Abstract
Nicotianamine synthase (NAS) is an enzyme that is critical for the biosynthesis of the mugineic acid family of phytosiderophores in graminaceous plants, and for the homeostasis of metal ions in nongraminaceous plants. We isolated one genomic NAS clone, ZmNAS3, and two cDNA NAS clones, ZmNAS1 and ZmNAS2, from maize (Zea mays cv Alice). In agreement with the increased secretion of phytosiderophores with Fe deficiency, ZmNAS1 and ZmNAS2 were positively expressed only in Fe-deficient roots. In contrast, ZmNAS3 was expressed under Fe-sufficient conditions, and was negatively regulated by Fe deficiency. This is the first report describing down-regulation of NAS gene expression in response to Fe deficiency in plants, shedding light on the role of nicotianamine in graminaceous plants, other than as a precursor in phytosiderophore production. ZmNAS1-green fluorescent protein (sGFP) and ZmNAS2-sGFP were localized at spots in the cytoplasm of onion (Allium cepa) epidermal cells, whereas ZmNAS3-sGFP was distributed throughout the cytoplasm of these cells. ZmNAS1 and ZmNAS3 showed NAS activity in vitro, whereas ZmNAS2 showed none. Due to its duplicated structure, ZmNAS2 was much larger (65.8 kD) than ZmNAS1, ZmNAS3, and previously characterized NAS proteins (30–38 kD) from other plant species. We reveal that maize has two types of NAS proteins based on their expression pattern and subcellular localization.
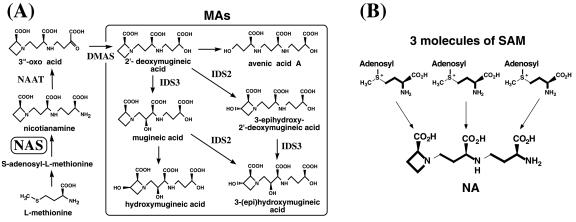
To acquire Fe, graminaceous plants secrete Fe chelators, known as mugineic-acid family phytosiderophores (MAs). MAs dissolve Fe in the rhizosphere, followed by reabsorption of the Fe(III)-MA complexes through YS1 transporters in the plasma membrane (Takagi, 1976; Curie et al., 2001). Only graminaceous plants use the MA mechanism of acquiring Fe(III), classified as the Strategy II mechanism (Marschner et al., 1986). Fe deficiency is a problem in crop production worldwide, especially in calcareous soils, where Fe is sparingly soluble due to the high soil pH. The ability of graminaceous plants to tolerate Fe deficiency is thought to depend on the quantity of MAs secreted during Fe deficiency (Takagi, 1976; Mori et al., 1987, 1988; Römheld, 1987; Kawai et al., 1988; Mihashi and Mori, 1989; Singh et al., 1993). The biosynthetic pathways of MAs (Fig. 1A) have been determined (Mori and Nishizawa, 1987; Kawai et al., 1988; Shojima et al., 1990; Ma et al., 1999), and almost all the genes involved have been isolated in our laboratory (Higuchi et al., 1999b; Takahashi et al., 1999; Nakanishi et al., 2000; Kobayashi et al., 2001). NAS is a key enzyme in MA biosynthesis, catalyzing the trimerization of SAM into one molecule of NA (Higuchi et al., 1999b). NAS activity in graminaceous plants is well correlated with tolerance to Fe deficiency. In maize (Zea mays), a plant susceptible to Fe deficiency, NAS activity is very low (Higuchi et al., 1996a), and maize secretes lower amounts of MAs than barley or oat (Avena sativa), cereals that are tolerant of low Fe supply (Römheld, 1987; Lytle and Jolley, 1991). NAS is also important for growth in nongraminaceous plants, which do not synthesize MAs (Higuchi et al., 1996a). In these plants, NA has been implicated in the internal transport of metal ions (Scholz et al., 1992; Stephan and Scholz, 1993; Pich et al., 1994; Stephan et al., 1994). Since Higuchi et al. (1999b) isolated the first NAS gene from barley, NAS genes have been isolated from barley again (Herbik et al., 1999), and from tomato (Lycopersicon esculentum; Ling et al., 1999), Arabidopsis (Suzuki et al., 1999), and rice (Oryza sativa; Higuchi et al., 2001). In this study, we focus on maize, an economically important cereal that is susceptible to Fe deficiency. We isolated and characterized three NAS genes in maize (ZmNAS), and demonstrated that there are two types of NAS genes with different expression patterns; ZmNAS1 and ZmNAS2 are expressed only in roots under Fe-deficient conditions, and ZmNAS3 is expressed under Fe-sufficient conditions. We discuss the role of NA in Fe-deficient roots and in Fe-sufficient leaves.
Figure 1.
A, Metabolic pathway for the synthesis of MAs, based on existing data. A dioxymugineic acid synthase gene was recently cloned from barley (Hordeum vulgare). B, Nicotianamine synthase (NAS) catalyzes S-adenosyl-Met (SAM) trimerization and ring formation to synthesize NA. Bold lines indicate the unit incorporated into NA.
RESULTS
Isolation of a Genomic Clone Containing a Full-Length NAS-Like Sequence
We screened a maize genomic library (Clontech, Palo Alto, CA) for NAS clones using the rice NAS gene (OsNAS1), and found one genomic clone with an NAS-like sequence. This clone, named ZmNAS3 (accession no. AB042551), contains a full-length NAS-like sequence with a putative open reading frame of 1,080 bp, with a calculated molecular mass for ZmNAS3 of 38.8 kD and a pI value of 5.43. Like the NAS genomic clones from barley and rice (Higuchi et al., 2001), ZmNAS3 contains no introns. It contains 5′-upstream and 3′-downstream regions of 9 and 3 kb, respectively.
Isolation of NAS cDNA Clones
We prepared a cDNA library from Fe-deficient maize roots and screened this library for NAS clones using ZmNAS3 as a probe. DNA sequence analysis of the isolated cDNA clones revealed that two had NAS-like sequences. The NAS-like insert of one clone, ZmNAS1, was 1,250 bp long; the other insert, ZmNAS2, was much longer (2,154 bp) than previously characterized NAS genes from other plant species.
