Abstract
The phytohormone cytokinin is an important regulator of plant growth and development; however, relatively few genes that mediate cytokinin responses have been identified. Genome-wide analyses of Arabidopsis seedlings using the approximately 8,300-element Affymetrix Arabidopsis GeneChips (Affymetrix, Santa Clara, CA) to examine cytokinin-responsive genes were conducted, revealing at least 30 genes whose steady-state level of mRNA was elevated and at least 40 that were down-regulated at multiple time points after application of cytokinin. The cytokinin up-regulated genes include the type-A Arabidopsis response regulators (ARRs), which had been shown previously to be cytokinin primary response genes, cytokinin oxidase, which encodes an enzyme that degrades cytokinins, and several transcription factors. Cytokinin down-regulated genes include several peroxidases and kinases and an E3 ubiquitin ligase. We identified a common sequence motif enriched in the upstream regions of the most consistently cytokinin up-regulated genes. This motif is highly similar to the optimal DNA-binding sites for ARR1/ARR2, type-B ARRs that have been implicated in the transcriptional elevation of the type-A ARRs. Additionally, genome-wide analyses of cytokinin receptor mutants (wol/cre1) revealed large-scale changes in gene expression, including down-regulation of the type-A ARRs and several meristem and cell cycle genes, such as CycD3. Mutations in CRE1 reduced but did not eliminate the effect of cytokinin on gene expression for a subset of cytokinin-responsive genes and had little or no effect on others, suggesting functional redundancy among the cytokinin receptors.
Cytokinins are a group of adenine derivatives that affect multiple aspects of plant growth and development, including cell division, vascular development, sink/source relationships, apical dominance, and leaf senescence (Binns, 1994; Mok and Mok, 1994, 2001). A pathway for cytokinin biosynthesis and metabolism is emerging from molecular and biochemical studies. This has been highlighted by the recent cloning of several genes encoding enzymes involved in cytokinin biosynthesis or metabolism, including ipt (Kakimoto, 2001; Takei et al., 2001), which catalyzes the first committed step in cytokinin biosynthesis; cytokinin oxidase (Houba-Hérin et al., 1999; Morris et al., 1999), which cleaves the N6 side chain from cytokinins; and several enzymes that catalyze the conjugation of sugar moieties to cytokinins (Martin et al., 1999a, 1999b, 2001). A model for cytokinin perception and signal transduction has emerged that is similar to prokaryotic two-component response pathways (Haberer and Kieber, 2001; Hutchison and Kieber, 2002; Lohrmann and Harter, 2002). A family of genes that are similar to bacterial two-component response regulators, the type-A Arabidopsis response regulators (ARRs), was identified as cytokinin primary response genes (Brandstatter and Kieber, 1998; Sakakibara et al., 1998; Taniguchi et al., 1998; D'Agostino et al., 2000). The cytokinin receptors (CRE1, AHK2, and AHK3) mediate the induction of these type-A ARRs and are similar to bacterial two-component His kinases (Inoue et al., 2001; Ueguchi et al., 2001; Yamada et al., 2001). These receptors act through other two-component elements, including Arabidopsis homologs of His phosphotransfer proteins (AHPs) and the type-B ARRs, involved in the up-regulation of type-A ARRs (Hwang and Sheen, 2001; Sakai et al., 2001).
CRE1 (also called WOL and AHK4) is the best characterized of the three cytokinin His kinase receptors. The signal transduction function of this gene in single-cell systems such as Escherichia coli, yeast (Saccharomyces cerevisiae), and transient Arabidopsis protoplasts has been shown to be dependent on the exogenous application of cytokinin. Furthermore, loss-of-function mutants of CRE1 result in reduced cytokinin sensitivity (Inoue et al., 2001; Ueguchi et al., 2001). Although the interaction between CRE1 and the other cytokinin receptors, AHK2 and AHK3, is unclear, examination of CRE1 mutants has revealed clues regarding its in vivo functions. The cre1-1 mutation was found to be allelic to the wol mutant previously identified in genetic screens for altered root patterning. The severely reduced root growth phenotype of the wol mutants is likely to be due to an insufficient number of vascular initial cells, resulting in a lack of phloem tissue (Scheres et al., 1995; Mähönen et al., 2000). CRE1 is also required for the periclinal division that increases the number of cell files in the vasculature of the root and hypocotyl. CRE1 gene expression is highest in roots and is localized in the vascular precursor cells of globular stage embryos, the hypocotyl procambium, the cotyledon shoulders, and the embryonic root (Mähönen et al., 2000). The wol mutation is a missense allele that disrupts the cytokinin binding domain of the protein; thus, the phenotype of wol appears to be the result of impaired cytokinin perception in tissues where CRE1 is expressed.
Several researchers have examined gene expression in response to application of exogenous cytokinin (Crowell and Amasino, 1994). In one study, 20 genes were identified as induced within 4 h of cytokinin treatment, including two genes encoding ribosomal proteins and a β-expansin (Crowell et al., 1990). Exogenous cytokinin has also been shown to up-regulate the cell cycle genes cdc2 and CycD3 (Menges et al., 2002). Additionally, both STM and KNAT1 genes, which are involved in meristem function, have elevated expression in Arabidopsis plants overexpressing the bacterial ipt gene (Rupp et al., 1999). Recently, a receptor-like kinase gene, CRK1, was found to be rapidly down-regulated by cytokinin (Schäfer and Schmülling, 2002). Several studies have also examined cytokinin-regulated gene expression during tissue differentiation including nodulation, senescence, floral development, lateral bud induction, and various aspects of light development (Mok and Mok, 2001). Genes identified in these studies include photosynthetic genes, ribosomal protein genes, nitrate reductase, and many novel genes. However, most of these genes are also induced by other stimuli, most notably light and auxin, and none of these genes are induced with kinetics, suggestive of an immediate early response.
In this study, we sought to identify genes regulated by cytokinin to better understand the mode of action of this hormone. Additionally, we examined the role of the cytokinin receptor CRE1 in mediating the effect of cytokinin on gene expression in Arabidopsis roots.
