaves than wild type at the same stage. C, A 15-d-old psd-5 plant with no true leaves. D, A 15-d-old psd-5 plant with a terminal leaf. E, A 14-d-old Columbia (Col) plant. F and G, 14-d-old psd-6 plants with fewer true leaves than Col. One cotyledon was absent in G. H, A 20-d-old psd-6 plant with a terminal leaf. I, A 20-d-old F1 plant from a cross between Col and Ler. J, A 20-d-old F1 plant from a cross between psd-5 and psd-6. Fewer rosette leaves are found than the corresponding wild type (I) at the same stage. K, A 13-d-old psd-5 hua2-1 (Ler) transgenic seedling containing pPZP211-HEN5p3/4 showing normal true leaf formation. L, A 13-d-old psd-5 hua2-1 (Ler) transgenic seedling containing pPZP-35S-green fluorescent protein (GFP) showing the psd-5 phenotype. The magnification in K and L is different from that in A through J.
psd Mutations Result in Floral Organ Identity and Floral Determinacy Defects
In the hua1-1 hua2-1 background, psd-5 resulted in stamen-to-petal and carpel-to-sepal transformation in the flower (Fig. 2, A–C), whereas hua1-1 psd-5 or hua2-1 psd-5 flowers were indistinguishable from psd-5 flowers, which were largely normal (Fig. 2G) except for reduced fertility. hua1-1 hua2-1 psd-5 flowers had petals in the third whorl and gynoecia with smaller and irregularly shaped ovary in the fourth whorl (Fig. 2, B and E). Occasionally in late-arising flowers, additional flowers appeared in the center (Fig. 2C), suggesting loss of floral determinacy. Scanning electron microscopy (SEM) showed that valve epidermal cells throughout hua1-1 hua2-1 psd-5 ovaries had epicuticular striations that resembled sepal instead of valve epidermal cells (Fig. 2, H and I), whereas cells with sepal characteristics were only found at the apical portion of hua1-1 hua2-1 valves (Chen et al., 2002; Western et al., 2002). Furthermore, although psd-5 single mutant flowers were apparently normal (Fig. 2G), occasional valve cells with epicuticular striations could be found in the apical portion of the psd-5 ovaries (Fig. 2K). Cells with such sepal characteristics were never found in wild-type ovaries (Fig. 2J). In summary, psd-5 exhibited extremely weak loss-of-C function defects on its own and enhanced the partial loss-of-C function defects of the hua1-1 hua2-1 mutants.
Figure 2.
Floral phenotypes of psd mutants and interactions with mutations in A, B, and C genes. A, A hua1-1 hua2-1 (Col) flower. B, An early hua1-1 hua2-1 psd-5 flower with petals instead of stamens in the third whorl (arrow). C, A late hua1-1 hua2-1 psd-5 flower with petals in the third whorl and an additional flower in the fourth whorl. D, A hua1-1 hua2-1 psd-6 (Col) flower showing stamen-to-petal transformation in the third whorl. E, Comparison of hua1-1 hua2-1 (1), hua1-1 hua2-1 psd-5 (2), and hua1-1 hua2-1 psd-6 (3 and 4) gynoecia. F, A hua1-1 hua2-1 psd-6 flower with an internal flower in the fourth whorl. G, A psd-5 flower with normal floral organ types. H and I, Abaxial surfaces of the base of a hua1-1 hua2-1 (H) and a hua1-1 hua2-1 psd-5 valve (I). J and K, Abaxial surfaces of the apical portion of an Ler (J) and a psd-5 valve (K). The psd-5 valve has a few cells with typical sepal epicuticular striations (arrow). L, A hua1-1 hua2-1 ag-1 flower. M, A hua1-1 hua2-1 psd-5 ag-1 flower. N, A hua1-1 hua2-1 ap2-2 flower. O, A hua1-1 hua2-1 psd-5 ap2-2 flower. P, A hua1-1 hua2-1 pi-3 flower with filaments (arrow) in the third whorl. Q, A hua1-1 hua2-1 psd-5 pi-3 flower with sepals in the third whorl. R, A hua1-1 hua2-1 ap1-1 flower. S, A hua1-1 hua2-1 psd-5 ap1-1 “flower” with leaf-like organs in a spiral phyllotaxy, and an internal terminal structure with carpel character. Size bars in H through K = 10 μm.
Another allele, psd-6, was isolated from the Salk T-DNA collection and was in the Col background. psd-6 single-mutant flowers did not display any floral homeotic phenotypes. The psd-6 allele was crossed into hua1-1 hua2-1 (Col), a strain in which the hua1-1 and hua2-1 mutations were introgressed into the Col background to determine if this psd allele also behaved similarly to psd-5. The flowers of hua1-1 hua2-1 (Col) usually had normal stamens (Fig. 2A) and occasionally had petaloid stamens in the third whorl. The hua1-1 hua2-1 psd-6 flowers had petals in the third whorl (Fig. 2D) and gynoecia with elongated gynophores or another flower in the fourth whorl (Fig. 2, E and F). We also crossed psd-6 into hua1-1 hua2-1 in the Ler background and the F2 triple mutants displayed floral phenotypes similar to those of hua1-1 hua2-1 psd-5 or hua1-1 hua2-1 psd-6 in the Col background (data not shown). Thus, the two psd alleles exhibit similar defects in organ identity and floral determinacy and reveal a requirement for PSD in floral patterning.