Nucleotide sequences of ZmNAS1 and ZmNAS2 and Deduced Amino Acid Sequences of ZmNAS1 and ZmNAS2
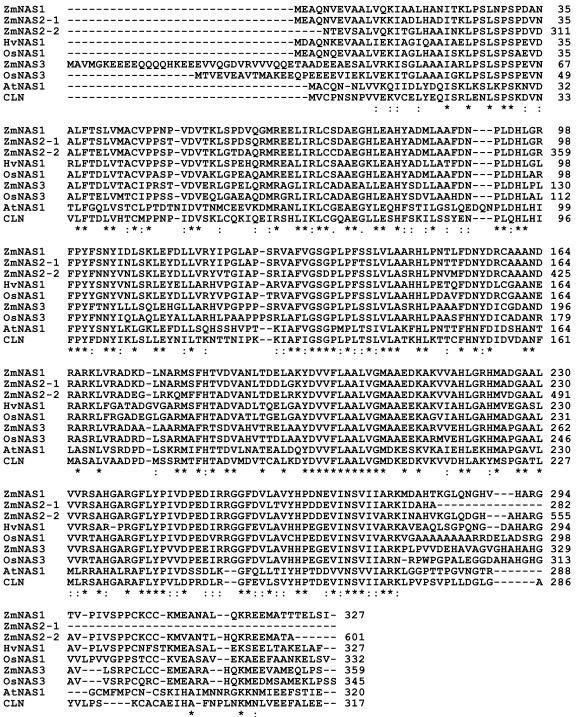
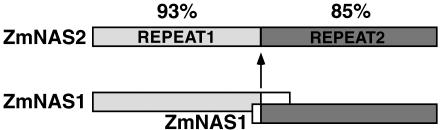
The ZmNAS1 and ZmNAS2 cDNAs have open reading frames encoding 327 and 601 amino acids, respectively. We calculated molecular mass and pI values of 35.6 kD and 6.11 for ZmNAS1, and 64.1 kD and 6.08 for ZmNAS2. Deduced amino acid sequences for ZmNAS1 and ZmNAS2 are shown with those of other plant NASs in Figure 2. Both proteins are highly homologous to other NASs. ZmNAS1 has nearly the same molecular mass as the others, whereas ZmNAS2 is approximately twice as large. Interestingly, the ZmNAS2 sequence partially duplicates that of ZmNAS1. The first and second NAS domains, comprising amino acids 1 to 281 and 282 to 601, are very similar to ZmNAS1 (with 93% and 85% amino acid identity, respectively; Fig. 3).
Figure 2.
Comparison of the putative amino acid sequences of ZmNAS1 to ZmNAS3 with those of other NASs. HvNAS, barley NAS; OsNAS, rice NAS; AtNAS, Arabidopsis NAS; and CLN, tomato NAS. Asterisks indicate identical amino acid residues in all sequences. ZmNAS2-1 and ZmNAS2-2 signify the first and second NAS-like sequences in ZmNAS2, respectively. The terminal amino acid of ZmNAS2-1 (A) is followed by the first amino acid of ZmNAS2-2 (N) (see Fig. 3).
Figure 3.
Schematic illustration of ZmNAS2 aligned with ZmNAS1. Repeats 1 and 2 indicate amino acids 1 to 281 and 282 to 601, respectively. The percentages of ZmNAS1 amino acids' identity to ZmNAS2 are indicated.
The expression of ZmNAS1, ZmNAS2, and ZmNAS3
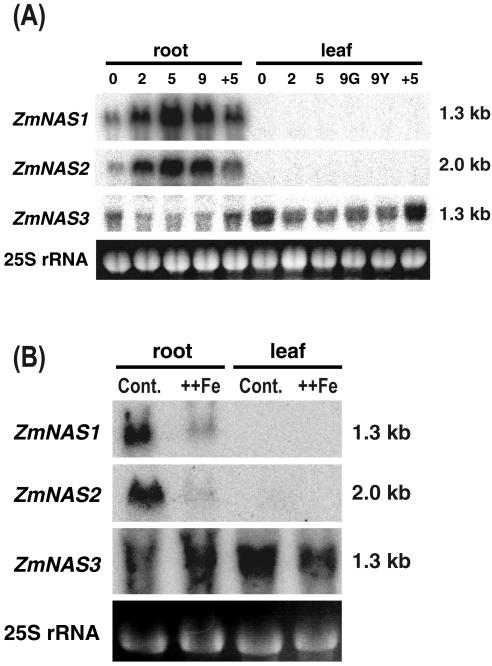
We confirmed the existence of longer NAS transcripts by northern-blot analysis. The ZmNAS2 probe detected transcripts of approximately 1.3 and 2.0 kb (data not shown), corresponding to ZmNAS1 and ZmNAS2, respectively, in Fe-deficient roots, but not in Fe-deficient or Fe-sufficient leaves. The amount of each transcript in Fe-deficient roots increased gradually until d 5, plateaued until d 9 with ZmNAS1 and ZmNAS2, and decreased 5 d after Fe was resupplied. Transcripts of both genes were present in low concentrations in Fe-sufficient roots (Fig. 4A). In contrast, ZmNAS3 was expressed in Fe-sufficient roots and leaves. Its expression was decreased by Fe deficiency and was induced by Fe resupply. The results of quantitative reverse transcription-PCR using specific primers for ZmNAS3 were consistent with the results of northern-blot analysis using specific probes for ZmNAS3 (data not shown). A supply of excess Fe suppressed the expression of ZmNAS1 and ZmNAS2, whereas the expression of ZmNAS3 was not changed by the Fe excess treatment (Fig. 4B).
Figure 4.
Northern-blot analysis of ZmNAS gene expression in maize during Fe deficiency and after resupply of Fe (A) and during Fe excess treatment (B).+5 indicates the number of days after Fe resupply. G and Y indicate green and yellow leaves of Fe-deficient plants, respectively. ++Fe indicates plants under Fe-excess conditions. A specific probe for each gene was used for hybridization. Equal loading of total RNAs was confirmed by ethidium bromide staining of 25S RNA.