RESULTS
Regulation of Gene Expression by Cytokinin
Light-grown Arabidopsis seedlings were treated with exogenous cytokinin for various times over a 24-h period, and gene expression was analyzed using the approximately 8,300-element Affymetrix Arabidopsis GeneChips (Affymetrix, Santa Clara, CA). Ten cytokinin treatments were conducted and normalized to control samples prepared and treated identically with dimethyl sulfoxide (DMSO; the solvent used for the cytokinin). For our analyses, we chose a 2-fold change in expression level compared with the control as a minimum for a gene to be called altered in response to cytokinin and a minimum raw value cutoff of 175 to 500, depending on the particular chip hybridization. These raw level cutoffs were assigned based on scatter plot analyses of a treated sample versus its control and used because expression levels of genes below these cutoff thresholds frequently showed variability greater than 2-fold and often had Affymetrix flag calls of Absent (data not shown). In addition, only the cytokinin-treated samples with an Affymetrix flag call of Present and the minimum raw cutoff level were used to identify up-regulated genes; likewise, only the control samples with the same restrictions were used to identify down-regulated genes. By using these restrictions, we sought to distinguish authentic changes in expression from background noise and false positives. Using these parameters, the steady-state levels of expression of more than 1,000 genes were altered by cytokinin alone in at least one treatment; however, the majority of these were affected at only a single treatment and generally not in replicate samples (see Supplemental Tables I and II at http://www.plantphysiol.org).
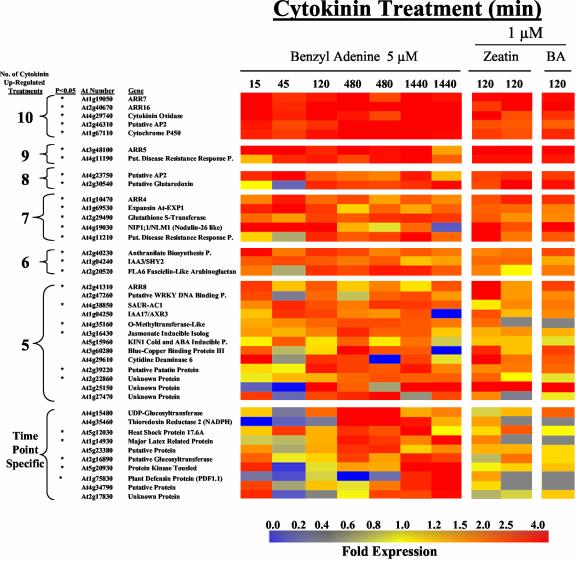
Thirty genes were found to be up-regulated at five or more of the 10 cytokinin time course treatments, and 17 were up-regulated in at least six independent treatments (Fig. 1A). The observation that these genes were reproducibly up-regulated by cytokinin suggests that they are cytokinin responsive. A Welch's t test confirmed that all of the top 17 genes showed significant changes in expression levels with cytokinin treatment. We chose to include other genes that fit the criteria described above for cytokinin regulation but that showed a P value ≥ 0.05 because these are still potential cytokinin-regulated genes. These results are further supported by the fact that four of the top 17 genes, ARR4, ARR5, ARR7, and ARR16, have been identified previously as cytokinin-responsive type-A ARRs (D'Agostino et al., 2000). Other reproducibly identified cytokinin up-regulated genes include AP2 transcription factors, a P450, and two putative disease resistance response proteins that are novel in their link to cytokinin (Fig. 1A).
Figure 1.
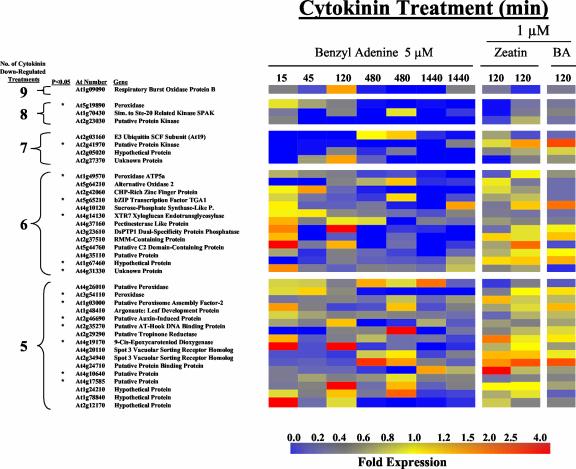
Cytokinin-regulated genes. Expression ratios of genes from wild-type-Arabidopsis seedlings treated under various conditions (compared in all cases with normal media grown seedlings treated with a DMSO control for an identical time) are shown as colored squares. The colors correspond to the relative fold change from the respective controls using the scale shown at the bottom of the figure. Genes with expression levels found to be altered by cytokinin more than 2.0-fold at five or more of the 10 cytokinin time course treatments or at specific time points with a 2.5-fold change and a minimum raw level are shown. The Arabidopsis thaliana (At) number and gene description are indicated to the left. The asterisk indicates that a gene is significantly different (P < 0.05) due to cytokinin treatment relative to the DMSO control by Welch's t test comparisons (see “Materials and Methods”). Treatments shown from right to left are with 5 μm benzyl adenine (BA) for 15, 45, 120, and two replicates of 480 and 1,440 min, 1 μm zeatin for two replicates of 120 min, and 1 μm BA for 120 min. A, Genes whose expression increased more than 2-fold relative to their cognate DMSO control. B, Genes whose expression decreased more than 2-fold relative to their cognate DMSO control. P. refers to Protein.
Approximately 40 genes were identified that showed greater than a 2-fold decrease in their expression level at five or more of the 10 treatments after cytokinin application (Fig. 1B). One group of genes that showed reductions in their steady-state mRNA levels was a set of genes involved with oxidation, including four peroxidases and two oxidases. Another group of down-regulated genes was three kinases, including an Ste-20 and SNF1-related kinase. Other reproducibly identified cytokinin down-regulated genes include an ubiquitin ligase SCF complex subunit, three transcription factors, and the leaf development protein Argonaute (Fig. 1B). In comparison with the set of induced genes, which contains only two unknowns, the cytokinin-repressed genes include more than one-quarter (11 genes) for which there is no known function (Fig. 1A,Fig. 1B).
A number of distinct induction profiles can be identified from this time course (Fig. 1A,Fig. 1B). There is a set of genes that is elevated or reduced at all time points examined. This includes most of the type-A ARRs, the cytokinin oxidase, the AP2s, and the P450 from the up-regulated list, and the respiratory burst oxidase and the three kinases from the down-regulated list. There is only a small set of genes that are induced rapidly (15, 45, and 120 min), but transiently (return to baseline at 480 and 1,440 min), such as the nodulin-26-like gene NIP1;1, whereas no down-regulated genes appear to fit this pattern. There is a larger set of genes that are up- or down-regulated only upon prolonged cytokinin treatment (>120 min). For the up-regulated genes, these include a glutaredoxin, a disease resistance response protein, and an unknown protein. For the down-regulated genes, these include a zinc finger protein and three of the four down-regulated peroxidases. Another class of genes displays alterations in gene expression at specific time points. Most of these genes are specifically induced or repressed at a single time point, such as a putative peroxidase that is primarily repressed at 120 min by both BA and zeatin cytokinin treatments, as seen in Figure 1B.
