Abstract
Mitogen-activated protein kinase (MAPK) cascades are known to transduce plant defense signals, but the downstream components of the MAPK have as yet not been elucidated. Here, we report an MAPK from rice (Oryza sativa), BWMK1, and a transcription factor, OsEREBP1, phosphorylated by the kinase. The MAPK carries a TDY phosphorylation motif instead of the more common TEY motif in its kinase domain and has an unusually extended C-terminal domain that is essential to its kinase activity and translocation to the nucleus. The MAPK phosphorylates OsEREBP1 that binds to the GCC box element (AGCCGCC) of the several basic pathogenesis-related gene promoters, which in turn enhances DNA-binding activity of the factor to the cis element in vitro. Transient co-expression of the BWMK1 and OsEREBP1 in Arabidopsis protoplasts elevates the expression of the β-glucuronidase reporter gene driven by the GCC box element. Furthermore, transgenic tobacco (Nicotiana tabacum) plants overexpressing BWMK1 expressed many pathogenesis-related genes at higher levels than wild-type plants with an enhanced resistance to pathogens. These findings suggest that MAPKs contribute to plant defense signal transduction by phosphorylating one or more transcription factors.
Mitogen-activated protein kinase (MAPK) cascades are known to play essential roles in the signal transduction pathways involved in numerous eukaryotic cellular processes from cell division to cell death (Davis, 2000; Ligterink and Hirt, 2001). In the last few years, it has become apparent that MAPK cascades also play vital roles in signal transduction pathways of plants, including plant defense signaling (Innes, 2001; Tena et al., 2001; Zhang and Klessig, 2001). The Arabidopsis genome sequence has revealed the presence of 23 MAPK genes in the genome, which suggests that the MAPK cascades in plants may be quite complex.
Accumulating lines of evidence indicate that plants rapidly activate MAPKs when exposed to a variety of abiotic and biotic stress stimuli (Ligterink et al., 1997; Zhang et al., 1998; Seo et al., 1999; Cardinale et al., 2000; Ichimura et al., 2000). These include pathogens, pathogen-derived elicitors, and defense-related second messengers. In tobacco (Nicotiana tabacum), two MAPKs, SIPK and WIPK, are activated by both various pathogen-related signals and diverse abiotic stresses, indicating that pathogen defense signaling is part of an integrated stress-signaling network in plants. Orthologs of SIPK and WIPK in Arabidopsis (AtMPK6 and AtMAPK3, respectively) and alfalfa (Medicago sativa; SIMK and SAMK, respectively) are also activated by both biotic and abiotic stresses (Seo et al., 1995; Zhang and Klessig, 1997; Nuhse et al., 2000). Recently, the MAPKK, NtMEK2, was identified to operate in the cascade upstream of SIPK and WIPK because a constitutively active NtMEK2 activates endogenous SIPK and WIPK molecules in transiently transformed tobacco cells. Furthermore, the constitutively active NtMEK2 induces hypersensitive cell death and the expression of defense genes (Yang et al., 2001; Zhang and Liu, 2001). However, other constitutively active tobacco MAPKKs neither activate SIPK or WIPK nor induce defense responses. This suggests that these two MAPKs are involved specifically in the defense response of tobacco plants (Yang et al., 2001). Another type of MAPK that plays negative regulatory roles in plant defense has been identified in Arabidopsis, namely MAPK4 (AtMPK4; Petersen et al., 2000). Thus, several groups have clearly shown that MAPK cascades are involved in plant defense mechanisms. However, less well defined are the downstream components of the MAPK signaling pathway associated with plant defense responses, including the substrates that are directly phosphorylated by MAPK.
Understanding the transcriptional regulation of defense-associated genes is important in helping improve the disease resistance mechanisms in plants (Dangl and Jones, 2001). It has been reported that potential substrates for protein kinases in response to fungal elicitors include DNA-binding proteins because a novel basic Leu zipper DNA-binding protein, G/HBF-1, shows enhanced DNA-binding activity to the Chs15 (chalcone synthetase15) promoter after being phosphorylated in vitro (Droge-Laser et al., 1997). In addition, elicitor-induced phosphorylation of the nuclear factor PBF-1 is required before it can bind to the promoter of the pathogenesis-related (PR) gene, PR10a (Despres et al., 1995). Furthermore, the resistance gene product Pto, a Ser/Thr protein kinase in tomato (Lycopersicon esculentum) plants, phosphorylates and thereby activates Pti4, a transcription factor that binds to GCC box elements of PR genes (although whether Pti is the true in vivo Pto kinase target involved in pathogen resistance remains to be elucidated; Thara et al., 1999; Gu et al., 2000). Moreover, an elicitor-responsive MAPK (ERMK) in parsley (Petroselinum crispum) is activated and transported to the nucleus upon stimulation with a pathogen-derived elicitor, which suggests that it may participate in the transcriptional activation of defense-responsive genes (Ligterink et al., 1997). Thus, several lines of evidence suggest that protein phosphorylation, particularly the phosphorylation of transcription factors, is involved in the regulation of plant defense responses. Recently, Asai et al. (2002) reported that MAP kinase cascade (MEKK1, MKK4/MKK5, and MPK3/MPK6) and WRKY22/WRKY29 transcription factors in Arabidopsis may function downstream of the flagellin receptor-like kinase (FLS2).
To date, some MAPKs have been identified and characterized from rice (Oryza sativa; He et al., 1999; Xiong et al., 2001; Agrawal et al., 2002; Huang et al., 2002; Song and Goodman, 2002; Wen et al., 2002). Despite these observations, however, there is no direct evidence that plant MAPKs phosphorylate transcription factor(s) and that this leads to the transcriptional activation of defense-responsive genes, although this is common in mammals.
Here, we report our molecular and functional analysis of a rice MAPK, BWMK1. This protein phosphorylates the rice transcription factor OsEREBP1 (rice ethylene-responsive element-binding protein 1; accession no. AF193803). Such EREBPs are known to bind to the GCC box DNA motif (AGCCGCC) that is located in the promoter of several PR genes. BWMK1 is localized in the nucleus and is activated by a fungal elicitor. In vitro phosphorylation of OsEREBP1 by BWMK1 enhanced its ability to bind to the GCC box. Transient co-expression of BWMK1 with OsEREBP1 in Arabidopsis protoplasts improved the expression of the β-glucuronidase (GUS) reporter gene driven by the GCC box fused to a minimal 35S promoter. Furthermore, ectopic expression of the BWMK1 in tobacco plant induced the expression of a broad spectrum of PR genes and was associated with increased resistance to pathogens. These observations provide new insights into the MAPK signaling pathways involved in plant defense responses.
RESULTS
BWMK1 Belongs to a New Family of Plant MAPKs
To isolate the rice MAPKs that are induced by a fungal elicitor, we screened the full-length cDNA from fungal elicitor-treated rice cDNA library using the MAP kinase fragment that resulted from reverse transcriptase-PCR using degenerate oligonucleotide primers corresponding to highly conserved regions found in plant Ser/Thr protein kinases, namely, LREIKLCRM and DVWSVGCIF. The longest cDNA consisted of 2,032 bp that contained full-length cDNA (EMBL/GenBank accession no. AF194415) encoding a 55.7-kD protein. The deduced amino acid sequence of the protein contains MAPK motifs, and we designated it as OsMAPK1 (rice MAPK isoform 1). After we registered the OsMAPK1 clone, an identical rice blast- and wounding-activated MAP kinase (BWMK1, accession no. AF177392) was reported by He et al. (1999), although this study did not characterize the clone biochemically and functionally. Thus, we refer to this gene as BWMK1 for further characterization.
BWMK1 is composed of an N-terminal kinase domain (KD) and an unusually long C-terminal extension domain (CD) that contains a putative Leu zipper motif (He et al., 1999). Unlike most other plant MAPKs, the KD region of BWMK1 carries a TDY phosphorylation motif instead of TEY, a sequence believed to be essential in MAPK activation. Thus, based on these common structural elements, we propose that BWMK1 is a member of a new family of plant MAPKs.