Western-Blot Analysis of Maize Roots
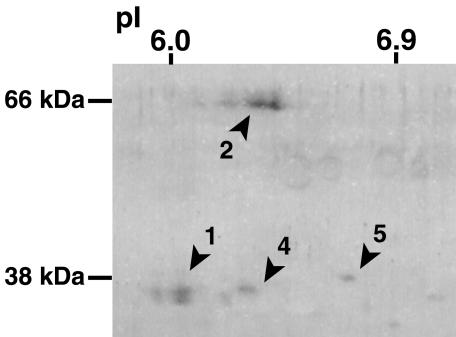
At least four protein spots were detected in Fe-deficient maize roots by two-dimensional-PAGE western-blot analysis (Fig. 5). We identified two spots corresponding to proteins derived from ZmNAS1 and ZmNAS2 from their calculated molecular mass and pI values (35.6 kD and 6.11 for ZmNAS1, and 64.1 kD and 6.08 for ZmNAS2). However, no spot corresponding to ZmNAS3 (38.8 kD and a pI value of 5.43) was detected in Fe-deficient roots. We designated the other spots ZmNAS4 and ZmNAS5. ZmNAS4 corresponds to the protein derived from an expressed sequence tag clone (GenBank accession no. Q619504) judging from its predicted molecular mass and pI value. Spots corresponding to ZmNAS1 and ZmNAS2 were larger than those of ZmNAS4 and ZmNAS5; therefore, we concluded that ZmNAS1 and ZmNAS2 are the most abundant known NAS isoforms in Fe-deficient maize roots.
Figure 5.
Western-blot analysis of NAS-like proteins in Fe-deficient maize roots. Figures indicate spots corresponding to putative ZmNAS gene products. 1, ZmNAS1; 2, ZmNAS2; 4, ZmNAS4; and 5, ZmNAS5
NAS Activity of ZmNAS1, ZmNAS2, and ZmNAS3 Gene Products
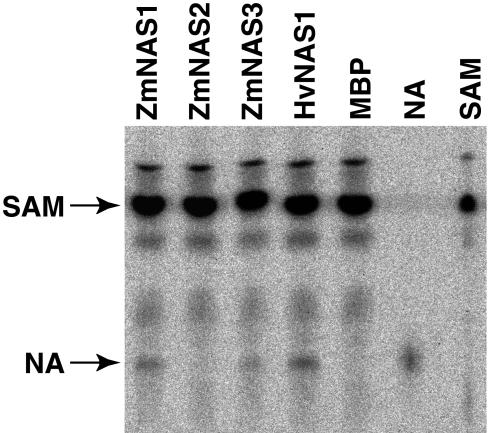
To confirm the enzymatic function of their gene products, ZmNAS genes were fused to the maltose-binding protein (MBP) gene and the resulting fusion proteins were produced in Escherichia coli. MBP-NASs were purified using amylose resin affinity columns, and 1 μg of each fusion protein was used for each enzyme assay. MBP-ZmNAS1 and MBP-ZmNAS3 displayed NAS activity (Fig. 6). In contrast, MBP-ZmNAS2 showed no NAS activity, even though ZmNAS2 has two NAS-like domains.
Figure 6.
Thin-layer chromatography analysis of NAS activity in assay mixtures of MBP-NAS fusion proteins using 1 μg of each fusion protein or MBP for the enzyme assay. Lane SAM contains standard SAM; lane NA contains standard NA.
Localization of ZmNAS-Synthesized Green Fluorescent Protein (sGFP) Fusion Protein in Onion (Allium cepa) Epidermal Cells
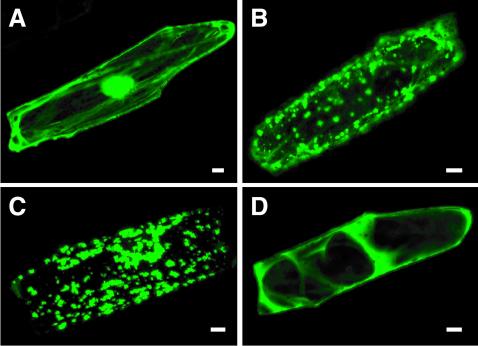
The ZmNAS protein fused to the N terminus of sGFP was transiently expressed under the control of the cauliflower mosaic virus (CaMV) 35S promoter in onion epidermal cells. The green fluorescence of sGFP was viewed with a confocal microscopy. When sGFP alone was expressed (Fig. 7A), the protein was localized in the cytoplasm and nucleus. The fluorescence of ZmNAS1-sGFP and ZmNAS2-sGFP was observed as spots in the cytoplasm of onion epidermal cells (Fig. 7, B and C). On the other hand, the fluorescence of ZmNAS3-sGFP was distributed throughout the cytoplasm of onion epidermal cells (Fig. 7D).
Figure 7.
Subcellular localization of the transiently expressed ZmNAS-sGFP fusion protein in onion epidermal cells observed by confocal laser scanning microscopy. A, Onion epidermal cells expressing sGFP alone. B, Onion epidermal cells expressing ZmNAS1-sGFP fusion protein. C, Onion epidermal cells expressing ZmNAS2-sGFP fusion protein. D, Onion epidermal cells expressing ZmNAS3-sGFP fusion protein. Bars = 20 μm.
DISCUSSION
Two Different Expression Patterns with Different Localization
The ZmNAS3 gene was isolated by screening a genomic library using a heterologous probe. Subsequently, using ZmNAS3 as a probe, two genes that encode NAS-like proteins, ZmNAS1 and ZmNAS2, were isolated from a cDNA library prepared from Fe-deficient maize roots. The molecular masses of ZmNAS1 and ZmNAS3 were similar to previously characterized NAS isoforms (Fig. 2); however, ZmNAS2 had a higher molecular mass and contained two NAS-like sequence domains (Fig. 3). ZmNAS1 and ZmNAS3 showed NAS activity in vitro, but ZmNAS2 did not (Fig. 6).
Although ZmNAS2 did not exhibit NAS activity, the expression of ZmNAS1 and ZmNAS2 was coordinated, with both genes being up-regulated by Fe deficiency and suppressed by a resupply of Fe (Fig. 4). Transcripts of both genes were detected in Fe-deficient roots, but not in Fe-deficient leaves, not even in chlorotic leaves. This is similar to the case in barley, in which there is no HvNAS gene expression in leaves (Higuchi et al., 2001). Interestingly, transcripts of OsNAS1 and OsNAS2, the expression of which is up-regulated by Fe deficiency, were detected not only in rice Fe-deficient roots, but also in chlorotic leaves (Higuchi et al., 2001). Therefore, although maize and rice are similarly susceptible to Fe deficiency, the expression patterns of NAS genes differ in response to Fe deficiency. In contrast to ZmNAS1 and ZmNAS2, ZmNAS3 was expressed under Fe-sufficient conditions, and its transcript level was decreased by Fe deficiency and was induced by an Fe resupply (Fig. 4). This is the first report of an NAS gene that is down-regulated by Fe deficiency in nongraminaceous or graminaceous plants. The decrease in NAS transcript levels in nongraminaceous plants in response to Fe deficiency was predicted based on the result that NAS enzyme activity is reduced in Fe-deficient roots of tobacco (Nicotiana tabacum; Higuchi et al., 1995). However, down-regulation of an NAS gene in graminaceous plants is unexpected because the biosynthesis of phytosiderophores is greatly enhanced by Fe deficiency.