PSD Is Most Probably the Ortholog of Human Exportin-t
To better understand the molecular basis of the psd mutant phenotypes, a map-based cloning approach was used to identify the PSD gene. PSD was mapped to a 27-kb region at the south end of chromosome I on the bacterial artificial chromosome (BAC) F28P22 (Fig. 3A). Sequencing four genes in this region revealed a single nucleotide deletion in the ninth exon of the gene At1g72560 (Fig. 3A). The deletion of a G is predicted to cause a premature stop codon 11 bp after the mutation site. Therefore, At1g72560 is likely the PSD gene. To confirm this, a 7-kb genomic fragment surrounding the coding region of At1g72560 was amplified by PCR and cloned into the plant expression vector pPZP211. The resulting construct pPZP211-HEN5p3/4 and another unrelated construct pPZP211-35S-GFP were transformed into psd-5 hua2-1 plants. All 30 transgenic plants containing pPZP211-HEN5p3/4 had normal true leaf initiation or appearance (Fig. 1K). In contrast, transgenic plants containing pPZP211-35S-GFP exhibited similar leaf initiation defects to those of psd-5 (Fig. 1L). Thus, At1g72560 complements the psd mutant phenotypes.
Figure 3.
The PSD gene and protein. A, Positional cloning of PSD. The black lines represent BACs with the BAC names indicated above. The recombination frequencies at the markers (vertical lines) are shown below the BACs as number recombination/number chromosome. The PSD genomic structure is shown with rectangles representing exons and lines representing introns. The psd-5 single nucleotide deletion is marked as a star in exon 9 and the psd-6 T-DNA insertion mutation is represented as an inverted triangle in exon 1. B, A clustalW alignment of PSD protein with exportin-t (human, Homo sapiens) and Los1p (Saccharomyces cerevisiae). The amino acids showing identity and ones showing similarity are outlined, and highlighted in dark and light gray, respectively. The lines below the alignment denote the Ran-binding domain.
To further confirm that At1g72560 is PSD, we obtained a T-DNA insertion line, which we later named psd-6, from the Salk T-DNA collection. We verified that this line had a T-DNA inserted in the first exon of the At1g72560 gene (Fig. 3A). Compared with wild-type Col plants, these plants had a slower rate of leaf appearance, fewer rosette leaves, and lighter colored leaves (Fig. 1, F–H), phenotypes also exhibited by psd-5 plants. This T-DNA insertion line was kanamycin sensitive, probably due to silencing of the NPTII gene in the T-DNA. To confirm that the phenotypes observed were due to a T-DNA insertion in PSD rather than another mutation in the background, we crossed this line with psd-5. The F1 plants showed the psd phenotypes, in contrast to the F1 plants that resulted from crosses between Col and Ler plants (Fig. 1, I and J). Therefore, the T-DNA mutant is another psd allele, which we named psd-6. When we introduced this mutation into the hua1-1 hua2-1 (Col) background, the flowers of the resulting triple-mutant plants had phenotypes similar to hua1-1 hua2-1 psd-5: petals in the third whorl instead of stamens, gynoecia with elongated gynophores, or another flower in the center (Fig. 2, D–F).
A PSD cDNA was isolated by reverse transcription-PCR from inflorescence tissue. The gene contained 13 exons and 12 introns (Fig. 3A). The predicted PSD protein has 988 amino acids and shares 27% identity and 48% similarity to human exportin-t, and 21% identity and 41% similarity to yeast Los1p throughout the proteins (Fig. 3B). Human exportin-t shares 19% identity and 41% similarity to Los1p. Thus, PSD and human exportin-t are more closely related to each other than either one is to Los1p. All three proteins have a conserved Ran-binding domain, a signature of the importin-β protein family, at the N terminus of the proteins (Fig. 3B). No other proteins with comparable similarity with exportin-t and Los1p exist in the Arabidopsis genome. Therefore, PSD is most likely a unique ortholog of exportin-t in Arabidopsis and possibly acts in the nucleocytoplasmic transport of tRNA in a Ran-dependent manner.
Expression of A, B, and C genes in hua1-1 hua2-1 psd-5
We examined the expression of a class A gene APETALA1 (AP1), the two class B genes APETALA3 (AP3) and PISTILLATA (PI), and the class C gene AG in hua1-1 hua2-1 and hua1-1 hua2-1 psd-5 flowers to begin to understand the molecular basis of the homeotic transformation in hua1-1 hua2-1 psd-5. Consistent with the observed stamen-to-petal transformation in the third whorl and carpel-to-sepal transformation in the fourth whorl of hua1-1 hua2-1 psd-5 flowers, AP1 RNA was present in the inner two whorls (Fig. 4, A and B). Despite the fact that hua1-1 hua2-1 psd-5 flowers showed loss-of-C function phenotypes, AG RNA was readily detected in the inner two whorls (Fig. 4, C and D). We did not detect any difference in the temporal or spatial patterns of AG expression between hua1-1 hua2-1 and hua1-1 hua2-1 psd-5 (Fig. 4, C and D, and data not shown). The class B gene PI also showed similar expression patterns in the two genotypes (data not shown). Interestingly, although AP3 RNA was found exclusively in whorls 2 and 3 in hua1-1 hua2-1 flowers (Fig. 4E), AP3 RNA was detected in some cells on the adaxial side of the ovary in hua1-1 hua2-1 psd-5 flowers (Fig. 4F). This suggests a factor that acts to restrict the expression of AP3 from ovary cells in the fourth whorl is affected by mutations in PSD, a putative exportin-t.
Figure 4.
