Abstract
The gene specifying plastid transketolase (TK) of maize (Zea mays) was cloned from a cDNA library by southern blotting using a heterologous probe from sorghum (Sorghum bicolor). A recombinant fusion protein comprising thioredoxin of Escherichia coli and mature TK of maize was expressed at a high level in E. coli and cleaved with thrombin, affording plastid TK. The protein in complex with thiamine pyrophoshate was crystallized, and its structure was solved by molecular replacement. The enzyme is a C2 symmetric homodimer closely similar to the enzyme from yeast (Saccharomyces cerevisiae). Each subunit is folded into three domains. The two topologically equivalent active sites are located in the subunit interface region and resemble those of the yeast enzyme.
Transketolase (TK) catalyzes the reversible transfer of two-carbon units from ketose phosphates to aldose phosphates (for review, see Schenk et al., 1998). In heterotrophic organisms, TK provides a link between glycolysis and the pentose phosphate pathway and provides precursors for nucleotide, aromatic amino acid, and vitamin biosynthesis. In addition, in plants, the enzyme plays a central role in the Calvin cycle and thereby participates in the fixation of approximately 5 × 1011 metric tons of carbon dioxide per year (Hess, 1999).
TKs from a wide variety of organisms show significant sequence similarity (Schenk et al., 1997). Thus, the enzymes of man and Escherichia coli comprise about 24% identical amino acid residues. Typically, the enzymes are homodimers of 70- to 74-kD subunits. They require thiamine pyrophosphate (TPP) and divalent cations, e.g. Mg2+, for activity.
TKs of animals and fungi are located in the cytoplasmic compartment. On the other hand, the TK activity in photosynthetic and in non-photosynthetic plant tissues seems to be restricted to the plastid (Schnarrenberger et al., 1995; Debnam and Emes, 1999; Henkes et al., 2001). The TKs of the plants Craterostigma plantagineum (Bernaccia et al., 1995) and pepper (Capsicum annuum; Bouvier et al., 1998) show 47% and 53% identical amino acid residues, respectively, as compared with TK from yeast. Except for two non-constitutively expressed isoforms in the resurrection plant C. plantagineum (Bernaccia et al., 1995), the plant TK genes studied to date specify N-terminal plastid-targeting sequences (Flechner et al., 1996; Bouvier et al., 1998; Henkes et al., 2001). The in vitro import of a TK precursor in isolated spinach (Spinacia oleracea) chloroplasts has been shown (Flechner et al., 1996), as well as the association of the mature TK with the thylakoid membrane of spinach chloroplasts (Teige et al., 1998).
The activity of plastid TK is a limiting factor for the maximum rate of photosynthesis. A reduced level of TK activity is conducive to a reduction of the primary and secondary metabolism of tobacco (Nicotiana tabacum; Henkes et al., 2001). A reduction of TK activity also leads to decreased growth and decreased levels of aromatic acids and compounds of the phenylpropane metabolism. Due to its crucial role in plant metabolism, plastid TK is a potential target for herbicide development.
The structure of TK from yeast (Saccharomyces cerevisiae) has been determined to a resolution of 2.0 Å by x-ray crystallography (Nikkola et al., 1994). More recently, the structure of a yeast enzyme intermediate complex has been refined to a resolution of 1.86 Å (Fiedler et al., 2002).
TK is an essential enzyme of the Calvin cycle and, thus, is an attractive herbicide target. The development of maize (Zea mays) cultivars with resistance against such a herbicide could benefit substantially from the structural information presented in this study. This paper describes the x-ray structure of plastid TK from maize as the first representative from the plant kingdom. The structure data could also be useful in the development of a potential herbicide structure.
RESULTS AND DISCUSSION
Cloning, Expression, and Protein Purification
A DNA probe comprising 947 bp of the plastid TK gene from sorghum (Sorghum bicolor; Wyrich et al., 1998) was used to screen a λ-phage cDNA library from maize. A 2,141-bp clone was sequenced and afforded a partial open reading frame extending from the end of the putative plastid targeting sequence to the poly(A+) tail. Eight overlapping clones were partially sequenced to cover the remaining 5′ part of the gene. The combined sequences afforded an open reading frame specifying 729 amino acids.
The N terminus of native mature TK from spinach chloroplasts has been determined earlier by peptide sequencing (Teige et al., 1998). Based on sequence alignment with this sequence, we constructed a plasmid specifying residues 55 to 729 of the maize TK followed by a hexa-His tag. That construct was poorly expressed in recombinant E. coli cells.
Then, we constructed a gene specifying a fusion protein with an N-terminal bacterial thioredoxin domain followed by a hexa-His sequence, a thrombin cleavage site, an S-Tag, an enterokinase cleavage site, and, finally, the amino acid residues 55 to 729 of maize TK. Under the control of a T7 promoter and lac operator, the recombinant gene was expressed efficiently in E. coli strain BL21trxB, affording a soluble fusion protein that represented about 40% of cell protein.
The fusion protein was efficiently purified by affinity chromatography on a column of nickel-chelating Sepharose. The purified protein was specifically cleaved with thrombin at its thrombin cleavage site. The resulting TK carried an artificial sequence of 34 amino acids preceding the maize enzyme. That recombinant protein showed a specific activity of 23.3 μmol min–1 mg–1 and was crystallized in two different crystal forms using polyethylene glycol as precipitating agent. Data sets of crystal form II were obtained but could not be interpreted because the crystals were invariably twinned.