The localization of ZmNAS1-sGFP fusion protein and ZmNAS2-sGFP fusion protein was different from that of ZmNAS3-sGFP fusion protein. ZmNAS1-sGFP and ZmNAS2-sGFP were localized as small spots in the cytoplasm, whereas ZmNAS3-sGFP was distributed throughout the cytoplasm. PSORT (http://psort.nibb.ac.jp/) predictions were determined using full-length predicted protein sequences. All of the ZmNAS proteins were predicted to be localized at the membrane of the endoplasmic reticulum. Therefore, the pattern of ZmNAS1-sGFP and ZmNAS2-sGFP localization is interpreted as vesicles derived from the endoplasmic reticulum. The prediction of the ZmNAS3 localization did not match the result from the ZmNAS3-sGFP fusion, which indicated cytosolic localization. We have previously shown that “particular vesicles” in the Fe-deficient barley root cell is related to the secretion of MAs (Nishizawa and Mori, 1987). These data are consistent with the idea that NAS proteins induced by Fe deficiency in roots are localized at the membrane of vesicles derived from the endoplasmic reticulum, and that the vesicle is the place for the synthesis of MAs. On the other hand, ZmNAS3, localized throughout the cytoplasm, appears to synthesize NA itself, not as the precursor of MAs in the vesicles. ZmNAS3 has a longer amino-terminal region (Fig. 2) and this region was hydrophilic, which may affect the subcellular localization of ZmNAS3.
NA is essential not only for Strategy II plants, but also for Strategy I plants (nongraminaceous plants). The chloronerva mutant of tomato (Lycopersicon esculentum), which lacks NAS activity (Higuchi et al., 1996b), is not able to use Fe correctly (Becker et al., 1995). This mutant shows symptoms typical of Fe deficiency (Stephan and Grün, 1989), including intercostal chlorosis in young leaves, although it accumulates more Fe in all tissues than do wild-type plants (Scholz et al., 1985; Becker et al., 1992). Grafting the chloronerva mutant onto the wild type, or vice versa, restores the normal phenotype (Böhme and Scholz, 1960), and exogenous application of NA is also able to revert the phenotype (Budesinsky et al., 1980). These facts indicate that NA is necessary for Fe homeostasis in nongraminaceous plants. The expression of ZmNAS3 under Fe-sufficient conditions implies that NA also plays an important role in Fe homeostasis in graminaceous plants. In support of this, Curie et al. (2001) recently isolated the YS1 gene from maize that encodes a MAs-Fe(III) complex transporter. Interestingly, Arabidopsis also possesses eight homologs to YS1, although it does not produce or use MAs because of the different strategy for Fe acquisition (Marschner et al., 1986; Guerinot and Yi, 1994; Briat and Lobréaux, 1997). These YS1-like genes have been proposed to encode NA-metal complex transporters (Walker, 2002). Rice also possesses at least 16 YS1-like genes, and some of their products may function as transporters of NA-metal complexes. These results imply that in nongraminaceous and graminaceous plants, NA is synthesized under Fe-sufficient conditions, and NA-Fe complexes are transported into and out of cells or subcellular organelles via Yellow Stripe-Like transporters. In contrast, NA synthesized in roots in response to Fe deficiency is used as a precursor in synthesis of MAs.
NA is also thought to play an important role in the detoxification of excess intracellular Fe (von Wirén et al., 1999). NA concentrations in tomato increase in response to Fe overload (Pich et al., 2001). Northern-blot analysis under Fe excess conditions revealed that the expression of ZmNAS1 and ZmNAS2 was repressed, and that of ZmNAS3 was not affected by excess Fe.
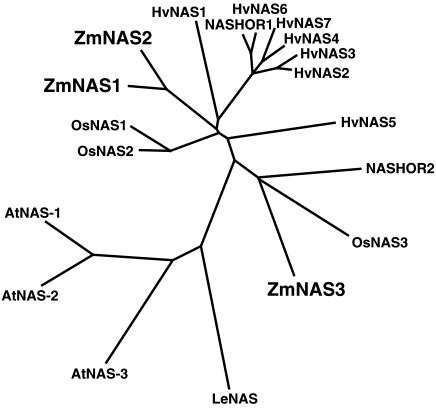
The unrooted phylogenic tree (Fig. 8) suggests that NASs in graminaceous plants are also classified into two groups as are ZmNAS proteins. OsNAS3 and NASHOR2 have a longer amino-terminal region and are closely related to ZmNAS3 in the unrooted phylogenic tree. In fact, OsNAS3 was expressed in leaves under Fe-sufficient conditions and was suppressed by Fe deficiency, as is ZmNAS3 (H. Inoue, personal communication). Presumably, NASHOR2 has similar characteristics to ZmNAS3 and OsNAS3 and may show a similar expression pattern in barley. Seven of nine HvNAS genes were isolated from an Fe-deficient root cDNA library (Higuchi et al., 1999a), and NASHOR2 was isolated from a seed-specific cDNA library (Herbik et al., 1999). These facts also support the idea that NASHOR2 is expressed in Fe-sufficient barley leaves and is suppressed by Fe deficiency.
Figure 8.
The unrooted phylogenic tree for NAS amino acid sequences for which open reading frames are confirmed. ZmNAS1 to 3 are from maize; HvNAS1 to 7 and NASHOR1 and 2 from barley; OsNAS1 to 3 from rice; AtNAS1 to 3 from Arabidopsis; and CLN from tomato. Calculations were performed using the CLUSTAL W neighbor-joining method and the tree was visualized with TreeView. Accession numbers of genes are: AB061270 (ZmNAS1), AB061271 (ZmNAS2), AB042551 (ZmNAS3), AB010086 (HvNAS1), AB011265 (HvNAS2), AB011264 (HvNAS3), AB011266 (HvNAS4), AB011268 (HvNAS5), AB011269 (HvNAS6), AB019525 (HvNAS7), AF136941 (NASHOR1), AF136942 (NASHOR2), AB021746 (OsNAS1), AB023818 (OsNAS2), AB023819 (OsNAS3), AB021934 (AtNAS1), AB021935 (AtNAS2), AB021936 (AtNAS3), and AJ242045 (CLN).