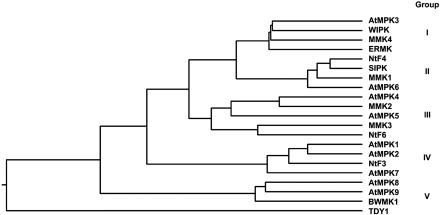
The phylogenetic tree resulting from comparisons of deduced amino acid sequences indicates that plant MAPKs can be grouped into at least five distinct families (Fig. 1). Among them, the MAPKs in families I and II are mostly involved in pathogen and abiotic stress signalings, whereas some family III MAPKs are involved in cell cycle regulation (Zhang and Klessig, 2001). Interestingly, AtMPK4, which is in family III, negatively regulates systemic acquired resistance (SAR; Petersen et al., 2000). BWMK1 belongs to family V, which includes the AtMPK8, AtMPK9, and TDY1, which have the TDY motif instead of TEY in KD and also have the CD. Recently, Schoenbeck et al. (1999) reported that the TDY1 gene is expressed in the leaf mesophyll-surrounding areas of mechanical wounding and pathogen invasion, which suggests that it may play a role in wound signaling. Furthermore, BWMK1 was also activated by wounding and rice blast fungus (He et al., 1999). Thus, the family V of MAPKs including BWMK1 is believed to be involved in pathogen and wounding signal transduction.
Figure 1.
BWMK1 belongs to an MAPK family in plants. Phylogenetic tree of plant MAPKs. Plant MAPKs included were alfalfa MMK1-4 (Jonak et al., 1995, 1996) and TDY1 (Schoenbeck et al., 1999); Arabidopsis AtMPK1-7 (Mizoguchi et al., 1993), AtMPK8 (accession no. BAA92222), and AtMPK9 (accession no. BAA92223); parsley ERMK (Ligterink et al., 1997); tobacco NtF3, 4, and 6 (Wilson et al., 1995), WIPK (Seo et al., 1995), and SIPK (Zhang and Klessig, 1997); and rice BWMK1 (He et al., 1999). The phylogenic tree was created using ClustalW program (Thompson et al., 1994).
The CD of BWMK1 Is Essential for Kinase Activity and Nuclear Localization
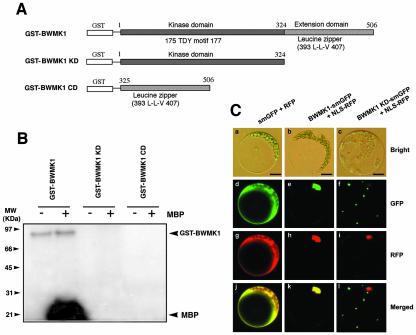
To characterize the BWMK1, we produced glutathione S-transferase (GST) fusion proteins containing full-length, KD or CD in Escherichia coli and then purified the GST fusion proteins (Fig. 2A). Before we performed the kinase activity assay with purified GST fusion proteins, amounts of purified proteins was checked by loading with 10% (w/v) SDS-PAGE (data not shown). As shown in Figure 2B, the full-length protein was able to phosphorylate both MBP and itself. However, both phosphorylation activities were completely destroyed when the CD was deleted, indicating that it is essential for the kinase activity of BWMK1. Similar observations have been made for other protein kinases, such as Nlk (Brott et al., 1998) and TSL (Roe et al., 1996), which also have an extension domain containing a Leu zipper.
Figure 2.
The CD of BWMK1 is essential for both kinase activity and nuclear localization. A, Schematic diagram of the GST fusion constructs of BWMK1. Amino acid numbers of domain boundaries are indicated. B, Role of the C-terminal domain in autophosphorylation and myelin basic protein (MBP) kinase activity. +, With MBP; –, without MBP. The arrows indicate the positions where GST-BWMK1 and MBP migrated. The apparent molecular masses (kilodaltons) are indicated at the left. C, Subcellular localization of BWMK1. Arabidopsis protoplasts were cotransfected with three sets of smGFP and RFP constructs, that is, smGFP and RFP (a, d, g, and j), smGFP-fused BWMK1 and RFP-fused NLS (b, e, h, and k), and smGFP-fused KD and RFP-fused NLS (c, f, i, and l). Bars = 20 μm.
To determine the subcellular localization of the MAPK in vivo, the green fluorescent protein (smGFP) gene (Davis and Vierstra, 1998) was fused to the C-terminal end of either the full-length BWMK1 (BWMK1-smGFP) or the KD (BWMK1 KD-smGFP), and these constructs were cotransfected with the red fluorescent protein gene fused to the nuclear localization signal peptide (NLS-RFP; Lee et al., 2001) as a positive control into Arabidopsis protoplasts. As shown in Figure 2C, the full-length BWMK1-smGFP fusion protein localized exclusively in the nucleus showing completely overlapping location with NLS-RFP originated from the simian virus 40 large T antigen (Dingwall and Laskey, 1991). In contrast, KD-smGFP was not translocated to the nucleus. Thus, the CD of BWMK1, which contains a putative Leu zipper motif, is believed to be essential for the kinase activities of the protein and its targeting to the nucleus.
BWMK1 Is Activated by Pathogen Signals
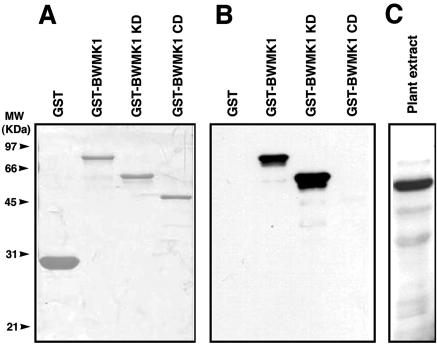
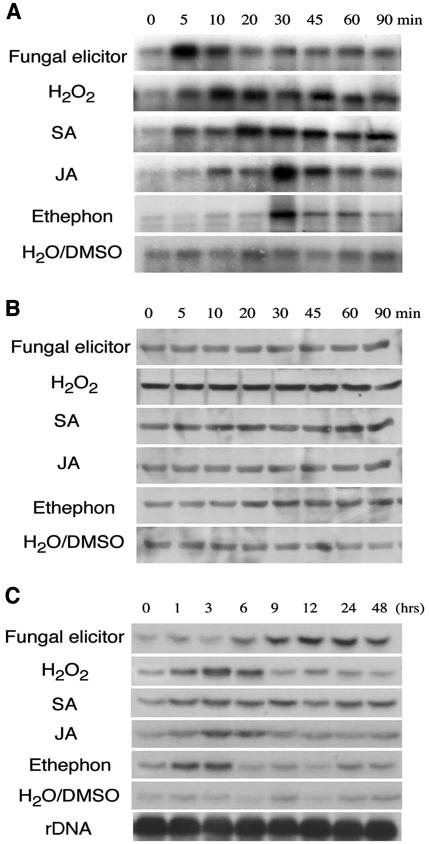
To determine whether BWMK1 is activated in response to pathogen signals, we performed immunocomplex kinase assays using Ab-pNBWMK1, an antibody that was raised against a synthetic peptide representing the N-terminal 15 amino acids of BWMK1. Before using the antibody for the further analysis, specificity of antibody was assessed by immunoblot analysis against a panel of different GST-fused BWMK1 proteins as described in Figure 2A. The anti-pNBWMK1 antibody recognized the GST-BWMK1 and GST-BWMK1 KD but not the GST-BWMK1 CD and GST protein (Fig. 3B). To verify the antibody specificity, we tested the immunoblot analysis using the plant crude extract from rice (Fig. 3C). As shown in Figure 3C, the major band (more than 90%) that was recognized with antibody was similar in molecular size, which was estimated by deduced amino acid of BWMK1. Thus, we used the antibody for immunocomplex kinase assay. The kinase activity of BWMK1 was rapidly and transiently activated by several defense signals, including a fungal elicitor, hydrogen peroxide (H2O2), salicylic acid (SA), jasmonic acid (JA), and ethephon (Fig. 4A). The activity peaked 5 to 30 min after treatment. Several plant MAPKs, including SIPK (Zhang and Klessig, 1997; Zhang et al., 1998), WIPK (Seo et al., 1995), and ERMK (Ligterink et al., 1997), have also been reported to be activated by such defense signals. The transcript levels of BWMK1 increased in a delayed manner after activation with the defense signals (Fig. 4C), but the BWMK1 protein levels did not change (Fig. 4B). This indicates that the BWMK1 protein is maintained at steady-state levels in the cell, which would permit the plant to respond rapidly to unpredictable pathogen attacks. The response consumes BWMK1 protein; thus, the plants produce new BWMK1 transcripts to maintain the baseline level of protein. Therefore, these observations suggest that BWMK1 activation is primarily achieved by post-translational modification, which is reminiscent of other plant MAPKs such as ERMK (Ligterink et al., 1997) and WIPK (Seo et al., 1995).