Our experimental design using both BA and zeatin treatments allowed us to examine the effect of these different classes of cytokinins on gene expression. At 120 min, seedlings were treated with either BA, a cytokinin with an aromatic-type side chain attached to the N6 position of the adenine ring, or trans-zeatin, a naturally occurring cytokinin with an isoprenoid-type side chain. Almost every gene found to be up-regulated by BA at both 120-min treatments was also up-regulated in at least one of the two zeatin treatments and vice versa. In contrast, there are many fewer genes down-regulated by zeatin as compared with BA, which suggests that some of these genes may be specifically down-regulated by BA or that they are not actually regulated by cytokinin.
It is important to note that for the genes that have altered expression levels in fewer numbers of cytokinin treatments, there is a decreased confidence that any particular gene is truly regulated by cytokinin. This is reflected in the statistical analysis of the data, which indicates that some but not all of the genes that we identified as cytokinin regulated were found to have significant P values. The genes identified as altered in both replicate treatments for a single time point are potentially transiently cytokinin-regulated genes and are more likely to be truly altered by cytokinin than those affected at only two distinct time points, but not in the replicates. Thus, we have included genes affected in both replicates of any single time point above a 2.5-fold change in expression in Figure 1A,Figure 1B. Nevertheless, it is possible that some of the genes identified as cytokinin regulated (Fig. 1A) are false positives, and it is likely that we have excluded genes that are weakly induced for a narrow window of time along with genes whose expression levels are relatively low. In summary, the set of genes presented in Figure 1A,Figure 1B represents the list of genes found in this study to most likely be regulated by cytokinin, with a decreasing confidence in this conclusion as a gene is identified as altered in fewer cytokinin-treated samples.
Type-B ARR-Binding Sites Are Enriched in the Upstream Regions of Cytokinin-Induced Genes
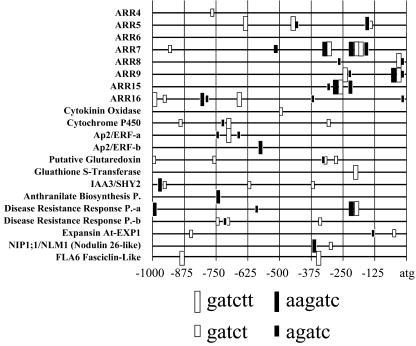
We used GeneSpring 5.0 software (Silicon Genetics, Redwood City, CA) to search for common motifs enriched within 1 kb upstream of the translational start site of the 17 genes that are consistently up-regulated by cytokinin (those genes induced in six or more of the cytokinin treatments as in Fig. 1A). The sequence GATCTT was identified as a core sequence motif found significantly enriched in the upstream regions of this set of genes. The relative positions of this motif upstream of the type-A ARRs and the other cytokinin-regulated genes are shown in Figure 2. White boxes represent this common upstream sequence motif GATCTT on the forward strand, and black boxes represent the complement of the motif AAGATC. Upstream regions of the cytokinin-regulated type-A ARRs showed the highest concentration of these motifs, generally clustering in the 500 bp proximal to the coding region. The sequence of this common upstream sequence motif closely matches (G/A)GAT(T/C), the optimal DNA-binding sequence identified in vitro for ARR1 and ARR2, two type-B ARRs (Sakai et al., 2000). Gel shift assays with various derivatives of the optimal type-B ARR-binding site NGAT(T/C) showed a slight preference for A at position 1 and a C at position 5, consistent with part of the common sequence motif we identified (Sakai et al., 2000).
Figure 2.
Putative cis-acting motifs in the promoters of cytokinin-regulated genes. The DNA sequences 1,000 bp upstream of the predicted translational start site of the 17 genes identified as cytokinin up-regulated at six or more of the 10 cytokinin time course treatments were analyzed for cis-acting sequence motifs that were significantly enriched in this region, representing potential sites of cytokinin regulation. The positions where the core sequence motif (GATCT) occurs are depicted as white boxes (black boxes represent the complement sequence), and the extended sequence (GATCTT) is depicted as a large white box, with its complement as a large black box.
The frequency of this common upstream sequence motif in the upstream regions of the type-A ARRs is positively correlated with the level of their induction by cytokinin. The most highly cytokinin-induced genes, ARR5, ARR7, and ARR16, have multiple copies of this motif. Additionally, genes that are either only weakly induced by cytokinin or not expressed at detectable levels, ARR3, ARR4, and ARR17, have one or no motifs (Fig. 2; data not shown). ARR6, which is moderately induced by cytokinin, does not fit this pattern because it has no consensus motif in its upstream region, although two such motifs are found within the fourth intron of this gene. The common upstream sequence motif (GATCTT) should randomly occur approximately 0.32 times per kilobase pair, taking into account the GC content observed in intergenic regions of Arabidopsis (approximately 33%). Our search of all genes present on the Affymetrix GeneChip that are expressed with a raw of at least 500 but not induced by cytokinin (950 genes) revealed that the sequence motif GATCTT is present a total of 358 times in the 1 kb upstream of the translational start site, or approximately 0.38 times per kilobase pair. In contrast, this core sequence motif is present 14 times among the upstream regions of the top 17 cytokinin up-regulated genes and only six times among the upstream regions of the 21 cytokinin down-regulated genes. This enrichment, coupled with the similarity to the previously defined ARR1/ARR2-binding sequence, suggests that this motif may play a role in the transcriptional regulation of these genes by cytokinin.
Effect of Cytokinin on Arabidopsis Two-Component Elements
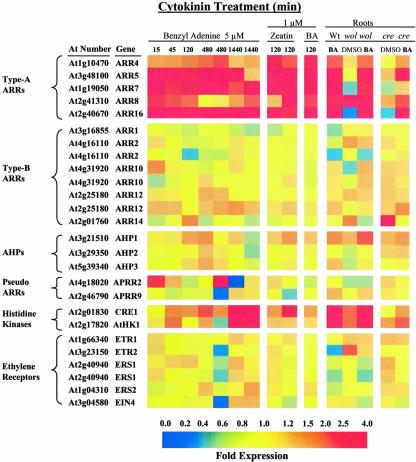
Previous studies have implicated Arabidopsis homologs of two-component signaling elements in the cytokinin response pathway. We analyzed the expression pattern of these genes under our experimental conditions (Fig. 3; see Supplemental Table III). As noted above, the five type-A ARR genes present on the chip were up-regulated at most of the cytokinin treatments. The three AHP genes on the chip are not affected by cytokinin, nor are the five type-B ARRs, consistent with previous results suggesting that the steady-state mRNA level of these classes of two-component genes are unaffected by cytokinin treatment (Imamura et al., 1998; Kiba et al., 1999; Lohrmann et al., 1999). The two pseudoresponse regulators, which lack invariant response regulator residues, including the Asp residue that is the target of phosphorylation, and have been implicated in circadian control (Matsushika et al., 2000; Strayer et al., 2000), are also not elevated in response to cytokinin. Interestingly, the cytokinin receptor CRE1 is moderately up-regulated by cytokinin over 24 h, suggesting that application of cytokinin may alter the sensitivity of seedlings to this hormone. The His kinase homolog AHK1, which lacks a CHASE cytokinin-binding domain and has been suggested to play a role in osmosensing (Urao et al., 2000), is also up-regulated by cytokinin. The expression of the ethylene receptor genes was not affected cytokinin.