Edman degradation of form II crystalline protein showed that 36 amino acid residues at the N terminus had been lost by spontaneous fragmentation. Therefore, we constructed a recombinant gene specifying an N-terminal thioredoxin domain, a hexa-His tag, and a thrombin cleavage site that was directly followed by the gene segment specifying amino acid residues 55 to 729 of maize TK (GenBank accession no. AY148193). That gene construct was expressed to high level and was purified and cleaved as described above to yield enzymatically active maize plastid TK (Table I) with a minimally modified N terminus. Although the engineered protein starts with Gly due to the introduction of the thrombin cleavage site, the wild-type protein carries Ala in the corresponding position. Partial Edman degradation of the recombinant protein afforded the expected sequence GAVETLQGKA. Electrospray mass spectrometry of the recombinant protein afforded an Mr value of 72,994.8 in good agreement with the predicted value of 72,993 for one subunit.
Table I.
Purification of engineered plastid TK from maize
| Procedure | Total Activity | Total Protein | Specific Activity | Purification | Yield |
|---|---|---|---|---|---|
| μmol min-1 | mg | μmol min-1 mg-1 | -fold | % | |
| Crude extract | 438 | 73 | 6.0 | 1 | 100 |
| Ni-chelating agarose | 388 | 38 | 10.2 | 1.7 | 89 |
| Superdex 200 | 380 | 15 | 25.3 | 4.2 | 87 |
The recombinant enzyme showed a specific activity of 25.3 μmol min–1 mg–1. The Km values for Rib-5-phosphate and xylulose-5-phosphate as substrates were 581 and 403 μm, respectively. The specific activity and the Km value for Rib-5-phosphate were similar to those reported earlier for recombinant pepper TK (specific activity of 30 μmol min–1 mg–1, Km value for Rib-5-phosphate of 750 μm; Bouvier et al., 1998) and native spinach TK (specific activity of 12 μmol min–1 mg–1, Km value for Rib-5-phosphate of 330 μm; Teige et al., 1998). For the second substrate, we found a 4- to 5-fold higher Km value compared with those of pepper (95 μm) and spinach (67 μm). The purified protein was crystallized under the conditions described in “Materials and Methods” affording crystals that diffract to a resolution of 2.3 Å.
Crystal Structure and Quality of the Model
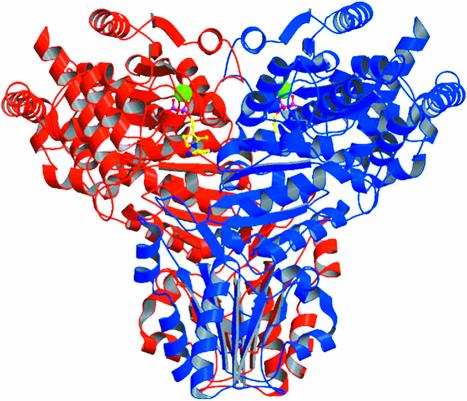
The crystal structure of recombinant maize TK was solved by Patterson search techniques (Hoppe, 1957; Rossman and Blow, 1962; Huber, 1965) using the coordinates of the homologous yeast enzyme (Nilsson et al., 1997). The calculations were carried out in two consecutive steps using diffraction data to 4.5-Å resolution because the asymmetric unit cell contains three TK subunits (solvent content of 50%). First, a dimeric yeast model was oriented and positioned with a correlation coefficient of 32.4% and a crystallographic R factor of 53.9%. To this fixed solution, a monomeric model could be added in a following Patterson search calculation with a correlation coefficient of 47.0% and a crystallographic R factor of 48.4%. This monomer forms the dimer (Fig. 1) by the 2-fold crystallographic symmetry axis.
Figure 1.
Dimeric maize TK with bound TPP cofactor (yellow, pyrophosphate moiety of TPP magenta) and Mg2+ (green). Subunits are shown in red and blue, respectively. Generated by Bobscript (Esnouf, 1997).
One subunit (Fig. 2A) consists of 675 residues that were well defined by their electron density map with the exception of the first 10 N-terminal residues, indicating their high degree of flexibility. The final model of the asymmetric unit comprises 1,998 amino acids (666 per monomeric subunit), 440 water molecules, three TPP cofactors, and three Mg2+. The structure was refined to 2.3-Å resolution with crystallographic R values of 16.6% (Rfree = 20.0%) with good stereochemistry (Table II). All 1,998 residues were found in the most favored and additional allowed regions of the Ramachandran plot (Ramachandran and Sasisekharan, 1968). Most of the side chains were clearly defined by their electron density map, except for some located at the surface of the protein. The temperature factors of bound TPP (B value of 20.7 Å2) were lower than those of the protein on average (B value of 24.6 Å2; Table II). Correspondingly, the cofactor molecules were well defined in the final electron density maps (Fig. 3A).
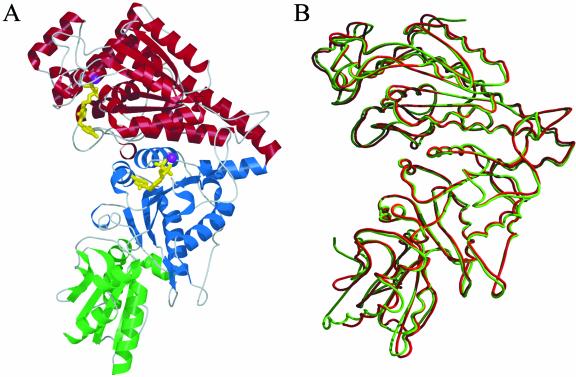
Figure 2.