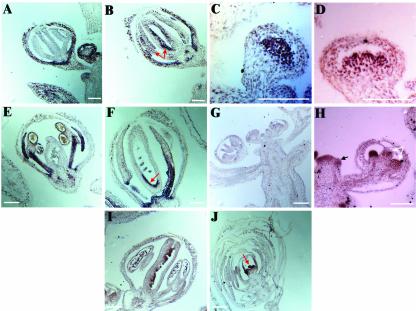
AP1, AG, AP3, and PSD RNA accumulation as determined by in situ hybridization. The brown/purple color represents positive hybridization signals. A, A stage 12 hua1-1 hua2-1 flower without AP1 expression in the inner two whorls. B, A hua1-1 hua2-1 psd-5 flower with ectopic AP1 expression in the inner two whorls (arrows). C, A stage 5–6 hua1-1 hua2-1 flower showing AG RNA in the center. D, A stage 6 hua1-1 hua2-1 psd-5 flower showing AG RNA in the center. E, A stage 12 hua1-1 hua2-1 flower with AP3 expression in the second and third whorls. F, A stage 12 hua1-1 hua2-1 psd-5 flower showing ectopic expression of AP3 on the adaxial side of the ovary (arrow). G, A longitudinal section of an Ler inflorescence hybridized to a PSD sense probe. H through J, Hybridization of Ler (H and I) and ag-1 (J) tissues to a PSD antisense probe. PSD expression can be detected in young stamens and carpels (white arrows) in a stage 10 flower and in young floral meristems (black arrow; H), in the ovules of a stage 12 flower (I) and in the central young floral organs and the meristem (arrow) in the ag-1 flower (J). Size bar = 50 μm.
PSD Is Required for Multiple Processes in Flower Development
We introduced mutations in the class A, B, or C genes into the hua1-1 hua2-1 psd-5 background to further determine whether PSD, as a putative exportin-t, specifically affects the class C pathway in flower development. Although the phenotypes of hua1-1 hua2-1 psd-5 ap2-2, hua1-1 hua2-1 psd-5 pi-3, or hua1-1 hua2-1 psd-5 ag-1 flowers indicate that primarily C function is affected in hua1-1 hua2-1 psd-5 flowers, the hua1-1 hua2-1 psd-5 ap1-1 combination revealed that psd-5 also affects an earlier step in floral patterning. The finding that psd-5 results in multiple developmental defects in the flower is consistent with the molecular role of PSD as a putative exportin-t.
hua1-1 hua2-1 psd-5 ap2-2
hua1-1 hua2-1 ap2-2 flowers resemble ap2-2 flowers in the outer three whorls and hua1-1 hua2-1 flowers in the fourth whorl (Chen and Meyerowitz, 1999; Fig. 2N). The hua1-1 hua2-1 psd-5 ap2-2 quadruple mutant differed from hua1-1 hua2-1 ap2-2 in that it had flowers with leaf-like organs in the outer three whorls and sepaloid carpels in the fourth whorl (Fig. 2O). This phenotype resembles that of ag-1 ap2-2 (Bowman et al., 1991) and is consistent with the assumption that hua1-1 hua2-1 psd-5 plants are compromised in class C function in the flower.
hua1-1 hua2-1 psd-5 pi-3
pi-3 is a week allele with sepals in the outer two whorls and carpels in the inner two whorls (Bowman et al., 1991). hua1-1 hua2-1 enhances the pi-3 phenotype such that the third whorl organs in hua1-1 hua2-1 pi-3 show reduced carpel character and develop into sepals, carpelloid sepals, and/or filaments (Chen and Meyerowitz, 1999; Fig. 2P). The third whorl phenotype of hua1-1 hua2-1 pi-3 was further enhanced by psd-5, such that only sepals were present in the third whorl of hua1-1 hua2-1 psd-5 pi-3 flowers (Fig. 2Q). The lack of carpel characteristics in the third whorl of hua1-1 hua2-1 psd-5 pi-3 flowers is consistent with the assumption that class C activity is greatly reduced in hua1-1 hua2-1 psd-5.
hua1-1 hua2-1 psd-5 ag-1
hua1-1 hua2-1 ag-1 flowers resemble those of ag-1 (Chen and Meyerowitz, 1999; Fig. 2L). hua1-1 hua2-1 psd-5 ag-1 quadruple mutants were indistinguishable from hua1-1 hua2-1 ag-1 triple mutants in terms of the homeotic transformation defects in the third whorl or the floral determinacy defects in the fourth whorl (Fig. 2M), indicating that ag-1 is epistatic to psd-5.
hua1-1 hua2-1 psd-5 ap1-1
ap1-1 is a severe loss of function mutation in the class A gene AP1 (Irish and Sussex, 1990; Bowman et al., 1993). In addition to its role in floral organ identity specification, AP1 also specifies floral meristem identity (Mandel et al., 1992; Mandel and Yanofsky, 1995). hua1-1 hua2-1 ap1-1 flowers resemble ap1-1 flowers (Chen and Meyerowitz, 1999; Fig. 2R). We generated the hua1-1 hua2-1 psd-5 ap1-1 quadruple mutant to determine whether the ectopic AP1 expression in the third whorl of hua1-1 hua2-1 psd-5 flowers was responsible for the stamen-to-petal transformation. The introduction of ap1-1 into hua1-1 hua2-1 hen4-1, which has petals in the third whorl, rescued the third whorl defect of hua1-1 hua2-1 hen4-1 such that stamens were found in the quadruple mutant (Cheng et al., 2003). However, hua1-1 hua2-1 psd-5 ap1-1 plants showed a novel phenotype. The quadruple mutant “flowers” resembled shoots in that they contained sepal/leaf-like organs with a spiral phyllotaxy and a few carpelloid organs in the center (Fig. 2S). The lack of recognizable third whorl organs in the quadruple mutants prevented the assessment of the role of AP1 in the stamen-to-petal transformation in the third whorl of hua1-1 hua2-1 psd-5 flowers. However, the novel phenotype suggests that psd-5 not only affects C function in flower development, but also affects an earlier activity, which specifies the identity of the floral meristem or to allow its proper development. This is consistent with the observation that psd-5 also enhanced the floral meristem defects of lfy mutants (see below).