Figure 3.
Regulation of two-component response regulator genes by cytokinin and other treatments. The expression ratios of two-component-like genes are depicted for the various samples relative to their cognate controls as described in Figure 1A,Figure 1B. The colors correspond to the relative fold change from the respective controls using the scale shown at the bottom of the figure. Genes shown are grouped into classes based on their similarity to different two-component elements, with their At number and gene description shown on the left. Cytokinin treatments shown from right to left are with 5 μm BA for 15, 45, 120, and two replicates of 480 and 1,440 min, 1 μm zeatin for two replicates of 120 min, 1 μm BA for 120 min, and root tissue of wild type (Landsburg erecta [Ler]) treated with 5 μm BA for 45 min; wol mutant roots and cre1-1 mutant roots treated with DMSO control or 5 μm BA for 45 min. All root tissue treatments were normalized to the wild-type root with 45 min of DMSO treatment.
Effect of CRE1 Mutations on Cytokinin-Regulated Gene Expression
We examined the effects that loss-of-function mutations of the cytokinin receptor CRE1 had on the pattern of gene expression by analyzing RNA isolated from the roots of two different cre1 mutant alleles, wol and cre1-1. Root tissue was examined because these mutations have been reported to principally affect root development (Scheres et al., 1995; Mähönen et al., 2000). The wol and cre1 mutations have dramatic effects on gene expression: Approximately 600 genes are elevated and 400 down-regulated in both wol treatments (+DMSO and +BA) relative to wild-type roots (Table I; data not shown). The observation that expression levels of these genes are decreased in both treatments of the wol background increases the confidence that many are genuine alterations in gene expression caused by this mutation. The large-scale changes in gene expression may largely be a secondary consequence of the wol mutation, caused by the lack of phloem tissue in the mutant roots. Consistent with this, several transporter genes, including an aquaporin, and a proline and two sugar transporters, are up-regulated in this mutant, potentially compensating for the reduced ability to uptake nutrients through the phloem (Table I). In addition, the cre1-1 allele, which has a much weaker root phenotype as compared with the wol allele, has a reduced effect on gene expression (approximately 150 genes are elevated and 75 genes down-regulated in both cre1-1 treatments compared with the larger numbers found in wol as described above).
Table 1.
wol- and cre1-regulated genes
Genes altered at least 2-fold for both wol and cre1 roots treated with DMSO are shown below by At no. and gene description. The expression level of each gene in wol and cre1 treated with 5 μm BA. All values are normalized to wild-type roots treated with DMSO.
|
Wol/cre1 Up-Regulated Genes
|
Wol/cre1 Down-Regulated Genes
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| At no. | Gene |
wol
|
cre1
|
At no. | Gene |
wol
|
cre1
|
||||
| DMSO | +BA | DMSO | +BA | DMSO | +BA | DMSO | +BA | ||||
| At2g46670 | Transcription factor ATHB-7 | 4.7 | 11.8 | 2.4 | 2.7 | At1g70210 | Cyclin Delta-1 | 0.1 | 0.2 | 0.3 | 0.3 |
| At2g28160 | Putative bHLH transcription factor | 2.6 | 0.8 | 2.3 | 1.0 | At4g34160 | Cyclin Delta-3 | 0.3 | 0.2 | 0.5 | 0.6 |
| At1g43160 | RAP2.6 AP2 protein | 2.3 | 1.0 | 3.2 | 1.5 | At4g20270 | CLV1 receptor kinase like | 0.1 | 0.2 | 0.5 | 0.7 |
| At2g38470 | WRKY DNA-binding protein | 4.6 | 5.7 | 2.0 | 1.4 | At4g20270 | CLV1 receptor kinase like | 0.2 | 0.3 | 0.5 | 0.7 |
| At3g46130 | R2R3-MYB transcription factor | 6.3 | 5.0 | 2.3 | 1.2 | At1g75820 | Receptor kinase | 0.2 | 0.1 | 0.4 | 0.6 |
| At1g67110 | Cytochrome P450 | 5.1 | 7.2 | 2.7 | 3.6 | At2g44940 | AP2 transcription factor | 0.4 | 0.2 | 0.4 | 0.8 |
| At4g40010 | Putative Ser/Thr kinase | 2.2 | 3.5 | 3.0 | 3.5 | At5g38430 | Rubisco | 0.4 | 0.3 | 0.3 | 0.4 |
| At2g26980 | Putative protein kinase | 2.5 | 5.5 | 2.2 | 1.8 | At4g10340 | Lhcb5 protein | 0.4 | 0.5 | 0.4 | 0.5 |
| At1g61590 | Putative protein kinase | 3.1 | 2.3 | 2.4 | 1.1 | At1g61520 | Chlorophyll a/b binding | 0.5 | 0.8 | 0.3 | 0.7 |
| At2g16750 | Putative protein kinase | 4.2 | 6.0 | 2.1 | 1.5 | At1g55670 | PSI subunit V precursor | 0.4 | 0.7 | 0.3 | 0.5 |
| At1g79570 | Putative protein kinase | 2.0 | 1.9 | 2.0 | 1.6 | At2g30570 | PSII reaction center protein | 0.4 | 0.8 | 0.4 | 0.7 |
| At1g72770 | PP2C | 2.0 | 5.4 | 2.4 | 2.1 | At2g30570 | PSII reaction center protein | 0.4 | 0.5 | 0.4 | 0.6 |
| At2g13640 | Putative nucleotide sugar transporter | 2.1 | 2.1 | 2.7 | 1.3 | At3g21055 | PSII protein | 0.3 | 0.4 | 0.5 | 0.6 |