Inactive ZmNAS2 in Spite of the Duplicated Structure
HvNAS1 showed NAS activity in vitro as a MBP fusion (MBP-HvNAS1; Fig. 6). Three MBP fusions of Arabidopsis NAS proteins (AtNAS1, AtNAS2, and AtNAS3) were also active in vitro (Suzuki et al., 1999). These results suggest that other MBP-NAS fusions should also exhibit NAS activity when produced in E. coli, supported by the finding that MBP-ZmNAS1 and MBP-ZmNAS3 were as active in vitro as MBP-HvNAS1. In contrast, MBP-ZmNAS2 was completely inactive under the same conditions (Fig. 6), despite the fact that ZmNAS2 contains partially duplicated NAS sequences. Båga et al. (2000) described a starch-branching enzyme I in wheat (Triticum aestivum) that contains partially repeated starch-branching enzyme I sequences and exhibits branching-enzyme activity. NAS catalyzes the complicated reaction in which three SAM molecules are conjugated into one NA molecule in a single step (Fig. 1B; Higuchi et al., 1999b). Therefore, it is conceivable that highly accurate protein conformation is required for adequate NAS activity. The chloronerva mutant harbors a single base mutation resulting in the change of one amino acid that is otherwise conserved among all other NAS family members. This mutation in the NAS protein strongly diminishes NAS activity in this mutant (Ling et al., 1999). Although ZmNAS2 possesses this conserved region in both of the NAS repeats, it has no NAS activity in vitro as a MBP fusion. The first NAS repeat in ZmNAS2 has one amino acid difference, L205I, in another highly conserved region, and this may prohibit NAS activity by this protein.
As described earlier, NAS is responsible for tolerance to low Fe availability in graminaceous plants. Many genes encoding NAS proteins have been isolated from graminaceous plants: nine from barley (Herbik et al., 1999; Higuchi et al., 1999b), three from rice (Higuchi et al., 2001), and three from maize, as reported in this study. In addition, we have identified many NAS-like expressed sequence tag clones from other graminaceous plants in public databases, including those for wheat, Sorghum bicolor, and winter rye. These data suggest that graminaceous plants, including maize, have increased the number of NAS genes, thus enhancing their tolerance of Fe deficiency. However, in the case of maize, it is possible that ZmNAS2 remains a fused gene because the two genes were not able to separate completely. Higuchi et al. (1999b) reported the presence of a NAS-like protein similar to ZmNAS2 in molecular mass and pI value in Fe-deficient S. bicolor roots. Such a large NAS-like protein has been detected only in maize and S. bicolor (subfamily Panicoideae). The failed NAS duplication event that produced ZmNAS2 thus probably occurred before maize and S. bicolor diverged.
It is possible that ZmNAS2 has some biological function. It is conceivable that a proteolytic cleavage takes place, producing an active enzyme when needed. We detected an uncloned NAS isoform (ZmNAS5) in Fe-deficient maize roots, possibly arising from the cleavage of ZmNAS2. In the absence of this cleavage, it is possible that the long polypeptide manages to titrate the substrate and lead to some kind of regulation.
A recent study showed that transgenic rice containing NAAT genes from barley is more tolerant than nontransgenic individuals to low Fe availability in calcareous soil (Takahashi et al., 2001). By introducing NAS genes such as HvNAS1, along with other genes involved in MAs synthesis (Fig. 1), we can expect to produce a transgenic maize that tolerates Fe deficiency by increasing the amount of secreted MAs. Increasing the productivity of maize in calcareous soils will help meet the increased demand for food that is expected owing to rapid population increases, increased biomass production for fuels, and reduce CO2 concentration by covering calcareous soils.
MATERIALS AND METHODS
Plant and Culture Conditions
Seeds of maize (Zea mays cv Alice) were germinated on paper towels soaked with distilled water in a tray covered with a sheet of aluminum foil for 5 d at 27°C. After germination, plants were transferred to a polythene net floating on standard culture solution (pH 5.5) and were placed in a growth chamber under conditions of 30°C/16 h of light and 25°C/8 h of dark. The standard culture solution consisted of 2 × 10–3 m Ca(NO3)2, 7 × 10–4 m K2SO4, 1 × 10–4 m KCl, 1 × 10–4 m KH2PO4, 5 × 10–4 m MgSO4, 1 × 10–5 m H3BO3, 5 × 10–7 m MnSO4, 5 × 10–7 m ZnSO4, 2 × 10–8 m CuSO4, 1 × 10–8 m (NH4)6Mo7O24, and 1.5 × 10–4 m Fe-EDTA. After 4 d, plants were transplanted into 20-liter plastic boxes and were cultured for 5 d in standard culture solution with adequate amounts of Fe. Plants were then kept in standard culture solution without Fe for 9 d. Finally, Fe was added to the culture solution in the form of 6.0 × 10–4 m Fe-EDTA. For preparing the Fe-excess plants, 1.0 × 10–3 m Fe-EDTA was added to the culture solution and plants were harvested after 10 d. Culture solutions were prepared with distilled water, adjusted to pH 5.5 every day using 1.0 m HCl, and renewed every 5 d. Roots and leaves were harvested at various stages, frozen in liquid nitrogen, and stored at –80°C until use.
Cloning of ZmNAS3 from a Maize Genomic Library
To isolate an NAS homolog, a genomic library (Corn Genomic Library, variety B73; CLONTECH) was screened using plaque hybridization techniques and a probe consisting of the open reading frame of the rice (Oryza sativa) NAS gene (OsNAS1; accession no. AB021746). The probe was labeled with [α-32P]dATP using a Random Primer Labeling kit (version 2; TaKaRa, Kusatsu, Japan) and was purified using a microcolumn (ProbeQuant G-50; Pharmacia, Uppsala). Of approximately 400,000 plaques, one positive clone was plaque purified and its insert was subcloned into the pBluescriptII (SK-) vector. This clone was designated ZmNAS3 (accession no. AB042551).
Screening a Maize cDNA Library
Total RNA was extracted from Fe-deficient maize roots using the SDS-phenol method. A cDNA library was constructed using the cloning vector pSPORT1 (Invitrogen, Carlsbad, CA) as described previously (Higuchi et al., 2001). Approximately 400,000 colonies of the cDNA library were screened for other NAS clones using colony hybridization with a probe consisting of a 0.7-kbp PstI/SalI fragment from ZmNAS3. Isolated cDNA clones were sequenced using a Thermo Sequenase Cycle Sequencing kit (Shimadzu, Kyoto) and a DNA sequencer (DSQ-2000L; Shimadzu).