PSD Is Required for the Specification or Proper Development of the Floral Meristem
psd mutants had slightly more cauline leaves than wild type (data not shown), a phenotype that suggests delayed transition of the SAM from generating shoots to making flowers. To elucidate the possible requirement for PSD in the process of shoot-to-flower transition, we generated psd-5 lfy-5 and psd-5 lfy-6 double mutants. LEAFY (LFY) confers floral meristem identity and transcriptionally activates various floral homeotic genes (Weigel et al., 1992; Mandel and Yanofsky, 1995; Weigel and Nilsson, 1995; Busch et al., 1999; Wagner et al., 1999; Lamb et al., 2002). lfy-6, a strong lfy allele, produces flowers subtended by bracts and early flowers with leaf-like organs arranged in a spiral instead of a whorled pattern, suggesting loss of floral identity and gain of inflorescence shoot characteristics (Weigel et al., 1992). Most of the floral organs in later flowers are sepaloid or carpelloid organs (Weigel et al., 1992; Fig. 5D). In contrast to lfy-6, the weak lfy-5 allele has flowers with more normal floral organs (Weigel et al., 1992; Fig. 5A) and is fertile. Nevertheless, lfy-5 also displays defects in shoot-to-flower transition (Weigel et al., 1992). psd-5 enhanced the phenotypes of lfy-5 and lfy-6 in terms of production of flowers from the SAM. psd-5 lfy-6 plants were similar to psd-5 until the stage of flower generation. After a few cauline leaves emerged, the primary inflorescence developed a number of lateral outgrowths with unknown identities (data not shown) and then terminated in a carpel-like structure with stigmatic tissue (Fig. 5E) or in filaments (Fig. 5F). The lateral outgrowths never developed further to flowers or any structures other than filaments (Fig. 5, E and F). Like in psd-5 lfy-6, the primary inflorescence in psd-5 lfy-5 plants also produced lateral outgrowths that lacked apparent differentiation (Fig. 5, B and C). The synergistic interaction of psd-5 and lfy and the absence of most floral organs in hua1-1 hua2-1 psd-5 ap1-1 plants (Fig. 2S) indicate a requirement for PSD in floral meristem identity specification or in the subsequent development of the floral meristem after its identity has been correctly specified.
Figure 5.
Interactions between psd and lfy. A, A lfy-5 inflorescence with flowers. B, A psd-5 lfy-5 primary inflorescence (arrow) without the formation of flowers. C, A magnification of the inflorescence stem in B. Outgrowths that appear undifferentiated (arrow) are found along the stem and the internodes are more compact than those in lfy-5. D, A lfy-6 inflorescence with abnormal flowers lacking petals and stamens. E, A psd-5 lfy-6 inflorescence, which produces filamentous organs at positions normally occupied by flowers and terminates in a structure with stigmatic tissues at the top. F, A psd-5 lfy-6 inflorescence terminating in filamentous organs that sometimes have two-branched trichomes (arrow), suggesting that the organs have leaf character.
PSD Is Required for AG Expression
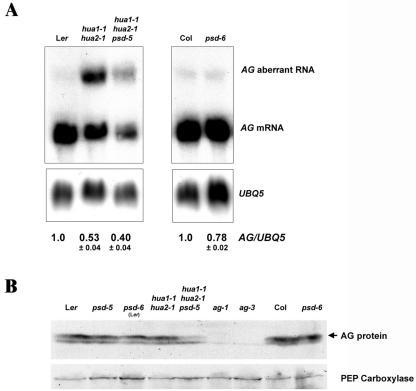
To determine the molecular basis of the floral phenotypes of hua1-1 hua2-1 psd-5 plants, we examined the expression of AG at the RNA and protein levels. HUA1 and HUA2 are required for the proper processing of AG pre-mRNA (Cheng et al., 2003). Aberrant AG RNAs with part of the AG second intron were produced in hua1-1 hua2-1 flowers (Cheng et al., 2003; Fig. 6A). The levels of AG mRNA and aberrant AG RNA were reduced approximately 25% in hua1-1 hua2-1 psd-5 as compared with hua1-1 hua2-1 (Fig. 6A). Similarly, AG mRNA accumulation was reduced by approximately 22% in psd-6 as compared with wild type (Col; Fig. 6A). Thus, we conclude that PSD is required for the accumulation of AG mRNA and likely acts independently of HUA1 and HUA2 because psd-5 did not result in an increase in the level of aberrant AG RNAs. Considering that PSD is the ortholog of exportin-t, it is likely that the expression of a positive regulator of AG is reduced below a threshold level by psd mutations. Previous studies showed that 40% to 50% of wild-type AG mRNA levels could result in flowers with floral homeotic and determinacy defects similar to those in hua1-1 hua2-1 psd flowers (Mizukami and Ma, 1995; Cheng et al., 2003). Therefore, the reduced AG mRNA level in hua1-1 hua2-1 psd-5 can be sufficient to result in its floral phenotypes.
Figure 6.
AG RNA and protein accumulation. A, AG RNA accumulation in various genetic backgrounds as determined by RNA filter hybridization (top panel). The same blot was hybridized with UBQ5 for comparison (bottom panel). The numbers shown below indicate the relative abundance of AG mRNA among different genotypes. These numbers were derived from statistical analysis of three independent experiments. Note that although some hybridization signals may appear saturated in this picture, the signal intensity was in the linear range of phosphorimager quantitation. B, AG protein accumulation in different genotypes (arrow, top panel) as determined by western blotting. As a loading and blotting control, phosphoenolpyruvate (PEP) carboxylase (Kandasamy et al., 2002) accumulation is shown in the bottom panel. The psd-6 (Ler) strain was obtained by two backcrosses of psd-6 (Col) to Ler.
We also examined AG protein accumulation in wild-type and various psd mutants. AG protein was readily detected in psd-5, psd-6 (Ler), and psd-6 (Fig. 6B), although the levels of AG, relative to PEP carboxylase, may be slightly reduced in the psd single mutants as compared with the wild-type controls. This was largely consistent with the slight reduction of AG mRNA levels in psd single mutants. However, a dramatic difference in AG protein levels was detected between hua1-1 hua2-1 psd-5 and hua1-1 hua2-1, although the difference at the AG mRNA level was not as great. This suggests that PSD may contribute to AG expression at the translational or posttranslational levels. Because PSD encodes a putative tRNA nuclear export receptor, it is likely that the low level of AG protein in hua1-1 hua2-1 psd-5 results from defects in AG RNA translation. The fact that the effect of psd mutations on AG protein level is only obvious in the hua1-1 hua2-1 background could be due to the reduced AG RNA level in hua1-1 hua2-1, which may cause AG RNA translation to be more sensitive to global perturbation in protein synthesis.