A, Domain arrangements (red, N-terminal, so-called PP domain; blue, middle domain, therefore called Pyr domain; and green, C-terminal domain) of one maize TK monomer with bound TPP cofactor (yellow) and one Mg2+ (magenta). B, Superposition of one subunit of TK from maize (red) and yeast (green). Generated by Bobscript (Esnouf, 1997).
Table II.
X-ray data processing and refinement statistics
| Structure Refinement | TK Data |
|---|---|
| Cell constants (Å) | a = b = 136.4; c = 203.7 |
| Space group | P3121 |
| Resolution range (last shell) (Å) | 33.52-2.3 (2.42-2.3) |
| Reflections, unique | 91,552 |
| Multiplicity | 3.5 |
| Rmergea overall (last shell) | 0.12 (0.36) |
| Completeness (%) overall (last shell) | 93.8 (93.8) |
| Non-hydrogen protein atoms | 15,246 |
| Solvent molecules | 440 |
| Non-hydrogen ligand atoms | 78 |
| Non-hydrogen ion atoms | 3 |
| Rvalueb (%) overall (Rfree) | 16.6 (20.0) |
| Root mean square deviations from ideality | 0.008 Å (bonds), 1.46° (angles) |
| Average B valuec (Å2), protein | 24.6 |
| Average B value (Å2), ligand | 20.7 |
| Average B value (Å2), solvent | 23.5 |
| Average B value (Å2), ion | 16.6 |
| Φ, Ψ angle distribution for residuesd | |
| Most favored regions (%) | 88.9 |
| Additional allowed regions (%) | 11.1 |
Rmerge = Σhkl [(Σi |Ii - (I)| Σi Ii].
Rvalue = Σhkl ∥Fobs∥ - ∥Fcalc∥/Σhkl |Fobs| is the cross-validation R factor computed for the test set of 5% of unique reflections. c Temperature factors. d Ramachandran statistics as defined by PROCHECK (Laskowski et al., 1993).
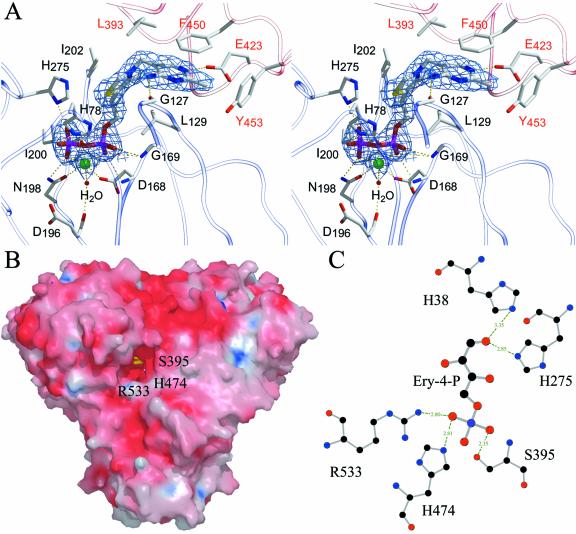
Figure 3.
A, Stereo drawings of the substrate binding site of TK from maize with bound TPP cofactor and Mg2+. The final 2FO-FC electron density map covering the ligand is contoured at 1.2 σ. Protein residues from the middle domain (Pyr domain) of the neighboring subunit are shown in red. Generated by Bobscript (Esnouf, 1997). B, Surface representation of the TK dimer. Generated by Grasp and Raster3D (Nicholls et al., 1993; Merritt and Murphy, 1994). C, Hydrogen bonding distances given in Å of a modeled sugar substrate, erythrose-4-phosphate, in the active site cleft.
Overall Structure and Subunit Fold
The crystal structure of TK from yeast has been studied in some detail (Lindquist et al., 1992; Nilsson et al., 1993, 1997; König et al., 1994; Nikkola et al., 1994; Meshalkina et al., 1997; Schneider and Lindqvist, 1998). The yeast enzyme is a homodimer in solution and crystalline state (Svergun et al., 2000). In close analogy with the yeast enzyme, the crystal structure shows the maize enzyme to be a c2 symmetrical homodimer. Each subunit contains one TPP cofactor and one Mg2+ in the active site cleft (Fig. 1) located at the dimer interface. The monomeric subunit is folded into three consecutive α-/β-domains (Fig. 2A). The N-terminal domain of maize TK is 332 amino acids long and comprises a central five-stranded parallel β-sheet with β1β2β3β5β4 topology surrounded by α-helices (α1 to α11) on both sides. The middle domain (residues from 333–542) consists of a central six-stranded parallel β-sheet (β8β7β9β10β12β11) embedded in helices from α12 to α18. These two domains of maize TK can be internally superimposed with a root mean square deviation value of 1.68 Å of 92 α-carbon atoms. Both domains participate in the dimer interface formation and in TPP cofactor binding.