Northern-Blot Analysis
Northern-blot analyses were conducted using specific probes for ZmNAS1, ZmNAS2, and ZmNAS3 labeled with [32P-]dATP. Specific primers for each gene were designed as follows and were used for preparing specific probes: ZmNAS1, 5′-GAGGAGATGGCGACCACGACAG-3′ and 5′-GAAGTGCATGAGAAATTCAGCA-3′; ZmNAS2, 5′-ATCGACGCCCATGCAAACAC-3′ and 5′-ATCCTCTGCGCGTCCGTGCC-3′; and ZmNAS3, 5′-GCCATGGCCGTCATGGGCAA-3′ and 5′-ATCTTGGCACCAGCGCCGACT-3′. Total RNA was extracted from maize roots and leaves using the SDS-phenol method. Ten micrograms of total RNA was separated, blotted, and hybridized with the probes at 42°C as described previously (Higuchi et al., 1999b).
Preparation of Polyclonal Antibodies to NAS (Higuchi et al., 1999a)
Two mice were immunized with a total of 100 μg of NAS peptides prepared from Fe-deficient barley (Hordeum vulgare) roots. For the first injection, the immunogen was emulsified in complete Freund's adjuvant. For the second and subsequent infections, incomplete Freund's adjuvant was used. After the fourth induction, whole blood was collected and the antiserum was stored at –80°C until use.
Western-Blot Analysis
Proteins were extracted from Fe-deficient maize roots, separated using two-dimensional PAGE, and blotted as described previously (Higuchi et al., 1999a). Western-blot analysis was performed using polyclonal NAS antibodies prepared by Higuchi et al. (1999a), along with a secondary antibody, goat anti-mouse IgG (H+L) conjugate and horseradish peroxidase (Wako, Osaka). The blot was stained with diaminobenzidine.
Expression of Recombinant ZmNAS Proteins in Escherichia coli
To subclone ZmNAS1 into pMAL-c2 (New England Biolabs, Beverly, MA), an annealed oligomer (5′-CCATGCGGAATTCCG-3′) was inserted into the NcoI site (including the first ATG) of ZmNAS1 cloned in pSPORT. This ZmNAS1 plasmid was then digested with EcoRI and HindIII, and the excised fragment containing the ZmNAS1 coding sequence was subcloned into pMAL-c2. To subclone ZmNAS2 into pMAL-c2, an EcoRI site was introduced close to the first ATG using PCR mutagenesis, using two primers: 5′-GAGACTCTGAATTCGCCATGGAGGCCCAGAACGTGGA-3′ and 5′-GAGACTCTAAGCTTCATATGAGTTCCATGCATCAGATGGACA-3′. The amplified fragments were digested with EcoRI and HindIII, and the excised fragment was cloned into pBluescriptII (SK-). This clone was designated ZmNAS2B. We exchanged the region between EcoRI and SmaI of ZmNAS2 in pSPORT with the EcoRI/SmaI ZmNAS2B fragment. The resulting ZmNAS2 construct was digested with EcoRI and HindIII, and the excised fragment containing the ZmNAS2 coding sequence was subcloned into pMAL-c2. To subclone ZmNAS3 into pMAL-c2, an EcoRI site was introduced close to the first ATG using PCR mutagenesis, using two primers: 5′-GAGACTCTGAATTCCATATGGCCGTCATGGGCAAGGAG-3′and 5′-GAGACTCTGGATCCCACTTAATACAATCACGGTGAC-3′. The amplified fragments were digested with EcoRI and BamHI, and the excised fragment was cloned into pBluescriptII (SK-). The resulting ZmNAS3 construct was digested with EcoRI and XbaI, and the excised fragment containing the ZmNAS3 coding sequence was subcloned into pMAL-c2. These pMAL-c2 plasmids, containing the coding sequences of ZmNAS, were introduced into E. coli XL1-Blue, which was induced to produce the recombinant fusion proteins. These proteins were purified as described previously (Higuchi et al., 1999b).
Assay of NAS Activity
NAS enzyme activity was assayed using the cell-free system modified by Higuchi et al. (1994) and Suzuki et al. (1999). One microgram of each fusion protein was added to the reaction buffer [50 mm Tris-HCl, 1 mm EDTA, 3 mm dithiothreitol, 10 μm (p-amidinophenyl) methanesulfonyl fluoride (p-APSMF), and 10 μm trans-epoxysuccinyl-l-leucylamido(4-guanidino) butane (E-64), pH 8.7] and was concentrated by ultrafiltration using a filter (Ultrafree C3LGC NMWL10000; Millipore, Bedford, MA). [14C]SAM was added to the concentrated enzyme solution to a final concentration of 20 μm. After a 20-min incubation at 25°C, 5 m HCl was added to a final concentration of 0.2 m to stop the enzyme reaction. [14C]NA was separated on thin-layer chromatography LK6 plates (Whatman, Clifton, NJ), which were developed with a phenol:n-butanol:formate:water solution (12:3:2:3, v/v). [14C]NA was then detected using an image analyzer (BAS2000; Fuji Film, Tokyo). Standard [14C] NA was synthesized from [14C]SAM using crude extracts of NAS prepared from Fe-deficient barley roots.
Construction of Plasmid ZmNAS-sGFP
Plasmid pUC18 containing the construct, CaMV 35S promoter-sGFP (S65T)-NOS3′, was kindly provided by Dr. Yasuo Niwa (University of Shizuoka, Shizuoka, Japan). The construct had SalI and NcoI sites on the 3′ side of the CaMV 35S promoter. The NcoI site “CCATGG” included the initiation codon for sGFP. An annealed oligomer (5′-TCGAGGGCCC-3′) was inserted into the SalI site of CaMV35S-sGFP (S65T)-NOS3′ to produce ApaI site “GGGCCC.” This modified plasmid was designated CaMV35S-ApaI-sGFP (S65T)-NOS3′. The open reading frame of ZmNAS1 was amplified using two primers: 5′-GAGACTCTGGGCCCATGGAGGCCCAGAACGTGGA-3′ and 5′-GAGACTCTGAATTCATGAAGATGGACAGCTCTGTCGTGG-3′. The amplified fragments were digested with ApaI and EcoRI, and the excised fragment was cloned into pBluescriptII (SK-). The resulting ZmNAS1 construct was digested with ApaI and BspHI, and the excised fragment containing the ZmNAS1 coding sequence was subcloned into CaMV35S-ApaI-sGFP (S65T)-NOS3′ digested with ApaI and NcoI. The open reading frame of ZmNAS2 was amplified using two primers: 5′-GAGACTCTGGGCCCATGGAGGCCCAGAACGTGG A-3′ and 5′-GAGACTCTGAATTCATGAACGCCGTTGCCATCTCCTCCCT-3′. The amplified fragments were subcloned into CaMV35S-ApaI-sGFP (S65T)-NOS3′ in the same way.