Expression of PSD in Plants
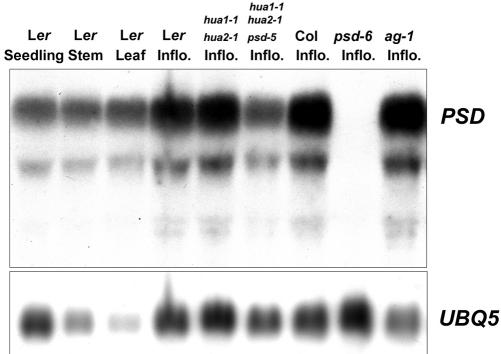
PSD was expressed in seedlings, leaves, stems, and inflorescences as determined by RNA filter hybridization (Fig. 7). Several forms of PSD RNA were detected. The major one corresponded in size to the full-length mRNA (approximately 3 kb) and constituted more than 75% of total PSD RNA. The molecular nature of the smaller RNA species, which were less abundant, was unclear. They were unlikely degraded PSD RNA from RNA isolation because the same RNA samples did not show degradation of UBQ5 RNA. We also examined the accumulation of PSD RNA in different mutants (Fig. 7). Similar levels of PSD RNA in Ler and hua1-1 hua2-1 were found, suggesting that PSD is not regulated by HUA1 or HUA2. PSD RNA abundance was increased in ag-1 inflorescences (Fig. 7), which may simply be due to the presence of more young floral organs in ag-1 flowers (see below). No PSD RNA species were detected in psd-6 inflorescence (Fig. 7), suggesting that psd-6 is a complete loss-of-function allele and indicating that all the RNA species detected in RNA filter hybridization correspond to PSD RNA. The fact that the psd-6 plants are viable and fertile suggests that there must be other proteins with a similar molecular function. Despite being a potentially more severe allele than psd-5, psd-6 results in less severe vegetative defects compared with psd-5. This is likely due to the presence of genetic modifiers in the two different ecotypes because psd-6 (Ler), which was derived from two backcrosses of psd-6 to Ler, appeared very similar to psd-5 in terms of the vegetative phenotypes.
Figure 7.
PSD RNA accumulation in total RNAs from different tissues and genotypes (top panel) as determined by RNA filter hybridization. The same blot was hybridized with a UBQ5 probe for comparison (bottom panel). Inflo., Inflorescences.
We examined PSD RNA expression patterns in flowers by in situ hybridization with a probe corresponding to the first exon of the PSD gene. PSD RNA was detected in all four types of young floral organs (Fig. 4, G–I and data not shown). Expression in stages one to two floral meristems (Fig. 4H) was also detected, consistent with the role of PSD in floral meristem identity. Expression of PSD RNA was also examined in ag-1 flowers. High levels of PSD RNA were detected in young internal flowers and in the meristems in the center of ag-1 flowers (Fig. 4J). The abundance of young floral organs expressing PSD in ag-1 flowers may explain the increased PSD RNA abundance in ag-1 flowers (Fig. 7).
DISCUSSION
PSD Is Required in the SAM and Floral Meristems
The hua1-1 hua2-1 psd-5 flowers have petals in the third whorl and sepaloid carpels or an internal flower in the fourth whorl, phenotypes that resemble those of ag mutants. psd-5 single mutants exhibit occasional sepal cells in the valves of the gynoecium. These phenotypes suggest that PSD is required for C function in the flower. Unlike HUA1 and HEN4, which seem to be specific for the class C pathway in flower development, PSD is required more broadly in flower development. This is reflected by the fact that AP3 is misexpressed in hua1-1 hua2-1 psd-5 but not hua1-1 hua2-1 flowers, and that psd-5 enhances the floral meristem identity defects of ap1-1 and lfy mutants. As a putative tRNA export receptor, it is unlikely that PSD acts directly to specify the identities of the floral meristem or floral organ primordia. Instead, it is more likely that PSD is required for the proper expression of genes in the regulatory networks that specify these identities. The delayed leaf appearance in psd mutants may reflect a requirement for PSD in cell division and/or differentiation in the SAM or in the lateral primordia originated from the SAM. The fact that psd-5 enhances lfy and ap1 in floral meristem identity defects suggests that PSD is required for lateral meristems to adopt their proper identities or to develop properly. In fact, almost all vegetative and floral defects of psd mutants point to a role for PSD in tissues undergoing cell division and/or differentiation, such as the SAM, lateral meristems, or organ primordia. Consistent with this, in transgenic plants containing the uidA gene driven by the PSD promoter, β-glucuronidase (GUS) activity can be detected at high levels in regions undergoing fast cell division and/or differentiation, such as young leaf, growing leaf blades, young floral organs, and root tips (Supplemental Fig. 1 available at http://www.plantphysiol.org). In contrast, little GUS expression was detected in mature organs (Supplemental Fig. 1). In light of the fact that PSD is likely a tRNA export receptor, the PSD expression patterns and the psd mutant phenotypes may reflect more active tRNA export in meristems and young organs.