The C-terminal domain comprises 133 amino acids (residues 543–675), forming a central five-stranded mixed β-sheet with four parallel strands (β16β17β15β18) and one antiparallel strand (β14). The biological function of this domain is still unclear (Schneider and Lindqvist, 1998) because no residues of the C-terminal domain are involved in the active dimer formation or in TPP cofactor binding.
TPP Cofactor Binding
As mentioned before, the homodimer is the functional unit of TK. The TPP cofactor is bound at the interface of two adjacent subunits in a conserved binding fold common to other TPP-dependent enzymes (Muller et al., 1993). The pyrophosphate moiety is bound by the N-terminal domain (so called PP domain) of one subunit, whereas the middle domain (Pyr domain) of the neighboring subunit binds the pyrimidine ring. These important subunit-subunit interactions form two deep, topologically equivalent active site clefts of approximately 19 Å in length with a width of 17 Å at the entrance and 7 Å at the interior. The cofactors bound to the active homodimer are separated by 18.2 Å (Fig. 1) and shielded from the solvent (Fig. 3B). The surface of the TK dimer is mostly negatively charged. Only the catalytically important C-2 atom of the thiazolium ring and the neighboring 4′-amino group are exposed to solvent and accessible for interaction with the substrate. The thiazolium ring of the cofactor is mainly bound in the active site via hydrophobic interactions with residues of both domains (Fig. 3A). The pyrimidine ring of the TPP cofactor is stacked in a mainly hydrophobic region, which is predominantly formed by the aromatic residues Tyr-443, Phe-447, Phe-448, Phe-450, and Tyr-453 and the aliphatic Val-449 of the Pyr domain. The side chains of Leu-392, Glu-423, Phe-450, and Tyr-453 are directly involved in binding of the pyrimidine ring, whereas the side chain oxygen of the strictly conserved Glu-423 is hydrogen bonded to the nitrogen ring atom at position 1. Phe-450 is in a stacking arrangement to the pyrimidine ring of TPP at a distance of 3.5 Å. On account of these protein-TPP interactions, the conformation of protein-bound TPP differs from the conformation of free TPP in its crystal structure (Pletcher and Sax, 1972). In the enzyme-bound V-shaped conformation, the catalytically active C-2 carbon atom of the thiazolium ring is in a close contact to the 4′-amino group of the pyrimidine ring at a distance of 3.0 Å. This distance is in the same range as for the yeast enzyme (3.1 Å).
The binding pocket of the pyrophosphate group of the cofactor is located at the turning point between the C-terminal parts of strands β1 and β2 of the β-sheet of the PP domain. The pyrophosphate and the metal binding site are exclusively built up by amino acids of the PP domain of one subunit. The pyrophosphate group is directly bound by hydrogen contacts to the conserved residues His-75, His-272, Asn-198, and via interactions to main chain nitrogen atoms of Asp-168 and Gly-169 (Fig. 3A). The Mg2+ has an important role for homodimerization (Wang et al., 1997) and for the catalytic activity (Heinrich et al., 1972). Other divalent cations such as Ca2+, Co2+, and Mn2+ can replace the Mg2+ and afford similar levels of catalytic activity. The Mg2+ is octahedrally coordinated by two diphosphate oxygen atoms, one main chain oxygen of Ile-200, two side chain oxygen atoms of Asp-168 and Asn-198, and a molecule, which is further stabilized in the binding site via hydrogen interaction to Asp-196.
Structural Comparison and Substrate Specificity
TKs from yeast, bacteria and plants are able to utilize a broad range of phosphorylated sugar substrates such as d-xylulose-5-phosphate, d-sedoheptulose-7-phoshate, d-Fru-6-phosphate, d-erythrose-4-phosphate, and hydroxypyruvate (Schenk et al., 1998). The affinity of the enzyme for unphosphorylated substrates is very low (Villafranca and Axelrod, 1971). Plant TKs have substrate specificity similar to that of the enzyme of yeast. Amino acid sequence comparison shows about 53% identity (about 60% similarity) of the enzymes of maize and yeast (Fig. 4), and the monomers can be superimposed with root mean square deviation values of only 0.91 Å for 638 α-carbon atoms (Fig. 2B).
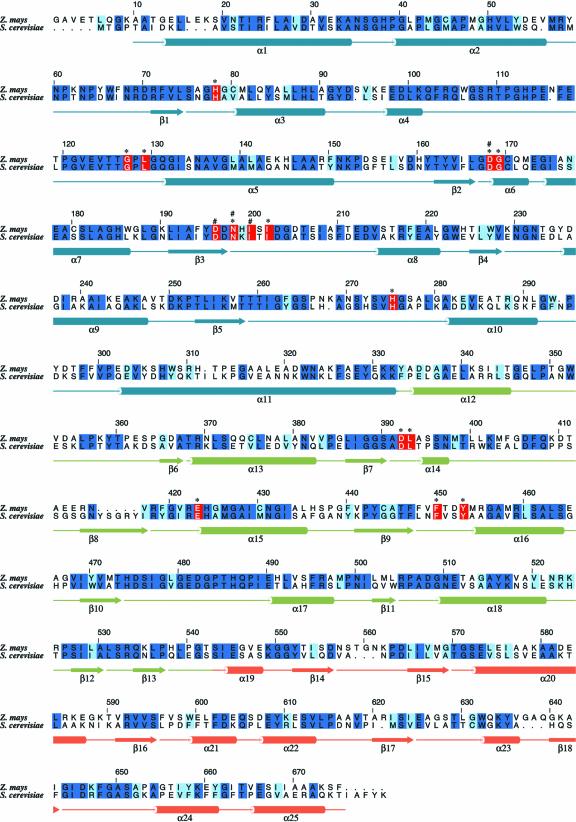
Figure 4.