Two annealed oligomers, 5′-TCGAACCATGGAGCAGCTGCCGTCCGG-3′ and 5′-CATGCCGGACGGCAGCTGCTCCATGGT-3′, which encoded five N-terminal amino acid residues of ZmNAS3, were inserted into the SalI/NcoI site of CaMV35S-sGFP(S65T)-NOS3′. The NcoI fragment excised from ZmNAS3 was inserted in to the NcoI site of the resulting plasmid.
Transformation and Microscopic Observation of Onion (Allium cepa) Epidermal Cells
Transformation of onion epidermal cells was carried out by the Biolistic PDS-1000/He Particle Delivery System (Bio-Rad, Hercules, CA). White onion bulbs were purchased locally and were stored in the dark until they were used. Inner epidermal layers were peeled and placed inside up on a Murashige and Skoog plate solidified with 2% (w/v) Gellan Gum. Gold particles with a diameter of 1.0 μm were coated with each plasmid DNA containing the different constructs and were prepared for bombardment according to the manufacturer's protocol. Plated onion epidermal layers were placed under the stopping screen at a distance of 8 cm and were bombarded in a vacuum of 28 inches of mercury using a helium pressure of 1,350 psi to accelerate the macrocarrier. Bombarded cells were kept in the dark at 28°C for 20 h. Bombarded onion cells were mounted on a slide glass, and fluorescence viewed with a laser-scanning confocal microscope (LSM510; Karl Zeiss, Jena, Germany) equipped with an argon laser and a GFP filter set.
Acknowledgments
We thank Dr. E. Yoshimura, Dr. P. Blamey for assistance with English expression, and an anonymous reviewer for a suggestion regarding the possible function of ZmNAS2.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.019869.
References
- Båga M, Nair RB, Repellin A, Scoles GJ, Chibbar RN (2000) Isolation of a cDNA encoding a granule-bound 152-kilodalton starch-branching enzyme in wheat. Plant Physiol 124: 253–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker R, Fritz E, Manteuffel R (1995) Subcellular localization and characterization of excessive iron in the nicotianamine-less tomato mutant chloronerva. Plant Physiol 108: 269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker R, Grün M, Scholz G (1992) Nicotianamine and the distribution of iron into the apoplasm and symplasm of tomato (Lycopersicon esculentum Mill.): determination of the apoplasmic and symplasmic iron pools in roots and leaves of the cultivar Bonner Beste and its nicotianamine-less mutant chloronerva. Planta 187: 48–52 [DOI] [PubMed] [Google Scholar]
- Böhme H, Scholz G (1960) Versuche zur Normalisierung des Phänotyps Mutante Chloronerva von Lycopersicon esculentum Mill. Kulturpflanze 8: 93–109 [Google Scholar]
- Briat J-F, Lobréaux S (1997) Iron transport and storage in plants. Trends Plant Sci 2: 187–193 [Google Scholar]
- Budesinsky M, Budzikiewicz H, Prochazka Z, Ripperger H, Römer A, Scholz G, Schreiber K (1980) Nicotianamine, a possible phytosiderophore of general occurrence. Photochemistry 19: 2295–2297 [Google Scholar]
- Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat J-F, Walker EL (2001) Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409: 346–349 [DOI] [PubMed] [Google Scholar]
- Guerinot ML, Yi Y (1994) Iron: nutritious, noxious, and not ready available. Plant Physiol 104: 815–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbik A, Koch G, Mock H-P, Dushkov D, Czihal A, Thielmann J, Stephan UW, Bäumlein H (1999) Isolation, characterization and cDNA cloning of nicotianamine synthase from barley. Eur J Biochem 265: 231–239 [DOI] [PubMed] [Google Scholar]
- Higuchi K, Kanazawa K, Nishizawa NK, Chino M, Mori S (1994) Purification an characterization of nicotianamine synthase from Fe deficient barley roots. Plant Soil 165: 173–179 [Google Scholar]
- Higuchi K, Kanazawa K, Nishizawa N-K, Mori S (1996a) The role of nicotianamine synthase in response to Fe nutrition status in Gramineae. Plant Soil 178: 171–177 [Google Scholar]
- Higuchi K, Nakanishi H, Suzuki K, Nishizawa NK, Mori S (1999a) Presence of nicotianamine synthase isozymes and their homologues in the root of graminaceous plants. Soil Sci Plant Nutr 45: 681–691 [Google Scholar]
- Higuchi K, Nishizawa N, Römheld V, Marschner H, Mori S (1996b) Absence of nicotianamine synthase activity in the tomato mutant “Chloronerva.” J Plant Nutr 19: 1235–1239 [Google Scholar]
- Higuchi K, Nishizawa NK, Yamaguchi H, Römheld V, Marschner H, Mori S (1995) Response of nicotianamine synthase activity to Fe-deficiency in tobacco plants as compared with barley. J Exp Bot 46: 1061–1063 [Google Scholar]
- Higuchi K, Suzuki K, Nakanishi H, Yamaguchi H, Nishizawa NK, Mori S (1999b) Cloning of nicotianamine synthase genes involved in the biosynthesis of phytosiderophores. Plant Physiol 119: 471–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi K, Watanabe S, Takahashi M, Kawasaki S, Nakanishi H, Nishizawa NK, Mori S (2001) Nicotianamine synthase gene expression differs in barley and rice under Fe-deficient conditions. Plant J 25: 159–167 [DOI] [PubMed] [Google Scholar]
- Kawai S, Itoh K, Takagi S, Iwashita T, Nomoto K (1988) Studies on phytosiderophore: biosynthesis of mugineic acid and 2′-deoxymugineic acid in Hordeum vulgare L. var Minorimugi. Tetrahedron Lett 29: 1053–1056 [Google Scholar]
- Kobayashi T, Nakanishi H, Takahashi M, Kawasaki S, Nishizawa N-K, Mori S (2001) In vivo evidence that Ids3 from Hordeum vulgare encodes a dioxygenase that converts 2′-deoxymugineic acid to mugineic acid in transgenic rice. Planta 864–871 [DOI] [PubMed]