PSD Is Required for Proper Expression of Multiple Genes in Plant Development
The PSD protein is most probably the Arabidopsis ortholog of exportin-t and Los1p, nuclear receptors of tRNA export in humans and yeast, respectively. Hunter et al. (C. Hunter et al., 2003) showed that PSD rescued the los1 mutant phenotype in yeast, suggesting that PSD can act in tRNA nuclear export. It is then reasonable to hypothesize that cytoplasmic tRNA abundance would decrease in psd mutants, leading to an overall reduced rate of translation and consequently lower level of protein accumulation. Such a potential molecular function of PSD is consistent with the fact that PSD is required in multiple processes in development. This model also predicts that expression of almost all proteins should be affected because the function of tRNA export is fundamental. However, although many developmental processes are affected in psd mutants, leaf formation and SAM organization in seedlings appear to be most sensitive to loss of PSD function. Floral organ identity defects are only found in the hua1-1 hua2-1 background. We hypothesize that the apparent differential sensitivity of various developmental processes to loss of PSD activity may be due to the following reasons. First, PSD may not be required to the same extent for the expression of different proteins. Proteins required for SAM organization and leaf initiation may be affected more than other proteins by psd mutations. It was shown that the nuclear export of some tRNAs is not affected by los1 mutations in yeast (Grosshans et al., 2000,Grosshans et al., 2000) and that all tRNA species may not be transported by human exportin-t (Bohnsack et al., 2002). Therefore, some proteins may be preferentially sensitive to defects in the nuclear export of a subset of tRNAs in psd mutants. This is consistent with our observation that AG abundance is more affected in hua1-1 hua2-1 psd-5 than that of PEP carboxylase. Second, all proteins may be affected by psd mutations, but different developmental processes may require different threshold levels of their key regulator proteins such that some processes are more sensitive to reduction of protein abundance. Finally, the somewhat limited developmental defects of psd mutants may not reflect a differential effect of psd mutations on different cellular proteins, but rather a stronger requirement for tRNA export in actively growing tissues.
It is unknown what proteins are responsible for the various psd phenotypes, as a result of presumably globally reduced protein synthesis in psd mutants. The psd early seedling phenotypes are similar to that of pinhead/zwille mutants. PINHEAD (PNH)/ZWILLE (ZLL) is important for SAM formation and/or maintenance such that mutations in PNH/ZLL result in seedlings with terminal leaves (Moussian et al., 1998; Lynn et al., 1999), phenotypes also found in some psd-5 plants. Therefore, decreased expression of PNH/ZLL in psd plants may cause the defects in seedling development. The translation of AG RNA is probably affected by psd mutations. This is supported by the observation that AG protein abundance is greatly reduced in hua1-1 hua2-1 psd-5 compared with hua1-1 hua2-1, whereas the difference at the AG mRNA level is not so great. Expression of other gene(s) upstream of AG may also be directly facilitated by PSD because the AG mRNA level is reduced in psd single mutants compared with wild type. The reduced accumulation of AG RNA and protein is likely causing the homeotic phenotypes in hua1-1 hua2-1 psd flowers. psd-5, hua1-1 psd-5, and hua2-1 psd-5 plants show reduced fertility. We presume that psd-5 affects the expression of a gene or genes involved in fertility.
Redundant tRNA Export Pathway(s) in Arabidopsis
Because tRNA is an essential element required for translation, it would be expected that efficient tRNA export from the nucleus to the cytoplasm is essential for viability. However, Los1p, a tRNA exportin from yeast (Hellmuth et al., 1998; Sarkar and Hopper, 1998) is nonessential (Hurt et al., 1987), indicating that Los1p is dispensable for tRNA export in yeast and that there must be other tRNA export pathways. Several Los1p-independent pathways have been reported in yeast (Grosshans et al., 2000,Grosshans et al., 2000; Feng and Hopper, 2002), although none of the proteins found in these pathways seems to be a transport receptor, like Los1p, which is a member of the importin-β protein family (Görlich et al., 1997). The role of a mammalian importin-β protein, exportin-5, in tRNA export was recently reported (Bohnsack et al., 2002; Calado et al., 2002). Exportin-5, known previously to export double-stranded RNA-binding proteins (Brownawell and Macara, 2002), binds eukaryotic elongation factor 1A (eEF1A) via aminoacylated tRNAs and exports them out of the nucleus (Bohnsack et al., 2002; Calado et al., 2002). Here, we showed that plants are still viable even when PSD RNAs are completely depleted (i.e. in psd-6). This suggests that in Arabidopsis, as in yeast and mammals, there may be redundant pathways for tRNA nuclear export. Arabidopsis has an exportin-5 homolog, namely HASTY, loss-of-function mutations in which cause defects in many different processes in plant development (Bollman et al., 2003). Although a role in nucleocytoplasmic transport has yet to be demonstrated, HASTY interacts with Ran in a yeast two-hybrid assay (Bollman et al., 2003), which is consistent with such a potential role. An alternative possibility is that passive diffusion of tRNAs through nuclear pores is sufficient for viability. Although not essential for viability, the defects of psd mutants indicate that PSD is necessary for multiple developmental processes in a multicellular organism.
MATERIALS AND METHODS
Plant Strains and Ethyl Methanesulfonate Mutagenesis
The strains used in this work are hua1-1, hua2-1, hua1-1 hua2-1, hua1-1 hua2-1 ap1-1, hua1-1 hua2-1 ap2-2, hua1-1 hua2-1 pi-3, hua1-1 hua2-1 ag-1 (Chen and Meyerowitz, 1999), lfy-5, and lfy-6 (Weigel et al., 1992) in the Ler background. psd-5, in the Ler background, and psd-6, in the Col background, were isolated in this study. hua1-1 and hua2-1 were introgressed into the Col background by crossing hua1-1 hua2-1 to Col four times, resulting in hua1-1 hua2-1 (Col). All plants were grown in Pro-mix BX (Premier, Quakertown, PA) under continuous light or long-day (16 h of light/8 h of dark) conditions at 23°C.
Ethyl methanesulfonate mutagenesis was performed as described previously (Chen et al., 2002). DH677 is one of the M2 lines isolated, containing psd-5, hua1-1, and hua2-1 mutations simultaneously. psd-6 was identified by searching the Salk T-DNA insertion database and was obtained from the Arabidopsis Biological Resource Center. psd-6 was backcrossed into the Ler background twice before using in the analysis of AG protein accumulation.