| At1g05030 | Putative sugar transporter | 2.1 | 2.3 | 2.0 | 1.2 | At1g20340 | Putative plastocyanin | 0.5 | 0.4 | 0.3 | 0.5 |
| At2g16990 | Putative tetracycline transporter | 2.7 | 2.8 | 2.2 | 1.3 | At5g13930 | Chalcone synthase | 0.5 | 0.4 | 0.5 | 0.9 |
| At2g36830 | Putative aquaporin | 4.2 | 4.5 | 2.0 | 1.6 | At4g16250 | Phytochrome D | 0.5 | 3.7 | 0.1 | 0.1 |
| At3g47730 | ABC transporter | 2.0 | 1.9 | 2.4 | 2.1 | At4g29080 | Phytochrome-associated protein 2 | 0.2 | 0.1 | 0.4 | 0.5 |
| At2g36590 | Putative Pro transporter | 2.0 | 1.2 | 2.0 | 1.8 | At2g34490 | Cytochrome P450 | 0.1 | 0.1 | 0.4 | 0.5 |
| At2g47160 | Putative anion exchange protein | 2.6 | 3.0 | 2.3 | 1.6 | At4g37430 | Cytochrome P450 | 0.5 | 0.5 | 0.4 | 0.5 |
| At1g30760 | Putative berberine bridge enzyme | 18.2 | 16.5 | 2.1 | 1.7 | At4g39510 | Cytochrome P450 | 0.1 | 0.6 | 0.3 | 0.4 |
| At2g21270 | Ubiquitin protein | 2.8 | 2.5 | 3.3 | 2.0 | At1g22710 | SUC2 Suc-proton symporter | 0.1 | 0.1 | 0.5 | 0.7 |
| At2g20750 | Beta-expansin | 13.8 | 11.7 | 3.1 | 1.7 | At2g38530 | Nonspecific lipid transfer protein | 0.3 | 0.3 | 0.3 | 0.3 |
| At2g40100 | Lhcb4:3 protein | 4.4 | 9.6 | 10.1 | 7.2 | At2g38530 | Nonspecific lipid transfer protein | 0.2 | 0.2 | 0.2 | 0.2 |
| At5g24140 | Squalene epoxidase homolog | 9.4 | 7.2 | 5.4 | 2.5 | At2g38540 | Nonspecific lipid transfer protein | 0.1 | 0.1 | 0.4 | 0.4 |
| At1g54580 | Acyl carrier protein | 7.4 | 5.1 | 2.4 | 1.5 | At2g36870 | Xyloglucan endo-transglycosylase | 0.2 | 0.2 | 0.4 | 1.0 |
| At4g13050 | Oleoyl hydrolase | 2.6 | 2.2 | 2.6 | 1.9 | At4g30270 | Endo-xyloglucan transferase | 0.3 | 0.2 | 0.3 | 0.2 |
| At1g23020 | Potential FROHC | 5.8 | 5.8 | 2.1 | 1.4 | At2g28950 | Expansin At-EXP6 | 0.3 | 0.2 | 0.4 | 0.5 |
| At2g47550 | Putative pectinesterase | 5.4 | 3.1 | 2.5 | 1.4 | At1g20620 | Catalase 3 | 0.3 | 0.3 | 0.4 | 0.3 |
| At3g47400 | Pectinesterase-like protein | 2.2 | 1.6 | 2.5 | 2.5 | At3g12500 | Basic endochitinase | 0.1 | 0.1 | 0.5 | 0.5 |
| At3g54430 | Class IV chitinase | 4.1 | 3.0 | 2.1 | 1.1 | At4g04840 | Putative Met sulfoxide reductase | 0.1 | 0.1 | 0.3 | 0.7 |
| At2g04430 | mutT domain protein | 3.0 | 2.3 | 7.3 | 0.7 | At4g27570 | UDP Rha | 0.1 | 0.1 | 0.4 | 0.3 |
| At2g26930 | Putative ripening-associated protein | 2.9 | 3.8 | 2.1 | 1.6 | At2g20340 | Tyr decarboxylase | 0.3 | 0.6 | 0.4 | 0.6 |
| At2g39410 | Putative phospholipase | 2.9 | 2.5 | 2.6 | 1.2 | At4g29020 | Gly-rich protein | 0.0 | 0.0 | 0.2 | 0.2 |
| At2g17420 | NADPH thioredoxin reductase | 41.3 | 36.4 | 3.1 | 1.9 | At3g16380 | Pro-rich protein APG isolog | 0.1 | 0.1 | 0.3 | 0.1 |
| At2g02990 | Ribonuclease | 2.4 | 3.7 | 2.4 | 1.5 | At4g39330 | CAD1 | 0.2 | 0.2 | 0.4 | 0.5 |
| At2g04150 | Subtilisin-like protease | 2.0 | 3.1 | 2.6 | 2.2 | At1g78830 | S locus gycoprotein | 0.3 | 0.5 | 0.5 | 0.6 |
| At1g64060 | Atrboh F | 2.2 | 2.1 | 2.1 | 1.5 | At2g32990 | Putative glucanase | 0.0 | 0.4 | 0.4 | 0.6 |
| At2g47490 | Putative mitochondrial carrier | 2.1 | 1.7 | 2.7 | 1.6 | At4g20420 | Putative Ser proteinase | 0.1 | 0.2 | 0.4 | 0.4 |
| At2g04160 | Subtilisin-like protease | 2.1 | 2.6 | 2.0 | 2.0 | At5g25610 | RD22 | 0.1 | 0.4 | 0.3 | 0.3 |
| At4g25070 | Putative protein | 2.0 | 2.2 | 3.0 | 1.5 | At4g33720 | Pathogenesis-related1 precursor | 0.1 | 0.0 | 0.1 | 0.1 |
| At4g36640 | Putative protein | 2.5 | 3.8 | 2.3 | 2.7 | At4g33720 | Pathogenesis-related1 | 0.0 | 0.0 | 0.1 | 0.0 |
| At4g26470 | Putative protein | 2.3 | 2.8 | 2.0 | 1.3 | At5g24780 | Vegetative storage Protein | 0.3 | 0.1 | 0.2 | 0.3 |
| At4g25870 | Putative protein | 4.0 | 3.7 | 2.6 | 0.9 | At2g42840 | Protodermal factor 1 | 0.0 | 0.0 | 0.5 | 0.3 |
| At4g35110 | Putative protein | 3.5 | 3.8 | 2.1 | 1.6 | At4g24360 | Putative protein | 0.0 | 0.0 | 0.1 | 0.1 |
| At3g47380 | Putative protein | 4.0 | 2.3 | 2.3 | 2.4 | At2g47560 | Hypothetical protein | 0.1 | 0.1 | 0.4 | 0.4 |
| At2g23890 | Hypothetical protein | 2.3 | 1.7 | 3.0 | 2.7 | At2g33850 | Unknown protein | 0.0 | 0.0 | 0.3 | 0.4 |
| At2g39690 | Hypothetical protein | 2.1 | 1.8 | 2.9 | 1.8 | At2g39710 | Unknown protein | 0.4 | 0.2 | 0.5 | 0.7 |
| At4g16350 | Hypothetical protein | 2.2 | 2.7 | 2.7 | 1.9 | At2g16850 | Unknown protein | 0.2 | 0.3 | 0.5 | 0.6 |
| At1g24310 | Hypothetical protein | 3.8 | 3.7 | 2.6 | 1.8 | At1g67740 | Unknown protein | 0.4 | 0.4 | 0.4 | 0.5 |
| At2g14530 | Hypothetical protein | 3.7 | 2.4 | 2.5 | 1.0 | At2g44670 | Unknown protein | 0.1 | 0.3 | 0.4 | 0.4 |
| At2g37210 | Hypothetical protein | 3.5 | 7.1 | 3.3 | 2.8 | ||||||
| At2g12550 | Hypothetical protein | 2.6 | 2.4 | 4.0 | 1.9 | ||||||