Figure 3.
Specificity of Ab-pNBWMK1 antibody. A, SDS-PAGE analysis of GST fusion recombinant proteins of BWMK1. Purified proteins were separated on a 10% (w/v) SDS-PAGE and stained with Coomassie Brilliant Blue R-250. The apparent molecular masses (kilodaltons) are indicated at the left. B, Immunoblot analysis using the recombinant protein. Ab-pNBWMK1 specially recognized GST-BWMK1 and GST-BWMK1 KD. C, Immunoblot analysis using plant extracts from rice. Protein (50 μg) was separated by SDS-PAGE, blotted, and probed with Ab-pNBWMK1 (1:3,000 [v/v] dilution). After incubation with a horseradish peroxidase-conjugated secondary antibody, the complex was visualized using enhanced chemiluminescence.
Figure 4.
Activation of BWMK1 by defense signals. A, Kinase activities of BWMK1. Protein extracts (100 μg) from the suspension cell cultures treated with above stresses for various times were immunoprecipitated with anti-pNBWMK1 antibody and the performed the kinase assay as described in “Materials and Methods.” B, Protein levels of BWMK1. Fifty micrograms of protein extracts used for kinase assay was analyzed for BWMK1 protein levels by immunoblot analysis using the anti-pNBWMK1 antibody. C, Transcription levels of BWMK1. Total RNAs (20 μg) isolated from rice suspension cells treated by stresses were separated by 1.5% (w/v) formaldehydeagarose gel, transferred onto a membrane, and then hybridized with 32p-labeled BWMK1 cDNA. Fungal elicitor, Crude extract (50 μg mL–1) prepared from the rice blast Magnaporthe grisea; H2O2, 1 mm H2O2; SA, 1 mm SA; JA, 0.1 mm JA; ethephon, 5 mm ethaphon; H2O/DMSO, water or dimethyl sulfoxide control.
Overexpression of BWMK1 in Transgenic Tobacco Plants Induces Cell Death Formation and Elevates the Expression of SAR Genes
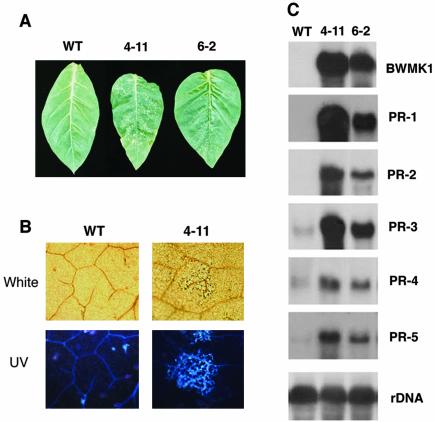
To examine the biological functions of the BWMK1 gene in plant defense responses, we constructed transgenic tobacco plants that constitutively express BWMK1 under the control of the cauliflower mosaic virus (CaMV) 35S promoter. Transgenic plant lines expressing BWMK1 were first selected by northern blotting and assayed for phenotypic changes and PR gene expression. We obtained eight BWMK1-expressed transgenic plants, and five lines among them showed the lesion mimic phenotype and the enhancement of PR gene expression. We then selected two lines for further analysis to test the resistance against pathogens. As shown in Figure 5A, hypersensitive response (HR)-like necrotic lesions formed on the rosette leaves of the transgenic lines. The earliest cell death in the transgenic plants was observed on the first rosette leaf 4 to 5 d after leaf emergence, and the succeeding true leaves also exhibited cell death. HR-like cell death is usually correlated with a variety of biochemical reactions, such as the accumulation of autofluorescent compounds, callose deposition, and lignification at and around the lesion sites (Dixon et al., 1994). To determine whether the lesions in the transgenic plants really are HR-like reactions, we assayed the leaves with lesions for autofluorescence. As shown in Figure 5B, the autofluorescence in the leaves with lesions was distributed in a pattern similar to that observed in leaves undergoing HR.
Figure 5.
Overexpression of BWMK1 in transgenic tobacco plant induces HR-like cell death and elevates SAR gene expression. A, Development of HR-like cell death in transgenic tobacco plants. Lesions formed in 6-week-old representative transgenic BWMK1 tobacco plants (4-11 and 6-2) and wild-type (WT) plant. B, Accumulation of autofluorescent materials in HR-like cell death lesions of transgenic plants. Fully expanded leaves from 6-week-old plants were used for microscopic analysis after clearing with lactophenol. Magnification is ×50. White, Lactophenol-cleaned leaves examined under a light microscope. UV, UV-stimulated autofluorescence in lactophenol-cleared tobacco leaves. C, Constitutive expression of PR genes in transgenic plants.
It has been shown that plants with spontaneous lesions often show an elevated expression of the genes that encode PR proteins and an increased resistance to pathogens (Dangl et al., 1996; Durner et al., 1997). Therefore, we assayed the transgenic plants for the expression of a variety of PR genes. As shown in Figure 5C, the transcript levels of all the PR genes examined were elevated in the transgenic plants relative to the wild-type plant, indicating that expression of BWMK1 induces the expression of a variety of PR genes. These observations clearly indicate that overexpression of BWMK1 causes HR-like cell death and enhances PR gene expression in the plant.
BWMK1 Transgenic Tobacco Plants Have an Enhanced Disease Resistance to Pathogens
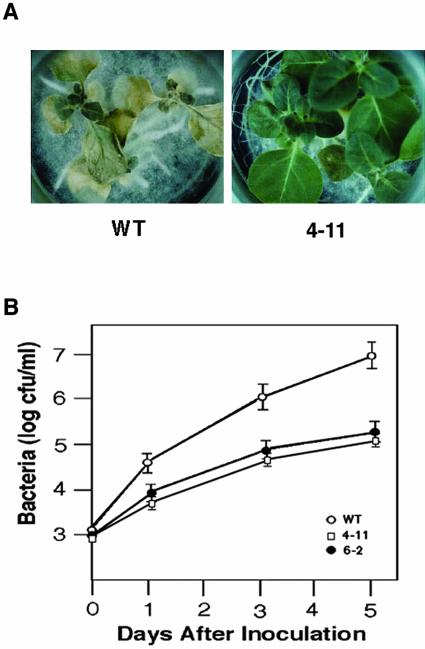
Because the transgenic plants showed elevated expression of PR genes with spontaneous lesions, we assayed whether the transgenic plants are better able to resist fungal and bacterial pathogens. We inoculated transgenic and wild-type plants with the oomycete pathogen Phytophthora parasitica var nicotianae. Five days later, the wild-type plants exhibited disease symptoms, but the transgenic plants did not. Seven days after inoculation, the wild-type plants had severe disease symptoms, including leaf wilting and stem rot, and most were dead 8 d after inoculation. However, the transgenic plants remained healthy without any appreciable disease symptoms (Fig. 6A). The transgenic plants also showed enhanced resistance to the virulent bacterial pathogen Pseudomonas syringae pv tabacci (Pst). At 5 d after inoculation, the in planta growth of Pst in transgenic plants was 100-fold less than in the wild-type plants (Fig. 6B). Thus, constitutive overexpression of BWMK1 appears to enhance resistance to a range of pathogens, probably because the expression of many PR genes is elevated.