Sequence alignment of engineered plastid TK from maize and TK from yeast. The numbering above the alignment corresponds to the enzyme from maize. Secondary structure elements found in maize TK are shown below the sequences. Identical residues (dark-blue background), conserved residues (light-blue background), and amino acid residues (red background) that are involved in cofactor binding (*) or metal binding (#) are indicated. The figure was drawn with ALSCRIPT (Barton, 1993).
The mechanistic properties of TK from yeast have been studied in some detail. Residues involved in cofactor binding and/or substrate binding are well characterized due to crystallographic and mutagenesis studies (Schneider and Lindqvist, 1998). Sequence comparison shows that the residues involved in TPP cofactor binding are strictly conserved in the maize enzyme (Fig. 4). Based on crystallographic studies of a complex of the yeast enzyme with the acceptor substrate erythrose-4-phosphate (Nilsson et al., 1997), a major role in substrate recognition and binding has been suggested for the invariant active site residues His-30 and His-263. Both residues are within hydrogen bonding distance to the aldehyde oxygen of the acceptor substrate. To identify the corresponding active site residues in the maize structure, we modeled the acceptor substrate, erythrose-4-phosphate, into the active site channel (Fig. 3C). Plausibly, His-38 and His-275 would form hydrogen bonds to the aldehyde group of the acceptor substrate.
Further, it can be seen that residues R533, S395, and H474, highly conserved in the TK sequences, are close to the phosphate group of the modeled substrate at the entrance of the substrate channel (Fig. 3B). This phosphate-binding site is consistent with the binding site (R359, R528, and H469) in the yeast structure. Replacement of any of these three residues by Ala (Nilsson et al., 1997) in the yeast TK substantially increases the Km values for phosphorylated substrates such as Rib-5-phosphate. This explains the preference of phosphorylated sugars over nonphosphorylated ones as substrate. The architecture of the substrate channel is determined by the phosphate-binding site, which is responsible for fitting of substrates varying in chain length from three to seven carbons. R533 can adjust the variations in the position of the phosphate group through its mobile side chain and, thus, allows the upper part of the substrate to approach the active site.
MATERIALS AND METHODS
Bacterial Strains and Growth Conditions
Bacterial strains and plasmids used in this study are summarized in Table III. Transformation of Escherichia coli was performed according to published procedures (Hanahan, 1983). Unless otherwise stated, bacteria were grown with shaking at 37°C in Luria-Bertani medium. Ampicillin (170 mg L–1) and kanamycin (15 mg L–1) were added as appropriate.
Table III.
Bacterial strains and plasmids
| Strain or Plasmid | Relevant Characteristics | Source |
|---|---|---|
| Bacterial strains | ||
| HMS174 (DE3) | F-recA1 hsdR(rK12- mK12+) RifR | Novagen (Madison, WI) |
| BL21trxB (DE3) | F-ompT hsdSB (rB- mB-) gal dcm trxB15::kan | Novagen |
| Plasmids | ||
| pET23a(+) | Expression vector providing a C-terminal hexahistidine tag | Novagen |
| pET23-Tkl | Plasmid expressing a C-terminally His-tagged pseudomature TK | This study |
| pET32a(+) | Expression vector providing a thioredoxin domain, hexahistidine tag, an S-Tag, and an enterokinase cleavage site | Novagen |
| pET32-Tkl7otp | Plasmid expressing a thioredoxin-TK-fusion protein | This study |
| pET32-ZMTK- | Plasmid expressing an engineered thioredoxin-TK-fusion protein | This study |
| Tb |
Cloning and Construction of Expression Constructs
A λ-phage cDNA library was constructed from maize (Zea mays cv KW 1071, KWS SAAT AG, Einbeck, Germany) mRNA using the cDNA Synthesis Kit (Amersham-Pharmacia Biotech, Uppsala) in combination with the Lambda ZAP II/EcoRI/Gigapack III Gold Cloning Kit (Statagene, La Jolla, CA) and was screened with a radioactive heterologous 957-bp probe from sorghum (Sorghum bicolor; Wyrich et al., 1998). A positive clone comprising 2,141 bp of the TK gene served as template for PCR amplification. For cloning of a C-terminally His-tagged TK, primers 5′-atagctacggcatatggccgccgtcgagacgctcca-3′ and 5′-tgcctagaacgcggccgcaaagctcttggcagctgcaa-3′ were used. The amplificate was digested with NdeI and NotI and ligated into the equally digested vector pET23a(+) (Novagen), yielding the plasmid pET23-Tkl. The oligonucleotides 5′-tatcgatatcgccgccgtcgagacgctc-3′ and 5′-cgtagcggccgcttaaaagctcttcgcagc-3′ were used as primers for the construction of an N-terminally thioredoxin-tagged TK. The amplificate was digested with EcoRV and NotI and ligated into an equally treated pET32a(+) (Novagen) yielding the plasmid pET32-Tkl7otp.