| At2g48080 | Unknown protein | 2.6 | 1.5 | 2.6 | 1.2 | ||||||
| At4g10970 | Unknown protein | 2.6 | 4.5 | 2.4 | 2.7 | ||||||
| At1g05170 | Unknown protein | 3.9 | 3.9 | 2.8 | 1.6 | ||||||
| At2g20320 | Unknown protein | 2.1 | 1.7 | 2.6 | 1.3 | ||||||
| At2g01650 | Unknown protein | 2.1 | 2.7 | 2.2 | 1.9 | ||||||
| At1g30900 | Unknown protein | 2.7 | 2.2 | 2.6 | 2.1 | ||||||
| At5g49440 | Unknown protein | 161.5 | 161.0 | 2.3 | 1.0 | ||||||
| At2g33220 | Unknown protein | 18.3 | 16.2 | 6.0 | 6.9 | ||||||
Of the genes that are down-regulated in both wol and cre1-1, one group of particular interest is those involved in regulation of cell division and meristem function, consistent with the primary defect in root development of this mutant. Interestingly, the cell cycle regulators CycD1 and CycD3 are down-regulated in both wol and cre1-1 relative to wild-type roots, independent of cytokinin treatment, and the cdc2 gene is down-regulated specifically in wol (see Supplemental Table IV). As in the shoots, we do not observe an induction of either of these genes in response to cytokinin in contrast to previous reports (Riou-Khamlichi et al., 1999; see Supplemental Table IV). The down-regulation of these cell cycle genes is consistent with the reduced meristem function of wol and cre1-1 mutants and could either be a cause (i.e. the lack of expression of the genes due to a reduced cytokinin response leads to reduced cell division in the vasculature precursor cells) or a consequence (the reduced number of dividing cells in the mutant leads to the lower expression of these cell cycle genes). Interestingly, a CLV1 receptor kinase-like gene is also down-regulated in both alleles under all treatments compared with wild type (Table I). CLV1 plays a role in regulating meristem size in the shoot apical meristem, and, perhaps, this homolog may play a similar role in the root meristem.
The type-A ARR genes downstream of the cytokinin receptor CRE1 were still induced by cytokinin in the wol and cre1-1 mutants, although the fold increase relative to untreated mutant roots was not always to the same level observed in wild-type roots (Fig. 3). Basal expression levels of the type-A ARR and several others of the top cytokinin-inducible genes were also lower in these cytokinin receptor mutants (Table II), as previously observed in the cre1-1 mutant for two type-A genes, ARR15 and ARR16 (Kiba et al., 2002). These CRE1 mutations reduced but did not eliminate the effect of exogenous cytokinin on gene expression on a subset of genes and had little or no effect on the expression of others. This suggests that other cytokinin receptors (e.g. AHK2 and AHK3) can largely compensate for the loss of CRE1 under conditions of high exogenous cytokinin, but the response to endogenous cytokinin levels is partially compromised.
Table II.
Cytokinin-regulated gene Expression in wol and cre1
The expression levels of cytokinin-regulated genes (from Fig. 1A,Fig. 1B) in either wild type (Ler) or wol- or cre1-treated roots are shown below either treated with DMSO or 5 μm BA for 45 min. All values are normalized to wild-type roots treated with DMSO.
| Cytokinin Up-Regulated Genes
|
Cytokinin Down-Regulated Genes
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| At no.
|
Gene
|
Ler
|
wol
|
cre1
|
At no.
|
Gene
|
Ler
|
wol
|
cre1
|
||||
| +BA | DMSO | +BA | DMSO | +BA | +BA | DMSO | +BA | DMSO | +BA | ||||
| At1g19050 | ARR7 | 5.6 | 0.5 | 2.8 | 0.6 | 1.4 | At1g09090 | Respiratory burst oxidase protein | 0.7 | 1.4 | 1.1 | 1.1 | 0.5 |
| At2g40670 | ARR16 | 20.6 | 0.4 | 4.4 | 0.5 | 3.7 | At5g19890 | Peroxidase | 0.4 | 1.4 | 1.1 | 1.5 | 0.8 |
| At4g29740 | Cytokinin oxidase | 8.2 | 30.5 | 88.4 | 0.8 | 1.8 | At1g70430 | Ste-20-related kinase SPAK | 0.5 | 1.8 | 2.6 | 1.2 | 1.6 |
| At2g46310 | Putative AP2 | 1.7 | 0.9 | 2.0 | 0.6 | 2.3 | At2g23030 | Putative protein kinase | 0.9 | 6.3 | 16.1 | 1.7 | 0.7 |
| At1g67110 | Cytochrome P450 | 6.0 | 5.1 | 7.2 | 2.7 | 3.6 | At2g03160 | E3 ubiquitin SCF subunit (At19) | 1.1 | 1.0 | 1.2 | 0.7 | 0.8 |
| At3g48100 | ARR5 | 4.0 | 0.8 | 6.0 | 1.4 | 5.5 | At2g41970 | Putative protein kinase | 0.7 | 0.9 | 0.8 | 2.1 | 1.7 |
| At4g11190 | Putative disease resistance response P. | 2.0 | 0.1 | 0.1 | 0.9 | 2.3 | At2g05020 | Hypothetical protein | 0.7 | 0.8 | 3.5 | 0.5 | 0.9 |
| At4g23750 | Putative AP2 | 0.7 | 1.2 | 1.2 | 0.9 | 0.9 | At2g27370 | Unknown protein | 0.8 | 0.9 | 1.0 | 1.9 | 1.2 |
| At2g30540 | Putative glutaredoxin | 5.3 | 0.1 | 1.3 | 0.5 | 1.0 | At1g49570 | Peroxidase ATP5a | 0.5 | 5.1 | 5.4 | 1.2 | 0.5 |
| At1g10470 | ARR4 | 2.4 | 1.2 | 2.3 | 1.0 | 1.8 | At5g64210 | Alternative oxidase 2 | 0.3 | 1.5 | 2.0 | 1.8 | 1.4 |