Figure 6.
Enhanced disease resistance of transgenic tobacco plants that constitutively overexpressed BWMK1. A, Disease responses to the virulent oomycete pathogen, P. parasitica var nicotianae, at 7 d after inoculation. B, In planta bacterial growth. Pst was inoculated into leaves of mature wild-type plants (WT) and two independent transgenic plant lines (4-11 and 6-2) at 105 colony-forming units (cfu) mL–1, and in planta bacterial growth was monitored over 5 d. Values represent average and sds of cfu extracted from leaf discs in three independent samplings.
BWMK1 Interacts with OsEREBP1, a Rice AP2/EREBP Family Transcription Factor
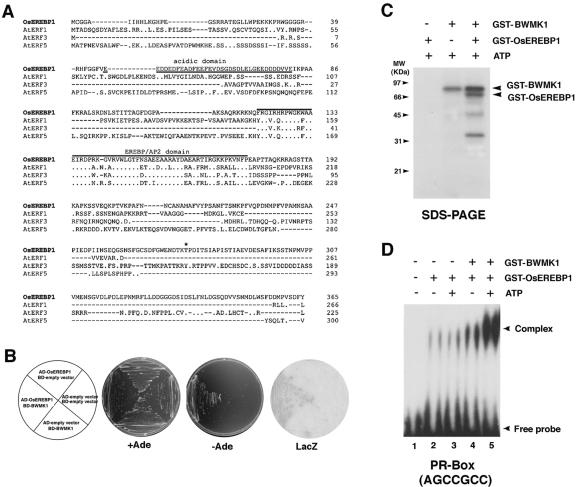
It has been suggested that potential substrates for the protein kinases activated in response to pathogen signals include DNA-binding proteins (Droge-Laser et al., 1997; Gu et al., 2000). Because BWMK1 is localized in the nucleus and is activated by several defense signals (Figs. 2 and 4), it may be able to interact with transcription factor(s) involved in the expression of defense-related genes. To determine whether BWMK1 can phosphorylate one or more transcription factors, we attempted to isolate cDNA clones encoding transcription factors that interact with BWMK1. To do so, we employed the yeast (Saccharomyces cerevisiae) two-hybrid screening method using full-length BWMK1 fused to the GAL4 DNA-binding domain as bait (BD-BWMK1). The bait was introduced into the yeast strain pJ69-4A together with a prey library containing rice cDNAs fused behind a GAL4 transcriptional activation domain. From 2 × 107 initial transformants, we obtained several positive colonies. The prey plasmids were isolated, and the insert cDNA fragments were sequenced. One of the positive clones had a high sequence homology with EREBPs. Thus, we designated this clone as OsEREBP1 (Fig. 7A; GenBank accession no. AF193803) and selected it for further study. The deduced amino acid sequence of OsEREBP1 shares only low overall sequence identity (around 20%) with other EREBPs, including AtERF1-5 (Fujimoto et al., 2000; Fig. 7A). However, the EREBP/AP2 domain of OsEREBP1 is highly homologous (60%–70%) to the corresponding domains in other plant EREBPs. Furthermore, OsEREBP1 contains a highly acidic domain in its N-terminal region (47–82 amino acids) and a region rich in basic amino acids (R-114-K-R-K-117) that could be a putative nuclear localization signal.
Figure 7.
BWMK1 phosphorylates OsEREBP1 and the phosphorylation enhances the ability of the transcription factor to bind to the cis-acting element. A, Alignment of the OsEREBP1 amino acid sequence with the EREBPs, AtERF1-5 (Fujimoto et al., 2000). Overlined sequences, Conserved EREBP/AP2 domain. Underlined sequences, N-terminal acidic domain. Identical residues are indicated with dots. A putative MAPK phosphorylation site is indicated by an asterisk. B, Yeast two-hybrid interaction of BWMK1 with OsEREBP1. Yeast strain pJ69-4A transformed with the constructs indicated in the left plant were grown on synthetic complete (SC) medium minus Trp and Leu (+Ade) or in SC medium minus Trp, Leu, and Ade (–Ade). β-Galactosidase activity in the colonies grown in +Ade medium (LacZ) was determined by filter-lift assay. BD, Fusion to a plasmid containing the GAL4 DNA-binding domain; AD, Fusion to a GAL4 transcriptional activation domain. C, BWMK1 phosphorylates OsEREBP1 in vitro. Arrows indicate the positions of autophosphorylated GST-BWMK1 and phosphorylated GST-OsEREBP1. The apparent molecular masses (kilodaltons) are indicated at the left. D, The DNA-binding activity of OsEREBP1 to the GCC box motif (AGCCGCC) is enhanced by BWMK1-mediated phosphorylation. The arrowheads mark the position of the protein-DNA complex and free probe.
To further confirm that BWMK1 interacts with OsEREBP1, OsEREBP1 was fused to the GAL4 activation domain of pGAD424 (CLONTECH Laboratories, Palo Alto, CA), and the prey construct, AD-OsEREBP1, was introduced into the yeast strain pJ69-4A carrying the bait construct, BD-BWMK1. Yeast cells could grow on synthetic drop-out plates lacking Trp, Leu, and adenine, and expression of the LacZ reporter gene was observed only when they were provided with both AD-OsEREBP1 and BD-BWMK1. Yeast cells expressing either AD-OsEREBP1 or BD-BWMK1 alone could not grow on selection media, indicating specific interaction between AD-OsEREBP1 and BD-BWMK1 (Fig. 7B).
BWMK1 Phosphorylates OsEREBP1 in Vitro and Induces Its Ability to Bind to the GCC Box Motif
Recombinant BWMK1 protein has both auto- and MBP substrate phosphorylation activities (Fig. 2) and directly interacts with OsEREBP1. To examine whether the MAPK can phosphorylate OsEREBP1, we produced a recombinant OsEREBP1 fused to GST in E. coli. We then assessed whether GST-BWMK1 could phosphorylate GST-OsEREBP1. As shown in Figure 7C, BWMK1 did phosphorylate OsEREBP1.
To test whether OsEREBP1 specifically binds to the GCC box (AGCCGCC) motif found in the promoter of several PR genes, we performed an electrophoresis mobility shift assay (EMSA) using a 32P-labeled GCC box (2xAGCCGCC) or a mutant GCC box (2xATC-CTCC) motif. OsEREBP1 specifically binds to the GCC box motif (Fig. 7D) but not to the GCC box mutant (data not shown). To determine if the phosphorylation of OsEREBP1 by BWMK1 affects its DNA-binding activity to the corresponding cis elements, we first phosphorylated OsEREBP1 with the BWMK1 protein and then examined its ability to bind to the GCC box (2xAGCCGCC) by EMSA. As shown in Figure 7D, the phosphorylation of OsEREBP1 by BWMK1 strongly enhanced its DNA-binding activity to the synthetic GCC box (2xAGCCGCC) motif in vitro. In addition, phosphorylation of GST-OsEREBP1 by the kinase in the presence of ATP also strongly enhanced its ability to bind to the GCC box motif (Fig. 7D, lane 5) relative to its activity in absence of ATP (Fig. 7D, lane 4). These observations strongly suggest that BWMK1 may regulate PR gene expression via the phosphorylation of one or more transcription factors.