Plasmid pET32-Tkl7otp was used as the first template in a series of three consecutive PCR amplifications, using the forward primer 5′-ataataatatctagaaataattttgtttaactttaagaagg-3′ and the reverse primers 5′-cgccggtggcggccttgccctggagcgtctcgacagcaccgcgtggcaccagaccagaagaatg-3′, 5′-cgatggccaggaaccggatcgtgttgaccgacttctcgagcagctcgccggtggcggccttgccctggag-3′, and 5′-tattattatcccgggtggccggagttggccttctcgacggcgtcgatggccaggaaccggatcgtgttg-3′, respectively. The final 566-bp fragment was digested with XbaI and AhyI and ligated into the equally digested pET32-Tkl7otp, yielding the plasmid pET32-ZMTK-Tb.
Expression and Purification of TK
E. coli strain BL21trxB (DE3) pET32-ZMTK-Tb was grown to an optical density of 0.6 (600 nm) at 37°C. The culture was cooled to 18°C, isopropyl-β-d-thiogalactopyranoside (Europa Bioproducts, Wicken, UK) was added to a concentration of 1 mm, and incubation was continued with shaking overnight. Cells were harvested by centrifugation and washed with 0.9% (w/v) NaCl. The pellet was suspended in 50 mm Tris hydrochloride (pH 7.5) containing 200 mm NaCl, 5 mm MgCl2, and 50 μm TPP (buffer A). The suspension was ultrasonically treated and centrifuged at 26,000g for 20 min. The supernatant was placed on a column of chelating agarose (6 × 1.6 cm, Amersham-Pharmacia Biotech) pretreated with NiSO4. The column was washed with five column volumes of buffer A followed by five column volumes of 5 mm imidazole in the same buffer. The enzyme was eluted with three column volumes of 200 mm imidazole in buffer A. Fractions were combined.
Partial Proteolysis
The combined fractions from the nickel affinity chromatography were supplemented with 2.5 mm CaCl2 and 2 units of thrombin (Sigma-Aldrich, Taufkirchen, Germany) per milligram of protein. The mixture was incubated at room temperature for 16 h. The solution was concentrated to 5 mL using an Amicon cell (Millipore, Bedford, MA) and placed on a Superdex 200 26/60 column (60 × 2.6 cm, Amersham-Pharmacia Biotech) that had been equilibrated with 10 mm Tris hydrochloride (pH 7.5) containing 150 mm NaCl, 2 mm MgCl2, and 50 μm TPP.
Enzyme Assay
Assay mixtures contained 10 mm Tris hydrochloride (pH 7.5), 4 mm MgCl2, 100 μm TPP, 400 μm NADH, 4 mm Rib-5-phosphate (Sigma-Aldrich), 4 mm Xylulose-5-phosphate (Sigma-Aldrich), 8 units of triosephosphate isomerase (Sigma-Aldrich), 8 units of α-glycerophosphate dehydrogenase (Sigma-Aldrich), and protein in a total volume of 1 mL. The mixtures were incubated at 25°C, and A340 was recorded.
Miscellaneous
Protein concentration was determined by published procedures (Bradford, 1976) using bovine serum albumin as standard. Peptide sequencing was performed on a 476A protein sequencer (PE-Applied Biosystems, Foster City, CA). Electrospray mass spectrometry experiments were performed as described by Mann and Wilm (1995) using a triple quadrupol ion spray mass spectrometer API365 (SciEx, Thornhill, ON, Canada). DNA was sequenced using the dideoxynucleotide method (Sanger and Coulson, 1974).
Crystallization and Data Collection
Crystallization was performed by the sitting drop method using 24-well Cryschem plates (Hampton Research, Laguna Niguel, CA). Protein solution was concentrated to 12 to 15 mg mL–1 and exchanged into 10 mm Tris hydrochloride (pH 7.5) containing 2 mm MgCl2 and 50 μm TPP using an Amicon cell (Millipore). Protein solution (2 μL) was mixed with the same amount of 13% (w/v) polyethylene glycol 3350, containing 130 mm ammonium acetate and 0.5 μL of 100 mm spermine (Hampton Research). Droplets were left to equilibrate with 400 μL of the mother liquor at 20°C. Crystals grew to an approximate size of 500 × 200 × 200 μm3 within 1 week. They belong to the space group P3121 with cell dimensions of a = b = 136.4 Å and c = 203.7 Å and angles of α = β = 90° and γ = 120°. The asymmetric unit contains three TK subunits with a resulting Matthews coefficient of 2.5 Å3 D–1 (Matthews, 1968) with a solvent content of 50% (w/v). Diffraction data were collected on a mar345 imaging plate detector system (mar research, Norderstedt, Germany) mounted on a RU-200 rotating anode (Rigaku, Kemsing, UK) operated at 50 mA and 100 kV with λ = CuKα = 1.542 Å. The native data up to 2.3 Å were integrated with MOSFLM (Leslie, 1998). The data collection statistics are summarized in Table II.