| At1g69530 | Expansin At-EXP1 | 1.7 | 1.9 | 1.4 | 1.2 | 1.2 | At2g42060 | CHP-rich zinc finger protein | 0.4 | 3.5 | 2.3 | 1.4 | 0.5 |
| At2g29490 | Glutathione S-transferase | 3.2 | 1.9 | 5.8 | 1.2 | 2.5 | At5g65210 | bZIP transcription factor TGA1 | 0.7 | 1.5 | 1.8 | 1.3 | 0.8 |
| At4g19030 | NIP1;1/NLM1 (nodulin-26 like) | 3.4 | 1.4 | 2.5 | 0.8 | 1.5 | At4g10120 | Suc-phosphate synthase like | 0.3 | 0.8 | 0.4 | 0.7 | 0.1 |
| At4g11210 | Putative disease resistance response P. | 3.8 | 0.1 | 0.1 | 3.0 | 4.2 | At4g14130 | Xyloglucan endotransglycosylase | 0.6 | 0.2 | 0.2 | 0.5 | 0.4 |
| At2g40230 | Anthranilate biosynthesis P. | 1.2 | 1.2 | 1.2 | 0.6 | 1.0 | At4g37160 | Pectinesterase-like protein | 0.9 | 1.2 | 1.0 | 1.3 | 1.2 |
| At1g04240 | IAA3/SHY2 | 4.9 | 0.1 | 0.5 | 0.6 | 1.9 | At3g23610 | Dual-specificity P. phosphatase | 0.3 | 1.5 | 2.5 | 4.0 | 5.8 |
| At2g20520 | FLA6 fasciclin-like arabinoglactan | 1.7 | 0.6 | 0.4 | 5.5 | 4.7 | At2g37510 | RMM-containing protein | 1.0 | 0.7 | 0.6 | 1.4 | 1.2 |
| At2g41310 | ARR8 | 3.1 | 5.1 | 8.8 | 1.6 | 2.5 | At5g44760 | Putative C2 domain-containing protein | 4.3 | 4.6 | 3.8 | 1.7 | 0.2 |
| At2g47260 | Putative WRKY DNA binding P. | 2.1 | 0.7 | 2.1 | 0.6 | 0.9 | At4g35110 | Putative protein | 1.6 | 3.5 | 3.8 | 2.1 | 1.6 |
| At4g38850 | SAUR-AC1 | 1.8 | 3.0 | 7.2 | 0.4 | 0.2 | At1g67460 | Hypothetical protein | 2.1 | 3.8 | 3.5 | 1.6 | 0.3 |
| At1g04250 | IAA17/AXR3 | 1.8 | 0.6 | 1.0 | 1.1 | 1.8 | At4g31330 | Unknown protein | 0.6 | 0.8 | 0.6 | 0.8 | 0.7 |
| At4g35160 | O-methyltransferase like | 2.5 | 0.3 | 0.4 | 0.9 | 1.4 | At4g26010 | Putative peroxidase | 0.4 | 0.4 | 0.2 | 1.5 | 1.4 |
| At3g16430 | Jasmonate-inducible isolog | 1.3 | 0.6 | 0.8 | 1.2 | 1.2 | At3g54110 | Peroxidase | 0.6 | 0.7 | 0.4 | 1.8 | 1.1 |
| At5g15960 | KIN1 cold- and abscisic acid-inducible P. | 0.8 | 0.8 | 1.0 | 1.7 | 1.4 | At1g03000 | Putative peroxisome assembly factor-2 | 0.9 | 0.8 | 0.8 | 1.7 | 1.1 |
| At3g60280 | Blue copper-binding protein III | 1.6 | 1.1 | 0.9 | 2.4 | 1.9 | At1g48410 | Argonaute | 0.5 | 2.9 | 5.7 | 1.6 | 1.6 |
| At4g29610 | Cytidine deaminase 6 | 1.2 | 0.1 | 0.4 | 0.3 | 0.3 | At2g46690 | Putative auxin-induced protein | 0.8 | 1.0 | 0.9 | 0.9 | 0.8 |
| At2g39220 | Putative patatin protein | 9.1 | 0.6 | 0.4 | 0.4 | 0.3 | At2g35270 | Putative AT-hook DNA-binding protein | 0.3 | 0.8 | 0.3 | 1.0 | 0.5 |
| At2g22860 | Unknown protein | 1.4 | 5.0 | 5.2 | 1.2 | 0.8 | At2g29290 | Putative tropinone reductase | 2.8 | 5.3 | 1.0 | 0.9 | 0.6 |
| At2g25150 | Unknown protein | 2.2 | 0.1 | 1.2 | 0.7 | 2.7 | At4g19170 | 9-Cis-epoxycaroteniod dioxygenase | 0.6 | 1.6 | 4.1 | 3.6 | 2.0 |
| At1g27470 | Unknown protein | 8.5 | 12.5 | 21.3 | 5.7 | 4.2 | At4g20110 | Spot 3 vacuolar-sorting receptor | 0.6 | 2.2 | 2.4 | 1.5 | 1.1 |
| At4g15480 | UDP-glucosyltransferase | 1.5 | 1.4 | 1.1 | 0.6 | 1.2 | At2g34940 | Spot 3 vacuolar-sorting receptor | 0.9 | 1.4 | 1.5 | 1.3 | 0.9 |
| At4g35460 | Thioredoxin reductase 2 (NADPH) | 0.9 | 1.2 | 1.7 | 1.0 | 1.2 | At4g24710 | Putative protein-binding protein | 1.6 | 1.2 | 0.3 | 2.0 | 1.0 |
| At5g12030 | Heat shock protein 17.6A | 0.4 | 1.4 | 1.6 | 1.0 | 1.5 | At4g10640 | Putative protein | 1.1 | 1.0 | 1.6 | 1.5 | 1.2 |
| At1g14930 | Major latex-related protein | 0.7 | 0.8 | 1.1 | 1.1 | 1.0 | At4g17585 | Putative protein | 1.6 | 1.2 | 1.3 | 0.8 | 1.3 |
| At5g23380 | Putative protein | 1.1 | 0.6 | 1.3 | 0.7 | 0.8 | At1g24210 | Hypothetical protein | 2.4 | 3.3 | 2.6 | 0.4 | 0.6 |
| At2g16890 | Putative glucosyltransferase | 1.5 | 0.3 | 0.5 | 0.9 | 0.9 | At1g78840 | Hypothetical protein | 1.2 | 0.8 | 1.0 | 0.9 | 0.8 |
| At5g20930 | Protein kinase tousled | 1.4 | 1.7 | 1.9 | 1.3 | 1.0 | At2g12170 | Hypothetical Protein | 0.3 | 0.2 | 0.2 | 1.3 | 0.5 |
| At1g75830 | Plant defensin protein (PDF1.1) | 0.9 | 4.0 | 10.4 | 0.5 | 2.6 | |||||||
| At4g34790 | Putative protein | 0.5 | 1.6 | 1.3 | 0.4 | 0.2 | |||||||
| At2g17830 | Unknown protein | 1.3 | 1.2 | 2.2 | 0.4 | 0.6 | |||||||
Effect of Inhibition of Protein Synthesis on Cytokinin-Regulated Gene Expression
Generally, genes that are involved in the primary response to a signal are still induced by that signal when protein synthesis is blocked. Furthermore, many primary response genes are elevated upon inhibition of protein synthesis, an effect that has been attributed to the rapid degradation of a negative regulator of transcription or the involvement of a short-lived RNase in the degradation of those transcripts. The effect of inhibition of protein synthesis on cytokinin up-regulated gene expression was examined in seedlings pretreated with cycloheximide, in the presence or absence of cytokinin (Table III).