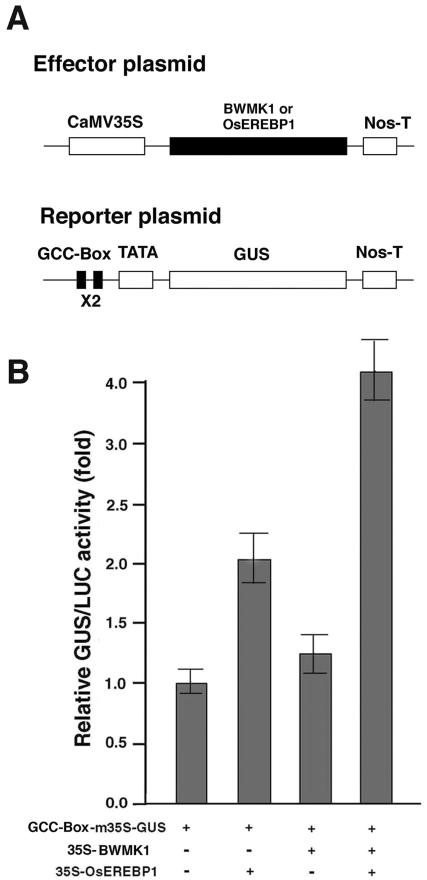
Transient Co-Expression of BWMK1 and OsEREBP1 in Arabidopsis Protoplasts Enhances the Expression of the GUS Reporter Gene Driven by a Minimal Promoter Containing a GCC Box
To demonstrate that the enhanced in vitro DNA-binding activity of the phosphorylated OsEREBP1 is also observed in vivo, we performed transient transactivation assays using Arabidopsis leaf mesophyll protoplasts (Abel and Theologis, 1994). The GUS reporter plasmid was constructed by fusing the GUS gene to a 68-bp synthetic DNA fragment containing a tandem dimer of the GCC box DNA-binding motif (2xAGCCGCC) and the CaMV 35S minimal promoter in pDel.151-8 (Sundaresan et al., 1995). The effector plasmids for BWMK1 and OsEREBP1 were constructed in the plant expression vector pHBT95 (Sheen, 1996), which contains the 35SC4PPDK promoter (Fig. 8A) and the nos terminator. Protoplasts were transfected with the reporter plasmid alone or in combination with one or both of the effector plasmids. As shown in Figure 8B, the expression of the GUS reporter gene was slightly elevated by cotransfection with BWMK1 alone (about 1.3-fold) and OsEREBP1 alone (2-fold). However, the expression level was elevated about 4-fold when the GUS reporter plasmid was cotransfected with both BWMK1 and OsEREBP1. These observations are consistent with those of the in vitro phosphorylation and EMSA experiments. Thus, BWMK1 phosphorylates OsEREBP1; this in turn enhances gene expression that is driven by a promoter containing a GCC box motif.
Figure 8.
Transactivation of the GCC-box-m35S-GUS fusion gene by BWMK1 and OsEREBP1 in transient assay using Arabidopsis protoplasts. A, Schematic diagram of the effector and reporter plasmids used in cotransfection experiments. B, BWMK1 enhances GCC box-driven gene expression by activating OsEREBP1. Bars = se of three replicates. The numbers show the fold of expression compared with the value obtained with the vector control.
DISCUSSION
Here, we report several important findings that provide new insights into the MAPK downstream signaling that is involved in plant defense mechanisms. First, we have isolated a unique MAPK, BWMK1, which is activated in rice by defense-related signals. Second, we have also isolated the substrate that BWMK1 probably phosphorylates in vivo, namely, OsEREBP1, which is a transcription factor that binds to the GCC box DNA motif found in the promoters driving several PR genes. The OsEREBP1 that is involved in plant defense signaling is supported by our experiments showing that BWMK1 phosphorylates OsEREBP1 in vitro and that this enhances the ability of OsEREBP1 to bind to the GCC box DNA motif.
The KD of BWMK1 contains a TDY phosphorylation motif instead of the more common TEY sequence and a putative Leu zipper motif in its CD (He et al., 1999). The CD was found to be essential for the protein kinase activity of BWMK1 and to be involved in the translocation of the protein into the nucleus (Fig. 2). Similar MAPKs that bear putative Leu zippers are AtMPK8 and AtMPK9 from Arabidopsis and alfalfa TDY1, which contain both the TDY motif and the CD. These similarities allow us to create a new subfamily of plant MAPKs.
The protein kinase activity of BWMK1 was rapidly and transiently activated by various pathogen signals, including a fungal elicitor, H2O2, SA, JA, and ethylene. Peak activity was reached 5 to 30 min after treatment, after which BWMK1 transcript levels increased. However, the protein levels of the kinase did not alter similarly during this period (Fig. 4). It is likely that the plant maintains a certain steady-state protein level of MAPKs to allow it to rapidly respond to an unexpected pathogen attack and that upon activation, the kinase is consumed, causing transcription of the kinase gene to be stepped up to maintain the protein levels. Therefore, these observations indicate that the pathogen signals activate the kinase by posttranslational modification, which is also observed for other plant MAPKs involved in defense responses, including SIPK, WIPK, and ERMK (Ligterink et al., 1997; Zhang and Klessig, 1997; Zhang et al., 1998; Seo et al., 1999). That BWMK1 is activated by a fungal elicitor is confirmed by the recent report of He et al. (1999), who showed that BWMK1 (which is identical to BWMK1) is activated by rice blast fungus attack. These authors also reported that wounding similarly activated BWMK1, which supports our observations that ethylene and JA, which are wounding-associated signals, activate BWMK1. That BWMK1 can be activated by SAR-associated signals such as SA and a fungal elicitor and by wounding signals suggests that the two signaling pathways might overlap through this kinase.
Because BWMK1 is activated in response to various defense signals (Fig. 4) and is localized to the nucleus (Fig. 2C), it is possible that this kinase is involved in the transcriptional regulation of PR gene expression by phosphorylating one or more transcription factors. We isolated one possible transcription factor that could interact with BWMK1 by yeast two-hybrid screening of a rice cDNA expression library. The transcription factor belongs to the EREBP family, and we designated it as OsEREBP1. The EREBP family consists of transcription factors that recognize the GCC box DNA element present in the promoters of several PR genes (Buttner and Singh, 1997; Fujimoto et al., 2000). Although OsEREBP1 shows relatively low overall amino acid sequence similarity to the other members of the EREBP family, it does recognize the synthetic GCC box motif (AGCCGCC) in vitro (Fig. 7D). Furthermore, BWMK1 could phosphorylate a recombinant OsEREBP1 in vitro (Fig. 7C), and this phosphorylation enhanced the ability of the transcription factor to bind to the synthetic GCC box motif in vitro (Fig. 7D). These observations support the hypothesis that BWMK1 transduces its signal(s) by phosphorylating one or more transcription factor(s).
To further verify the relationship between BWMK1-mediated phosphorylation of the OsEREBP1 and PR gene expression, we transiently transfected Arabidopsis leaf mesophyll protoplasts with a GUS reporter gene fused to a synthetic tandem dimer of the GCC box DNA motif and the minimal promoter pDel.151-8. Co-expression with the OsEREBP1 effector plasmid driven by the CaMV 35S promoter transactivated GUS reporter gene expression about 2-fold. When the protoplasts were cotransfected with the reporter construct and both the OsEREBP1 and BWMK1 effector plasmids, the reporter gene expression was further elevated by 4-fold. These observations strongly suggest that BWMK1 enhances the expression of GCC box-driven PR genes by phosphorylating the OsEREBP1 transcription factor.
The GCC box motif is present in the PR2 and PR5 gene promoter that is stimulated by both ethylene and JA (Zhou et al., 1997). Because BWMK1 is activated by the SAR-associated SA signal and by the ethylene and JA signals mainly associated with wounding, it cannot be ruled out that BWMK1 could phosphorylate another transcription factor(s), one that is exclusively associated with SA-mediated gene expression and that the ethylene/JA and SA-mediated defense signaling pathways may cross through this mechanism.