- Ling H-Q, Koch G, Bäumlein H, Ganal MW (1999) Map-based cloning of chloronerva, a gene involved in iron uptake of higher plants encoding nicotianamine synthase. Proc Natl Acad Sci USA 96: 7098–7103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle CM, Jolley VD (1991) Iron deficiency stress response of various C-3 and C-4 grain crop genotypes: strategy II mechanism evaluated. J Plant Nutr 14: 341–361 [Google Scholar]
- Ma JF, Taketa S, Chang Y-C, Iwashita T, Matsumoto H, Takeda K, Nomoto K (1999) Genes controlling hydroxylations of phytosiderophores are located on different chromosomes in barley (Hordeum vulgare L.). Planta 207: 590–596 [Google Scholar]
- Marschner H, Römheld V, Kissel M (1986) Different strategies in higher plants in mobilization and uptake of iron. J Plant Nutr 9: 695–713 [Google Scholar]
- Mihashi S, Mori S (1989) Characterization of mugineic acid-Fe transporter in Fe-deficient barley roots using the multicompartment transporter box method. Biol Metals 2: 146–154 [Google Scholar]
- Mori S, Hachisuka M, Kawai S, Takagi S, Nishizawa NK (1988) Peptides related to phytosiderophore secretion by Fe-deficient barley roots. J Plant Nutr 11: 653–662 [Google Scholar]
- Mori S, Nishizawa N (1987) Methionine as a dominant precursor of phytosiderophores in Graminaceae plants. Plant Cell Physiol 28: 1081–1092 [Google Scholar]
- Mori S, Nishizawa N, Kawai Y, Takagi S (1987) Dynamic state of mugineic acid and analogous phytosiderophores in Fe deficient barley. J Plant Nutr 10: 1003–1011 [Google Scholar]
- Nakanishi H, Yamaguchi H, Sasakuma T, Nishizawa NK, Mori S (2000) Two dioxygenase genes, Ids3 and Ids2, from Hordeum vulgare are involved in the biosynthesis of mugineic acid family phytosiderophores. Plant Mol Biol 44: 199–207 [DOI] [PubMed] [Google Scholar]
- Nishizawa N, Mori S (1987) The particular vesicle appearing in barley root cells and its relation to mugineic acid secretion. J Plant Nutr 10: 1012–1020 [Google Scholar]
- Pich A, Manteuffel R, Hillmer S, Scholz G, Schmidt W (2001) Fe homeostasis in plant cells: Does nicotianamine play multiple roles in the regulation of cytoplasmic Fe concentration? Planta 213: 967–976 [DOI] [PubMed] [Google Scholar]
- Pich A, Scholz G, Stephan UW (1994) Iron-dependent changes of heavy metals, nicotianamine, and citrate in different plant organs and in the xylem exudate of two tomato genotypes: nicotianamine as possible copper translocator. Plant Soil 165: 189–196 [Google Scholar]
- Römheld V (1987) Different strategies for iron acquisition in higher plants. Plant Physiol 70: 231–234 [Google Scholar]
- Scholz G, Becker R, Pich A, Stephan UW (1992) Nicotianamine: a common constituent of strategies I and II of iron acquisition by plants: a review. J Plant Nutr 15: 1647–1665 [Google Scholar]
- Scholz G, Schlesier G, Seifert K (1985) Effect of nicotianamine on iron uptake by the tomato mutant chloronerva. Physiol Planta 63: 99–104 [Google Scholar]
- Shojima S, Nishizawa NK, Fushiya S, Nozoe S, Irifune T, Mori S (1990) Biosynthesis of phytosiderophores: in vitro biosynthesis of 2′-deoxymugineic acid from l-methionine and nicotianamine Plant Physiol 93: 1497–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Chino M, Nishizawa NK, Mori S (1993) Genotypic variation among Indian graminaceous species with respect to phytosiderophore secretion. In PJ Randall, E Delhaize, RA Richards, R Munns, eds, Genetic Aspects of Plant Mineral Nutrition. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 335–339
- Stephan UW, Grün M (1989) Physiological disorders of the nicotianamineauxotroph tomato mutant chloronerva at different levels of iron nutrition: iron deficiency response and heavy metal metabolism Biochem Physiol Pflanz 185: 189–200 [Google Scholar]
- Stephan UW, Schmidke I, Pich A (1994) Phloem translocation of Fe, Cu, Mn, and Zn in Ricinus seedlings in relation to the concentrations of nicotianamine, an endogenous chelator of divalent metal ions, in different seedling parts. Plant Soil 165: 181 [Google Scholar]
- Stephan UW, Scholz G (1993) Nicotianamine: mediator of transport of iron and heavy metals in the phloem? Physiol Planta 88: 522–529 [Google Scholar]
- Suzuki K, Higuchi K, Nakanishi H, Nishizawa NK, Mori S (1999) Cloning of nicotianamine synthase genes from Arabidopsis thaliana. Soil Sci Plant Nutr 45: 993–1002 [Google Scholar]
- Takagi S (1976) Naturally occurring iron-chelating compounds in oat- and rice-root washings: activity measurement and preliminary characterization. Soil Sci Plant Nutr 22: 423–433 [Google Scholar]
- Takahashi M, Nakanishi H, Kawasaki S, Nishizawa NK, Mori S (2001) Enhanced tolerance of rice to low iron availability in alkaline soils using barley nicotianamine aminotransferase genes. Nat Biotechnol 19: 466–469 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Yamaguchi H, Nakanishi H, Shioiri T, Nishizawa N-K, Mori S (1999) Cloning two genes for nicotianamine aminotransferase, a critical enzyme in iron acquisition (Strategy II) in Graminaceous plants. Plant Physiol 121: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wirén N, Klair S, Bansal S, Briat J-F, Khodr H, Shioiri T, Leigh RA, Hider RC (1999) Nicotianamine chelates both FeIII and FeII: implications for metal transport in plants. Plant Physiol 119: 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker EL (2002) Functional analysis of the Arabidopsis Yellow Stripe-Like (YSL) family: heavy metal transport and partitioning via metal-nicotianamine (NA) complexes. Plant Physiol 129: 431–432 [Google Scholar]