Structure Solution and Refinement
The crystal structure of maize TK was solved by molecular replacement using the program MOLREP (Collaborative Computational Project Number 4, 1994) in the resolution range between 19.92 and 4.5 Å of the diffraction data. The structure of the yeast (Saccharomyces cerevisiae) enzyme (Nilsson et al., 1997; Protein Data Bank entry 1NGS) was used as a Patterson search model. Rotation search and subsequent translational search with a yeast TK dimer resulted in a prominent solution with a correlation coefficient of 32.4% and a crystallographic R factor of 53.9%. A further TK subunit could then be correctly placed with a correlation coefficient of 47.0% and a crystallographic R factor of 48.4% in the asymmetric unit building up a dimer via the 2-fold crystallographic symmetry axis. Examination of the packing of the enzymes within the crystal lattice indicated reasonable crystal contacts between the TK dimer without overlap of symmetry-related molecules. The initial model was subjected to rigid body and positional refinement using the CNS software (Brünger et al., 1998) with the Engh and Huber parameters (Engh and Huber, 1991). The crystallographic R factor was used for monitoring the stage of refinement, omitting 5% of the structure factors to calculate Rfree values. Model building was performed with the program MAIN (Turk, 1992). The initial electron density maps for all models showed well-defined FO-FC electron density for the TPP cofactor and one Mg2+. After several cycles of manual rebuilding, positional and B factor refinement, and two rounds of simulated annealing, 440 water molecules were incorporated automatically into the model. Restrained non-crystallographic symmetry of 30 kcal mol–1 Å–2 was applied in all subsequent steps of refinement. In later stages of crystallographic refinement, the maximum-likelihood algorithm implemented in the program CNS was used. A concluding geometry check using the program PROCHECK (Laskowski et al., 1993) revealed that 88.9% and 11.1% of all non-Gly residues lie within the most favored and additionally allowed regions of the Ramachandran plot (Ramachandran and Sasisekharan, 1968). The final structure refinement statistics are shown in Table II.
Coordinates
The atomic coordinates of the refined maize TK model have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank (entry 1ITZ).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.020982.
This work was supported by the Deutsche Forschungsgemein-schaft and by the Fonds der Chemischen Industrie.
References
- Barton GJ (1993) ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng 6: 37–40 [DOI] [PubMed] [Google Scholar]
- Bernaccia G, Schwall G, Lottspeich F, Salamini F, Bartels D (1995) The transketolase gene family of the resurrection plant Craterostigma plantagineum: differential expression during the rehydration phase. EMBO J 14: 610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier F, d'Harlingue A, Suire C, Backhaus RA, Camara B (1998) Dedicated roles of plastid transketolases during the early onset of isoprenoid biogenesis in pepper fruits. Plant Physiol 117: 1423–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye-binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges N, Pannu NS et al. (1998) Crystallography and NMR system (CNS): a new software system for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 (1994) The CCP4 Suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Debnam PM, Emes MJ (1999) Subcellular distribution of enzymes of the oxidative pentose phosphate pathway in root and in leaf tissues. J Exp Bot 50: 1653–1661 [Google Scholar]
- Engh RA, Huber R (1991) Accurate bond and angle parameters for x-ray protein-structure refinement. Acta Crystallogr A 47: 392–400 [Google Scholar]
- Esnouf RM (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J Mol Graph Model 15: 112–113, 132–134 [DOI] [PubMed] [Google Scholar]
- Fiedler E, Thorell S, Sandalova T, Golbik R, König S, Schneider G (2002) Snapshot of a key intermediate in enzymatic thiamin catalysis: crystal structure of the alpha-carbanion of (alpha,beta-dihydroxyethyl)-thiamin diphosphate in the active site of transketolase from Saccharomyces cerevisiae. Proc Natl Acad Sci USA 99: 591–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flechner A, Dressen U, Westhoff P, Henze K, Schnarrenberger C, Martin W (1996) Molecular characterization of transketolase (EC 2.2.1.1) active in the Calvin cycle of spinach chloroplasts. Plant Mol Biol 32: 475–484 [DOI] [PubMed] [Google Scholar]
- Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166: 557–580 [DOI] [PubMed] [Google Scholar]
- Heinrich PC, Steffen H, Janser P, Wiss O (1972) Studies on the reconstitution of apotransketolase with thiamine pyrophosphate and analogs of the coenzyme. Eur J Biochem 30: 533–541 [DOI] [PubMed] [Google Scholar]
- Henkes S, Sonnewald U, Badur R, Flachmann R, Stitt M (2001) A small decrease of plastid transketolase activity in antisense tobacco transformants has dramatic effects on photosynthesis and phenylpropanoid metabolism. Plant Cell 13: 535–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess D (1999) Pflanzenphysiologie, Ed 10. Ulmer, Stuttgart-Hohenheim, Germany, p 92
- Hoppe W (1957) Die Faltmolekülmethode: Eine neue Methode zur Bestimmung der Kristallstruktur bei ganz oder teilweise bekannten Molekülstrukturen. Acta Crystallogr A 10: 750–751 [Google Scholar]