The Pto kinase in tomato plants phosphorylates the Pti4 protein. The phosphorylation of Pti4 enhances its ability to bind to the GCC box contained in several PR gene promoters (Gu et al., 2000). Although this suggests that plant MAPKs phosphorylate transcription factor(s) and that this in turn triggers PR gene expression, no direct evidence of this has been obtained thus far. To provide this evidence, we created transgenic tobacco plants that overexpress BWMK1 under the control of the CaMV 35S promoter. Significantly, the plants had higher levels of PR gene expression and an enhanced resistance to virulent bacterial and fungal pathogens (Figs. 5 and 6). These observations strongly support the notion that BWMK1 is involved in triggering PR gene expression. Another observation made with the transgenic tobacco plants that overexpress BWMK1 is that the old leaves of transgenic plants had HR-like lesions. Recently, Zhang and Liu (2001) and Yang et al. (2001) also reported that overexpression of SIPK and WIPK resulted in both plant defense gene activation and enhanced programmed cell death, suggesting the two processes are linked.
In summary, BWMK1, which can be classified into family V MAPK, is activated by SA-associated defense signals and by ethylene/JA-associated plant defense signals. The MAPK phosphorylates the OsEREBP1, which in turn enhances the DNA-binding activity of the factor to the corresponding cis-acting element, GCC box motif (AGCCGCC), in several basic PR gene promoters. Transient cotransfection assays using Arabidopsis leaf protoplast support the in vitro DNA-binding result by elevating expression level of the GUS reporter gene about 4-fold when the reporter plasmid was cotransfected with both BWMK1 and OsEREBP1. Ectopic overexpression of the BWMK1 in transgenic tobacco plants causes HR-like cell death and increased resistance to pathogens with elevated level of PR gene expression. Thus, these results suggest that BWMK1 is involved in MAPK cascades in plant defense signal transduction via direct phosphorylation of a transcription factor(s).
MATERIALS AND METHODS
Rice (Oryza sativa L. Milyang 117) Suspension Cell Culture and Treatments
Suspension cell lines of rice were cultured and maintained as described by Kyozuka et al. (1990). Treatment with an elicitor or chemicals was performed in the dark. The fungal elicitor was prepared from the rice blast-causing fungus Magnaporthe grisea as described (Simmons et al., 1992) and was added to the cell culture at a final concentration of 50 μg mL–1 in terms of total reducing sugars. The chemicals used were SA (used at 1 mm), H2O2 (1 mm), ethephon (5 mm), and JA (0.1 mm). The cells were harvested by filtration at various time periods after treatment, quickly frozen in liquid nitrogen, and stored at –80°C until analysis.
Yeast (Saccharomyces cerevisiae) Two-Hybrid Screening
The yeast two-hybrid screening method was employed to isolate transcription factors that interact with BWMK1. We digested the BWMK1 cDNA with SmaI and BclI and ligated the fragments into the pGBT9 plasmid, which contains the Trp1 selection marker (CLONTECH). The prey library containing cDNA from rice suspension cells was constructed in plasmid pAD-GAL4 (Stratagene, La Jolla, CA), which harbors the Leu2 selection marker. The yeast strain pJ69-4A (James et al., 1996) was used to express these plasmids. The two-hybrid screenings and assays were performed as described in CLONTECH's Yeast Protocols. Positive interactions were verified by the β-galactosidase assay.
Northern-Blot Analysis
Total RNA was isolated as described by Lee et al. (1995), and 20 μg of the RNA was subjected to electrophoresis through a 1.5% (w/v) formaldehydeagarose gel, transferred onto a GeneScreen Plus membrane (New England Nuclear, Boston), and hybridized to random primer-labeled full-length BWMK1 cDNA or PR gene probes (Ryals et al., 1996) under the conditions described previously (Lee et al., 1995).
Expression of Fusion Proteins
For expression in bacteria, full-length BWMK1, the KD (BWMK1 KD), and the CD (BWMK1 CD) were fused to the C terminus of GST. The GST-BWMK1 fusion construct was generated by digesting the full-length BWMK1 in pBluescript SK– with SmaI/BclI and inserting the excised fragment into the corresponding sites of the GST expression vector pGEX-2T (Amersham, Buckinghamshire, UK). An EcoRI fragment of BWMK1 in pBluescript SK– that encodes only the KD of BWMK1 was subcloned into the GST expression vector to generate GST-BWMK1 KD, whereas an EcoRI-XhoI fragment encoding only the CD was subcloned into the same expression vector to create GST-BWMK1 CD. The OsEREBP1 cDNA in pBluescript SK– was also digested with SmaI/XhoI and ligated into the pGEX-5X vector to generate GST-OsEREBP1. The resulting constructs were then introduced into Escherichia coli strain BL21 (pLysS), and the GST fusion proteins were expressed and purified using glutathione-agarose beads according to the manufacturer's instructions (Amersham).
In Vitro Kinase Assay
Autophosphorylation activities of the full-length, KD and CD of BWMK1 were assayed at different protein concentrations in 20 μL of reaction buffer (20 mm Tris-HCl [pH 7.5], 1 mm dithiothreitol, 10 mm MgCl2, and 100 μm ATP) containing 5 μCi [γ-32P] ATP (6,000 Ci mmol–1; Amersham). The substrate kinase activity of GST-BWMK1 was assayed at room temperature for 20 min in a final volume of 20 μL using one of the substrates, MBP or GST-OsEREBP1, at a final protein concentration of 0.1 mg mL–1. Reactions were terminated by the addition of 4× SDS sample buffer. The samples were then analyzed by 12.5% (w/v) SDS-PAGE and subsequent autoradiography.
Antibody Production and Immunoblot Analysis
A polyclonal antibody recognizing BWMK1 (Ab-pNBWMK1) was raised by immunizing rabbits with a synthetic peptide representing the NH2 terminus (MEFFTEYGEAASQYQ) of BWMK1. For immunoblot analysis, 50 μg of total protein per lane was resolved by 10% (w/v) SDS-PAGE. The separated proteins were then transferred to a nitrocellulose membrane (Amersham) by semidry electroblotting. After blocking the membrane at room temperature for 1 h in Tris-buffered saline containing 0.1% (v/v) Tween 20 buffer (20 mm Tris [pH 7.5], 150 mm NaCl, and 0.1% [v/v] Tween 20) with 6% (w/v) nonfat dry milk (Carnation, Glendale, CA), the membrane was incubated with 0.2 μg mL–1 Ab-pNBWMK1 for 1 h. The blot was then washed four times in TTBS buffer, incubated with a horseradish peroxidase-conjugated secondary antibody (1:5,000 [v/v] dilution), and developed by using an enhanced chemiluminescence kit (Amersham).
Preparation of Protein Extracts and Immunocomplex Kinase Assay
Preparation of protein extracts from rice suspension cells treated with fungal elicitor or chemicals and immunocomplex kinase assay were performed as described by Zhang and Klessig (1997). Protein extracts (100 μg) were incubated with Ab-pNBWMK1 (2 μg) in immunoprecipitation buffer (20 mm Tris [pH 7.5], 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm Na3VO4, 1 mm NaF, 10 mm β-gylcerophosphate, 5 μg mL–1 antipain, 5 μg mL–1 aprotinin, 5 μg mL–1 leupeptin, and 0.1% [v/v] Tween 20) at 4°C for 4 h on a rocker. Approximately 20 μL packed volume of 50% (v/v) protein A-agarose washed with immunoprecipitation buffer was added, and the incubation was continued for another 4 h. Agarose bead-protein complexes were pelleted by brief centrifugation and washed three times with 1.5 mL of immunoprecipitation buffer, once with immunoprecipitation buffer plus 1 m NaCl, and three times with 1 mL of kinase reaction buffer. Kinase activity in the complex was assayed at room temperature for 20 min in a final volume of 20 μL containing 0.1 mg mL–1 MBP and 100 μm ATP with 5 μCi of [γ-32P-ATP]. The reaction was stopped by the addition of SDS-PAGE sample loading buffer. After electrophoresis on 12.5% (w/v) SDS-PAGE, the phosphorylated MBP was visualized by autoradiography.