RNA Filter and In Situ Hybridization
Total RNA was isolated with TRI-REAGENT solutions (Molecular Research Center, Cincinnatti). Approximately 50 μg of total RNA was loaded for each sample. RNA blotting and hybridization were carried out according to Li et al. (2001). Quantitation was performed with a Storm PhosphorImager (Molecular Dynamics, Sunnyvale, CA). The probe for detecting AG RNA was described previously as probe 1, which contains the AG full-length genomic region (Cheng et al., 2003). The PSD probe was amplified from the PSD cDNA with Exp1 (5′-atactaattcaaggcattgtttgacttgtatg-3′) and Exp2 (5′-ggttcgagtcggagaacatgattatg-3′).
In situ hybridization was carried out as described (Li et al., 2001). AG, AP1, PI, and AP3 probes were as described (Chen et al., 2002). The plasmids making PSD probes were generated by amplifying a 1-kb PSD cDNA with HEN5p5 (5′-gcgcggagctcatggatgaccttgaacaggcaatagta-3′) and HEN5p18 (gcgcggagctcttcgttcagcaaatccatggaga-3′), and cloning them into pCR2.1-TOPO (Invitrogen, Carlsbad, CA) in two orientations. The resulting pCR2.1-TOPO-HEN5AS and pCR2.1-TOPO-HEN5S plasmids were in vitro transcribed to generate the antisense and sense probes.
SEM
Tissue fixation, critical point drying, and image acquisition for SEM were performed as described previously (Western et al., 2002).
Immunological Detection of Plant Proteins
Plant proteins were isolated and resolved in SDS-PAGE as described (Riechmann et al., 1999). Rabbit antisera that specifically recognize Arabidopsis AG protein were used (1:3,000) in the immunological detection of AG. Rabbit anti-PEP carboxylase (maize [Zea mays] leaf) polyclonal antibody (Rockland, Gilbertsville, PA) was used as a loading and electroblotting control. Blotting and detection were performed according to instructions from the enhanced chemiluminescence plus Western Blotting Detection System (Amersham Pharmacia Biotech, Piscataway, NJ).
Map-Based Cloning of PSD, Genomic Complementation, and cDNA Isolation
hua1-1 hua2-1 psd-5/+ plants were crossed with hua1-1hua2-1 (Col), and the F2 population segregating the hua1-1 hua2-1 psd-5 floral phenotype was used as the mapping population, in which approximately 1,500 triple-mutant plants were identified. Genomic DNA was isolated as described (Edwards et al., 1991) from these F2 mutant plants. SSLP markers were used to initially locate PSD to the bottom of chromosome I between markers nga280 and AthATPASE. SSLP and cleaved-amplified polymorphic sequence markers based on the Cereon Ler/Col SNP database were used to further map PSD to a 27-kb region on the BAC of F28P22. Sequencing four candidate genes in this region revealed a single-nucleotide deletion mutation in At1g72560.
Approximately 7 kb of At1g72560 genomic region was amplified with HEN5p3 (5′-cggggtacctgatttgtagtctcatacgtgcgaatatacatt-3′) and HEN5p4 (5′-cggggtacccaattgtcaaaagaacaatctgtgtttgg-3′), and was cloned into the pPZP211 binary vector. The plasmid pPZP211-HEN5p3/4, or the control plasmid pPZP211-35S-GFP, was transformed into psd-5 hua2-1 plants by the Agrobacterium tumefaciens-mediated infiltration method. The T1 transgenic plants were selected on medium containing 50 μg mL–1 kanamycin.
Reverse transcription-PCR was performed on the total RNA isolated from Ler inflorescence with HEN5p8 (5′-aaaactgcagctaatttgatctggatggtgaaagcga-3′) and HEN5p11 (5′-aaaactgcagccatggatgaccttgaacaggcaatagtaatt-3′), which are located in the 5′- and 3′-untranslated regions, respectively. The amplified cDNA was cloned into pCR2.1-TOPO (Invitrogen) and sequenced. The sequence is in GenBank under the accession number AY288073.
PSD-GUS Expression Analysis
A 1.8-kb PSD genomic fragment upstream of ATG was amplified with HEN5p7 (5′-aaaactgcagtgatttgtagtctcatacgtgcgaatatacatt-3′) and HEN5p9 (5′-aaaactgcagtgtgttaacttaaccccactacaaaaaccc-3′), and was cloned into a pPZP211-GUS plant expression vector (Wang and X. Chen, unpublished data). The resulting pPZP211-PSD-GUS plasmid was transformed into Ler plants. Transgenic plants were selected as described above. GUS staining was performed as described (Jefferson et al., 1987).
Supplementary Material
Acknowledgments
We thank Yulan Cheng for the isolation of psd-5 and for help with the initial mapping of PSD, Dr. Tamara western for introgressing hua1-1 hua2-1 into Columbia, and Jiqun Zhao and Junhong Sun for assistance in the mapping and cloning of PSD. We also thank Drs. Nilgun Tumer, Randall Kerstetter, and Terri Kinzy for valuable suggestions on this work and Drs. Randall Kerstetter, Jun Liu, and Wonkeun Park for helpful comments on the manuscript. We acknowledge Cereon for the release of Ler/Col SNP database and Salk Institute Genomic Analysis Laboratory (SIGnAL) and the Arabidopsis Biological Resource Center (ABRC) for the isolation and distribution of T-DNA insertion lines.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.023291.
This work was supported by the National Institute of Health (grant no. 1–R01–GM–61146 to X.C.).
The online version of this article contains Web-only data. The supplemental material is available at http://www.plantphysiol.org.