- Huber R (1965) Die automatisierte Faltmolekülmethode. Acta Crystallogr A 19: 353–356 [Google Scholar]
- König S, Schellenberger A, Neef H, Schneider G (1994) Specificity of coenzyme binding in thiamin diphosphate-dependent enzymes: crystal structures of yeast transketolase in complex with analogs of thiamin diphosphate. J Biol Chem 269: 10879–10882 [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26: 283–291 [Google Scholar]
- Leslie AGW (1998) MOSFLM (CCP4: Supported Program). MRC Laboratory of Molecular Biology, Cambridge, UK
- Lindquist Y, Schneider G, Ermler U, Sundström M (1992) Three-dimensional structure of transketolase, a thiamine diphosphate dependent enzyme, at 2.5 Å resolution. EMBO J 11: 2373–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M, Wilm M (1995) Electrospray mass spectrometry for protein characterization. Trends Biochem Sci 20: 219–224 [DOI] [PubMed] [Google Scholar]
- Matthews BW (1968) Solvent content of protein crystals. J Mol Biol 33: 491–497 [DOI] [PubMed] [Google Scholar]
- Merritt EA, Murphy MEP (1994) Raster3D version 2.0: a program for photorealistic molecular graphics. Acta Crystallogr D 50: 869–873 [DOI] [PubMed] [Google Scholar]
- Meshalkina L, Nilsson U, Wikner C, Kostikowa T, Schneider G (1997) Examination of the thiamin diphosphate binding site in yeast transketolase by site-directed mutagenesis. Eur J Biochem 244: 646–652 [DOI] [PubMed] [Google Scholar]
- Muller YA, Lindqvist Y, Furey W, Schulz GE, Jordan F, Schneider G (1993) A thiamin diphosphate binding fold revealed by comparison of the crystal structures of transketolase, pyruvate oxidase and pyruvate decarboxylase. Structure 1: 95–103 [DOI] [PubMed] [Google Scholar]
- Nicholls A, Bharadwaj R, Honig B (1993) GRASP: graphical representation and analysis of surface properties. Biophys J 64: A166 [Google Scholar]
- Nikkola M, Lindqvist Y, Schneider G (1994) Refined structure of transketolase from Saccharomyces cerevisiae at 2.0 A resolution. J Mol Biol 238: 387–404 [DOI] [PubMed] [Google Scholar]
- Nilsson U, Lindqvist Y, Kluger R, Schneider G (1993) Crystal structure of transketolase in complex with thiamine thiazolone diphosphate, an analogue of the reaction intermediate, at 2.3 A resolution. FEBS Lett 326: 145–148 [DOI] [PubMed] [Google Scholar]
- Nilsson U, Meshalkina L, Lindqvist Y, Schneider G (1997) Examination of substrate binding in thiamin diphosphate-dependent transketolase by protein crystallography and site-directed mutagenesis. J Biol Chem 272: 1864. [DOI] [PubMed] [Google Scholar]
- Pletcher J, Sax M (1972) Crystal and molecular structure of thiamine pyrophosphate hydrochloride. J Am Chem Soc 94: 3998–4005 [DOI] [PubMed] [Google Scholar]
- Ramachandran GN, Sasisekharan V (1968) Conformation of polypeptides and proteins. Adv Protein Chem 23: 283–437 [DOI] [PubMed] [Google Scholar]
- Rossman MG, Blow DM (1962) The detection of subunits within the crystallographic asymmetric unit. Acta Crystallogr A 15: 24–31 [Google Scholar]
- Sanger F, Coulson AR (1974) A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol 94: 441–448 [DOI] [PubMed] [Google Scholar]
- Schenk G, Duggleby RG, Nixon PF (1998) Properties and functions of the thiamin diphosphate dependent enzyme transketolase. Int J Biochem Cell Biol 30: 1297–1318 [DOI] [PubMed] [Google Scholar]
- Schenk G, Layfield R, Candy JM, Duggleby RG, Nixon PF (1997) Molecular evolutionary analysis of the thiamine-diphosphate-dependent enzyme, transketolase. J Mol Evol 44: 552–572 [DOI] [PubMed] [Google Scholar]
- Schnarrenberger C, Flechner A, Martin W (1995) Enzymatic evidence for a complete oxidative pentose phosphate pathway in chloroplasts and an incomplete pathway in the cytosol of spinach leafs. Plant Physiol 108: 609–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G, Lindqvist Y (1998) Crystallography and mutagenesis of transketolase: mechanistic implications for enzymatic thiamin catalysis. Biochim Biophys Acta 1385: 387–398 [DOI] [PubMed] [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MH, König S (2000) Crystal versus solution structures of thiamine diphosphate-dependent enzymes. J Biol Chem 275: 297–302 [DOI] [PubMed] [Google Scholar]
- Teige M, Melzer M, Süss KH (1998) Purification, properties and in situ localization of the amphibolic enzymes d-ribulose 5-phosphate 3-epimerase and transketolase from spinach chloroplasts. Eur J Biochem 252: 237–244 [DOI] [PubMed] [Google Scholar]
- Turk D (1992) Development and usage of a macromolecular graphics program. PhD thesis. Technical University of Munich, Garching, Germany
- Villafranca JJ, Axelrod B (1971) Heptulose synthesis from nonphosphorylated aldoses and ketoses by spinach transketolase. J Biol Chem 246: 3126–3131 [PubMed] [Google Scholar]
- Wang J-L, Martin PR, Singleton CK (1997) Aspartate 155 of human transketolase is essential for thiamine diphosphate-magnesium binding, and cofactor binding is required for dimer formation. Biochim Biophys Acta 1341: 165–172 [DOI] [PubMed] [Google Scholar]
- Wyrich R, Dressen U, Brockmann S, Streubel M, Chang C, Qiang D, Paterson AH, Westhoff P (1998) The molecular basis of C4 photosynthesis in sorghum: isolation, characterization and RFLP mapping of mesophyll- and bundle-sheath-specific cDNAs obtained by differential screening. Plant Mol Biol 37: 319–335 [DOI] [PubMed] [Google Scholar]