Construction of Transgenic Plants
For construction of transgenic tobacco (Nicotiana tabacum cv Xanthi-nc) plants, BWMK1 cDNA was ligated into the plant binary vector pGA643 (An et al., 1988). The recombinant plasmids were introduced into Agrobacterium tumefaciens EHA101, and transgenic tobacco plants were generated by a standard leaf disc transformation method (Horsch et al., 1988) and selected by kanamycin resistance. T2 progeny of transgenic plants expressing high levels of BWMK1 were grown at 25°C (day) and 20°C (night) temperatures, a 16-h photoperiod, and 65% relative humidity, and used for the assays.
Histochemistry and Microscopy
The cell death phenotype of tobacco leaves was photographed by using a dissecting microscope. Autofluorescent materials and callos deposition were detected using an UV epifluorescence microscope as described by Dietrich et al. (1994)
Pathogen Infections and Resistance Assay
A bacterial pathogen (Pseudomonas syringae pv tabacci) was cultured overnight at 30°C in Kings B medium containing 50 μg mL–1 rifampicin and 50 μg mL–1 kanamycin. The bacterial culture was washed twice with 10 mm MgCl2 and resuspended in 10 mm MgCl2. Bacterial density was determined by absorbance at OD600 nm. Bacteria were diluted to the desired concentrations in 10 mm MgCl2 for inoculation. Six-week-old plants were inoculated by vacuum infiltration, and the inoculated plants were kept in a greenhouse. Leaf discs were ground in 10 mm MgCl2 and plated on appropriate Kings B plates. The number of bacteria in the leaves was calculated by counting cfu. Infection with Phytophthora parasitica pv nicotianae was performed as described by Mittler et al., (1995). The oomycete was subcultured at 23°C on one-quarter-strength potato dextrose agar. Conidia from 3-week-old cultures on potato dextrose agar were used for inoculation (Heo et al., 1999).
In Vitro DNA-Binding Assay
A synthetic GCC box (CAT AAG AGC CGC CAC TAA AAT AAG ACC GAT CAA ATA AGA GCC GCC AT) and GCC box mutant (CAT AAG ATC CTC CAC TAA AAT AAG ACC GAT CAA ATA AGA TCC TCC AT; Ohme-Takagi and Shinshi, 1995) were end labeled with 32P as described previously (Cheong et al., 1998). The probe (4 fmol) was mixed with each of the purified GST fusion proteins in a buffer containing 2 μg of poly (dA-dT).(dA-dT), 25 mm HEPES (pH 7.5), 40 mm KCl, 0.1 mm EDTA, 10% (v/v) glycerol, and 1 mm dithiothreitol. After incubation at room temperature for 20 min, the reaction mixtures were separated in a 5% (w/v) polyacrylamide gel using 0.5× Tris-borate/EDTA buffer. The gel was then dried and exposed to x-ray film.
Localization and Transient Expression Assay
To observe the cellular localization of BWMK1, we prepared BWMK1-smGFP or BWMK1 KD-smGFP fusion constructs. The BamHI (1–506 amino acids) or EcoRI (1–324 amino acids) fragment of BWMK1 was fused to the coding region of smGFP under the control of the CaMV 35S promoter. The fusion construct of nuclear localization signal from simian virus 40 large T antigen and red fluorescent protein (NLS-RFP) was used as a positive control (Lee et al., 2001). Polyethylene glycol-mediated cotransfection was performed to introduce the constructs into Arabidopsis protoplasts (Abel and Theologis, 1994). Expression of the fusion constructs was monitored at various times after transfection by an Axioplan 2 fluorescence microscope (Zeiss, Jena, Germany), and images were captured with a Zeiss Axiocam HR camera using XF116-2 (exciter, 475AF20; dichroic, 500DRLP; and emitter, 510AF23) and XF33 (exciter, 535AF35; dichroic, 570DRLP; and emitter, 605DF50) filter sets (Omega, Inc., Brattleboro, VT) for GFP and RFP, respectively. For transient expression, a GCC box dimer (2xAGCCGCC) was fused to a minimal –46 CaMV 35S promoter-GUS reporter gene (GCC-box-m35S-GUS; Jefferson et al., 1987; Zhou et al., 1997). To construct the two effector plasmids, full-length BWMK1 and OsEREBP1 cDNA were inserted into a plant expression vector (pHBT95) containing the 35SC4PPDK promoter and the nos terminator (Sheen, 1996). Transient expression of these constructs was carried out in Arabidopsis mesophyll protoplasts as described by Abel and Theologis (1994). In each transfection, 5 × 105 protoplasts were transfected with 10 μg of reporter construct alone or together with 15 μg of each effector construct or a vector DNA control (pHBT95). The transfected protoplasts were incubated in W5 medium for 16 h in the dark. A construct carrying the LUC (luciferase) gene driven by the 35S promoter was used as an internal control in each transfection. The GUS activity of the cell lysate was divided by the corresponding LUC activity to standardize the data. The results are expressed as means of three independent replicate transfections with sds.
Acknowledgments
We thank Drs. Cris Lamb and John A. Ryals for generously providing us PR protein cDNAs. We also thank Dr. Jen Sheen for providing pHBT95 plasmid and Dr. Inhwan Hwang for providing NLS-RFP plasmid.
Article, publication date, and citation information can be found at http://www.plantphysiol.org/cgi/doi/10.1104/pp.103.023176.
This work was supported by Korea Science and Engineering Foundation (grant no. 2000–2–20900–001–1), by Crop Functional Genomic Center (grant no. CG1512), by National Research Laboratory (grant no. 2000–N–NL–01–C–236), and by the BK21 program from the Ministry of Education to M.J.C.
References
- Abel S, Theologis A (1994) Transient transformation of Arabidopsis leaf protoplasts: a versatile experimental system to study gene expression. Plant J 5: 421–427 [DOI] [PubMed] [Google Scholar]
- Agrawal GK, Rakwal R, Iwahashi H (2002) Isolation of novel rice (Oryza sativa L.) multiple stress responsive MAP kinase gene, OsMSRMK2, whose mRNA accumulates rapidly in response to environmental cues. Biochem Biophys Res Commun 294: 1009–1016 [DOI] [PubMed] [Google Scholar]
- An G, Prebert AM, Ha SB (1988) Binary vectors. In SB Gelvin, RA Schilperoort, eds, Plant Molecular Biology Manual. Academic Press, Dordrecht, The Netherlands, pp 1–19
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Brott BK, Pinsky BA, Erikson RL (1998) Nlk is a murine protein kinase related to Erk/MAP kinases and localized in the nucleus. Proc Natl Acad Sci USA 95: 963–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner M, Singh KB (1997) Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc Natl Acad Sci USA 94: 5961–5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale F, Jonak C, Ligterink W, Niehaus K, Boller T, Hirt H (2000) Differential activation of four specific MAPK pathways by distinct elicitors. J Biol Chem 275: 36734–36740 [DOI] [PubMed] [Google Scholar]
- Cheong YH, Yoo CM, Park JM, Ryu GR, Goekjian VH, Nagao RT, Key JL, Cho MJ, Hong JC (1998) STF1 is a novel TGACG-binding factor with a zinc-finger motif and a bZIP domain which heterodimerizes with GBF proteins. Plant J 15: 199–209 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Dietrich RA, Richberg MH (1996) Death don't have no mercy: cell death programs in plant-microbe interactions. Plant Cell 8: 1793–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG (2001) Plant pathogens and integrated defense responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Davis R (2000) Signal transduction by the JNK group of MAP kinases. Cell 103: 239–252 [DOI] [PubMed] [Google Scholar]
- Davis SJ, Vierstra RD (1998) Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol 36: 521–528 [DOI] [PubMed] [Google Scholar]
- Despres C, Subramaniam R, Matton DP, Brission N (1995) The activation of the potato PR-10a gene requires the phosphorylation of the nuclear factor PBF-1. Plant Cell 7: 639–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL (1994) Arabidopsis mutants simulating disease resistance response. Cell 77: 565–577 [DOI] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA (1991) Nuclear targeting sequences: a consensus? Trends Biochem Sci 16: 478–481 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Harrison MJ, Lamb CJ (1994) Early events in the activation of plant defense responses. Annu Rev Phytopathol 32: 479–501 [Google Scholar]