References
- Arts GJ, Fornerod M, Mattaj IW (1998) Identification of a nuclear export receptor for tRNA. Curr Biol 8: 305–314 [DOI] [PubMed] [Google Scholar]
- Bohnsack TM, Regener K, Schwappach B, Saffrich R, Paraskeva E, Hartmann E, Görlich D (2002) Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J 21: 6205–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollman KM, Aukerman MJ, Park M-Y, Hunter C, Berardini TZ, Poethig RS (2003) HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130: 1493–1504 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119: 721–743 [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM (1989) Genes directing flower development in Arabidopsis. Plant Cell 1: 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM (1991) Genetic interactions among floral homeotic genes of Arabidopsis. Development 112: 1–20 [DOI] [PubMed] [Google Scholar]
- Brownawell AM, Macara IG (2002) Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J Cell Biol 156: 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch MA, Bomblies K, Weigel D (1999) Activation of a floral homeotic gene in Arabidopsis. Science 285: 585–587 [DOI] [PubMed] [Google Scholar]
- Calado A, Treichel N, Müller E, Otto A, Kutay U (2002) Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J 21: 6216–6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Liu J, Cheng Y, Jia D (2002) HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Meyerowitz EM (1999) HUA1 and HUA2 are two members of the floral homeotic AGAMOUS pathway. Mol Cell 3: 349–360 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Kato N, Wang W, Li J, Chen X (2003) Two RNA binding proteins, HEN4 and HUA1, act in the processing of AGAMOUS pre-mRNA in Arabidopsis thaliana. Dev Cell 4: 53–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR (2000) Nuclear RNA export pathways. Mol Cell Biol 20: 4181–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Hopper AK (2002) A Los1p-independent pathway for nuclear export of intronless tRNAs in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 99: 5412–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Dabrowski M, Bischoff FR, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E (1997) A novel class of RanGTP binding proteins. J Cell Biol 138: 65–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Kutay U (1999) Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15: 607–660 [DOI] [PubMed] [Google Scholar]
- Grosshans H, Hurt E, Simos G (2000) An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev 14: 830–840 [PMC free article] [PubMed] [Google Scholar]
- Grosshans H, Simos G, Hurt E (2000) Transport of tRNA out of the nucleus-direct channeling to the ribosome. J Struct Biol 129: 288–294 [DOI] [PubMed] [Google Scholar]
- Hellmuth K, Lau DM, Bischoff FR, Kunzler M, Hurt E, Simos G (1998) Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol 18: 6374–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C, Aukerman MJ, Sun H, Fokina M, Poethig RS (2003). PAUSED encodes the Arabidopsis exportin-7 orthologue. Plant Physiol 132: 2135–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt DJ, Wang SS, Li Y, Hopper AK (1987) Cloning and characterization of LOS1, a Saccharomyces cerevisiae gene that affects tRNA splicing. Mol Cell Biol 7: 1208–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish VF, Sussex IM (1990) Function of the apetala1 gene during Arabidopsis floral development. Plant Cell 2: 741–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy MK, Mckinney EC, Meagher RB (2002) Functional nonequivalency of actin isovariants in Arabidopsis. Mol Biol Cell 13: 251–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuersten S, Ohno M, Mattaj IW (2001) Nucleocytoplasmic transport: Ran, β and beyond. Trends Cell Biol 11: 497–503 [DOI] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Gorlich D (1998) Identification of a tRNA-specific nuclear export receptor. Mol Cell 1: 359–369 [DOI] [PubMed] [Google Scholar]
- Lamb RS, Hill TA, Tan QK, Irish VF (2002) Regulation of APETALA3 floral homeotic gene expression by meristem identity genes. Development 129: 2079–2086 [DOI] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jürgens G, Laux T (2001) Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105: 805–814 [DOI] [PubMed] [Google Scholar]
- Li J, Jia D, Chen X (2001) HUA1, a regulator of stamen and carpel identities in Arabidopsis, codes for a nuclear RNA binding protein. Plant Cell 13: 2269–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D (2001) A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105: 793–803 [DOI] [PubMed] [Google Scholar]
- Lohmann JU, Weigel D (2002) Building beauty: the genetic control of floral patterning. Dev Cell 2: 135–142 [DOI] [PubMed] [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK (1999) The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126: 469–481 [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360: 273–277 [DOI] [PubMed] [Google Scholar]
- Mandel MA, Yanofsky MF (1995) A gene triggering flower formation in Arabidopsis. Nature 377: 522–524 [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Ma H (1995) Separation of AG function in floral meristem determinacy from that in reproductive organ identity by expressing antisense AG RNA. Plant Mol Biol 28: 767–784 [DOI] [PubMed] [Google Scholar]
- Moussian B, Schoof H, Haecker A, Jurgens G, Laux T (1998) Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J 17: 1799–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Ito T, Meyerowitz EM (1999) Non-AUG initiation of AGAMOUS mRNA translation in Arabidopsis thaliana. Mol Cell Biol 19: 8505–8512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Hopper AK (1998) tRNA nuclear export in Saccharomyces cerevisiae: in situ hybridization analysis. Mol Biol Cell 9: 3041–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A, Bollman KM, Poethig RS (1997) Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124: 645–654 [DOI] [PubMed] [Google Scholar]
- Theissen G, Saedler H (2001) Floral quartets. Nature 409: 469–471 [DOI] [PubMed] [Google Scholar]
- Wagner D, Sablowski RW, Meyerowitz EM (1999) Transcriptional activation of APETALA1 by LEAFY. Science 285: 582–584 [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859 [DOI] [PubMed] [Google Scholar]
- Weigel D, Nilsson O (1995) A developmental switch sufficient for flower initiation in diverse plants. Nature 377: 495–500 [DOI] [PubMed] [Google Scholar]
- Western TL, Cheng Y, Liu J, Chen X (2002) HUA ENHANCER 2, a putative DExH-box RNA helicase, maintains homeotic B and C gene expression in Arabidopsis. Development 129: 1569–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.