- Droge-Laser W, Kaiser A, Lindsay WP, Halkier BA, Loake GJ, Doerner P, Dixon RA, Lamb C (1997) Rapid stimulation of a soybean protein-serine kinase that phosphorylates a novel bZIP DNA-binding protein, G/HBF-1, during the induction of early transcription-dependent defenses. EMBO J 16: 726–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durner J, Shah J, Klessig DF (1997) Salicylic acid and disease resistance in plants. Trends Plant Sci 2: 266–274 [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YQ, Yang C, Thara VK, Zhou J, Martin GB (2000) Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 12: 771–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Fong SH, Yang D, Wang GL (1999) BWMK1, a novel MAP kinase induced by fungal infection and mechanical wounding in rice. Mol Plant-Microbe Interact 12: 1064–1073 [DOI] [PubMed] [Google Scholar]
- Heo WD, Lee SH, Kim MC, Kim JC, Chung WS, Chun HJ, Lee KJ, Park CY, Choi JY, Cho MJ (1999) Involvement of specific calmodulin isoforms in salicylic acid-independent activation of plant disease resistance responses. Proc Natl Acad Sci USA 96: 766–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Fry J, Hoffmann N, Neidermeyer J, Rogers SG, Fraley RT (1988) Leaf disc transformation. In SB Gelvin, RA Schilperoot, eds, Plant Molecular Biology Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–9
- Huang HJ, Fu SF, Tai YH, Chou WC, Huang DD (2002) Expression of Oryza sativa MAP kinase gene is developmentally regulated and stress-responsive. Physiol Plant 114: 572–580 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K (2000) Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J 24: 655–665 [DOI] [PubMed] [Google Scholar]
- Innes RW (2001) Mapping out the roles of MAP kinases in plant defense. Trends Plant Sci 6: 392–394 [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 20: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C, Kiegerl S, Ligterink W, Barker PJ, Huskisson NS, Hirt H (1996) Stress signaling in plants: a mitogen-activated protein kinase pathway is activated by cold and drought. Proc Natl Acad Sci USA 93: 11274–11279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C, Kiegerl S, Lloyd C, Chan J, Hirt H (1995) MMK2, a novel alfalfa MAP kinase, specifically complements the yeast MPK1 function. Mol Gen Genet 248: 686–694 [DOI] [PubMed] [Google Scholar]
- Kyozuka J, Izawa T, Nakajima M, Shimamoto K (1990) Effect of the promoter and the first intron of maize. Adh1 on foreign gene expression in rice Maydica 35: 353–357 [Google Scholar]
- Lee SH, Kim JC, Lee MS, Heo WD, Seo HY, Yoon HW, Hong JC, Lee SY, Bahk JD, Hwang I et al. (1995) Identification of a novel divergent calmodulin isoform from soybean which has a different ability to activate calmodulin-dependent enzymes. J Biol Chem 270: 21806–21812 [DOI] [PubMed] [Google Scholar]
- Lee YJ, Kim DH, Kim Y-W, Hwang I (2001) Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligterink W, Hirt H (2001) Mitogen-activated protein (MAP) kinase pathways in plant: versatile signaling tools. Int Rev Cytol 201: 209–275 [DOI] [PubMed] [Google Scholar]
- Ligterink W, Kroj T, Nieden U, Hirt H (1997) Receptor-mediated activation of a MAP kinase in pathogen defense of plants. Science 276: 2054–2057 [DOI] [PubMed] [Google Scholar]
- Mittler R, Shulaev V, Lam E (1995) Coordinated activation of programmed cell death and defense mechanisms in transgenic tobacco plants expressing a bacterial proton pump. Plant Cell 7: 29–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Hayashida N, Yamaguchi-Shinozaki K, Kamada H, Shinozaki K (1993) ATMPKs: a gene family of plant MAP kinases in Arabidopsis thaliana. FEBS Lett 336: 440–444 [DOI] [PubMed] [Google Scholar]
- Nuhse TS, Peck SC, Hirt H, Boller T (2000) Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. J Biol Chem 275: 7521–7526 [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7: 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE et al. (2000) Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Roe JL, Durfee T, Zupan JR, Repetti PP, McLean BG, Zambryski PC (1996) TOUSLED is a nuclear serine/threonine protein kinase that requires a coiled-coil region for oligomerization and catalytic activity. J Biol Chem 272: 5838–5845 [DOI] [PubMed] [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner H-Y, Hunt MD (1996) Systemic acquired resistance. Plant Cell 8: 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenbeck MA, Samac DA, Fedorova M, Gregerson RG, Gantt JS, Vance CP (1999) The alfalfa (Medicago sativa) TDY1 gene encodes a mitogen-activated protein kinase homologue. Mol Plant-Microbe Interact 12: 882–893 [DOI] [PubMed] [Google Scholar]
- Seo S, Okamoto M, Seto H, Ishizuka K, Sano H, Ohashi Y (1995) Tobacco MAP kinase: a possible mediator in wound signal transduction pathways. Science 270: 1988–1992 [DOI] [PubMed] [Google Scholar]
- Seo S, Sano H, Ohashi Y (1999) Jasmonate-based wound signal transduction requires activation of WIPK, a tobacco mitogen-activated protein kinase. Plant Cell 11: 289–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J (1996) Ca2+-dependent protein kinases and stress signal transduction in plants. Science 274: 1900–1902 [DOI] [PubMed] [Google Scholar]
- Simmons CR, Litts JC, Huang N, Rodriguez RL (1992) Structure of a rice beta-glucanase gene regulated by ethylene, cytokinin, wounding, salicylic acid and fungal elicitors. Plant Mol Biol 18: 33–45 [DOI] [PubMed] [Google Scholar]
- Song F, Goodman RM (2002) OsBIMK1, a rice MAP kinase gene involved in disease resistance response. Planta 215: 997–1005 [DOI] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JDG, Dean C, Ma H, Martienssen R (1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Tena G, Asai T, Chiu W, Sheen J (2001) Plant mitogen-activated protein kinase signaling cascades. Curr Opin Plant Biol 4: 392–400 [DOI] [PubMed] [Google Scholar]
- Thara VK, Tang X, Gu YQ, Martin GB, Zhou JM (1999) Pseudomonas syringae pv tomato induces the expression of tomato EREBP-like genes Pti 4 and Pti 5 independent of ethylene, salicylate and jasmonate. Plant J 20: 475–483 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J-Q, Oono K, Imai R (2002) Two novel mitogen-activated proteinsignaling components, OsMEK1 and OsMAP1, are involved in a moderate low-temperature signaling pathway in rice. Plant Physiol 129: 1880–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C, Anglmayer R, Vicente O, Heberle-Bors E (1995) Molecular cloning, functional expression in Escherichia coli, and characterization of multiple mitogen-activated-protein kinases from tobacco. Eur J Biochem 233: 249–257 [DOI] [PubMed] [Google Scholar]
- Xiong L, Lee MW, Qi M, Yang Y (2001) Identification of defense-related rice genes by suppression subtractive hybridization and differential screening. Mol Plant-Microbe Interact 14: 685–692 [DOI] [PubMed] [Google Scholar]
- Yang KY, Liu Y, Zhang S (2001) Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc Natl Acad Sci USA 98: 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Du H, Klessig DF (1998) Activation of the tobacco SIP kinase by both a cell wall-derived carbohydrate elicitor and purified proteinaceous elicitins from Phytophthora spp. Plant Cell 10: 435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Klessig DF (1997) Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9: 809–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Klessig DF (2001) MAPK cascades in plant defense signaling. Trends Plant Sci 6: 520–527 [DOI] [PubMed] [Google Scholar]
- Zhang S, Liu Y (2001) Activation of salicylic acid-induced protein kinase, a mitogen-activated protein kinase, induces multiple defense responses in tobacco. Plant Cell 13: 1877–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Tang X, Martin GB (1997) The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J 16: 3207–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]