Table III.
Cytokinin-regulated gene expression in the presence of cycloheximide
The expression levels of cytokinin-regulated genes (from Figure 1A,Figure 1B) are shown below for seedlings in the presence of 50 μm cycloheximide either treated with DMSO or 5 μm BA for 45 min. All values are normalized to wild-type seedlings treated only with DMSO for 45 min.
| Cytokinin Up-Regulated Genes
| |||
|---|---|---|---|
| At no.
|
Gene
|
Cyclohexamide
|
|
| DMSO | +BA | ||
| At1g19050 | ARR7 | 16.4 | 20.8 |
| At2g40670 | ARR16 | 7.2 | 39.3 |
| At4g29740 | Cytokinin oxidase | 19.4 | 47.1 |
| At2g46310 | Putative AP2 | 7.2 | 10.7 |
| At1g67110 | Cytochrome P450 | 1.4 | 3.6 |
| At3g48100 | ARR5 | 9.6 | 14.0 |
| At4g11190 | Putative disease resistance response P. | 0.8 | 1.3 |
| At4g23750 | Putative AP2 | 7.1 | 8.5 |
| At2g30540 | Putative glutaredoxin | 2.2 | 7.0 |
| At1g10470 | ARR4 | 2.0 | 2.4 |
| At1g69530 | Expansin At-EXP1 | 3.0 | 4.8 |
| At2g29490 | Glutathione S-Transferase | 4.2 | 5.5 |
| At4g19030 | NIP1; 1/NLM1 (nodulin-26 like) | 0.0 | 1.8 |
| At4g11210 | Putative disease resistance response P. | 1.0 | 2.4 |
| At2g40230 | Anthranilate biosynthesis P. | 1.8 | 4.9 |
| At1g04240 | IAA3/SHY2 | 20.8 | 23.7 |
| At2g20520 | FLA6 fasciclin-like arabinoglactan | 0.6 | 0.2 |
| At2g41310 | ARR8 | 7.6 | 11.7 |
| At2g47260 | Putative WRKY DNA-binding P. | 8.7 | 11.1 |
| At4g38850 | SAUR-AC1 | 5.3 | 8.2 |
| At1g04250 | IAA17/AXR3 | 1.4 | 2.2 |
| At4g35160 | O-methyltransferase like | 2.0 | 2.4 |
| At3g16430 | Jasmonate-inducible isolog | 1.8 | 1.7 |
| At5g15960 | KIN1 cold- and abscisic acid-inducible P. | 1.1 | 1.1 |
| At3g60280 | Blue copper-binding protein III | 0.8 | 0.9 |
| At4g29610 | Cytidine deaminase 6 | 19.2 | 42.2 |
| At2g39220 | Putative patatin protein | 5.8 | 8.7 |
| At2g22860 | Unknown protein | 2.7 | 3.1 |
| At2g25150 | Unknown protein | 0.0 | 0.3 |
| At1g27470 | Unknown protein | 3.0 | 1.8 |
| At4g15480 | UDP-glucosyltransferase | 0.5 | 0.9 |
| At4g35460 | Thioredoxin reductase 2 (NADPH) | 1.3 | 1.7 |
| At5g12030 | Heat shock protein 17.6A | 0.9 | 0.9 |
| At1g14930 | Major latex-related protein | 1.0 | 1.1 |
| At5g23380 | Putative protein | 1.5 | 2.4 |
| At2g16890 | Putative glucosyltransferase | 4.5 | 4.5 |
| At5g20930 | Protein kinase tousled | 1.1 | 1.8 |
| At1g75830 | Plant defensin protein (PDF1.1) | 1.0 | 0.9 |
| At4g34790 | Putative protein | 1.5 | 3.2 |
| At2g17830 | Unknown Protein | 4.4 | 3.4 |
A large number (236) of genes was found to be elevated greater than 3-fold in response to cycloheximide (data not shown), including many genes previously shown to be up-regulated by cycloheximide treatment such as indole-3-acetic acids (IAAs; Koshiba et al., 1995), type-A ARRs (Brandstatter and Kieber, 1998), and ACS genes (Liang et al., 1992). Approximately one-half of the cytokinin up-regulated genes showed more than a 2-fold alteration in expression levels by cycloheximide treatment alone (Table III), a common feature of primary response and regulatory genes. An additive response to cytokinin and cycloheximide is observed for many of the cytokinin up-regulated genes, including the type-A ARRs that had been reported previously (D'Agostino et al., 2000). This suggests that the majority of the genes that are rapidly induced by exogenous cytokinin are primary response genes.
DISCUSSION
We have analyzed the effect of exogenous cytokinin on gene expression in Arabidopsis seedlings to identify novel cytokinin-regulated genes and to better understand the role of cytokinin in plant developmental processes. Our analysis revealed over 70 genes whose expression is altered by application of exogenous cytokinin in at least one-half of the treatments examined. The genes that are affected by cytokinin and their kinetics of induction provide some clues as to how seedlings respond to elevated cytokinin levels. One reaction to elevated cytokinin at the transcriptional level appears to be a reduction of both the level of active hormone and the sensitivity of the response pathway. The cytokinin signaling response is desensitized by up-regulation of type-A ARRs, which act as negative regulators of the cytokinin response pathway (J. To, G. Haberer, and J. Kieber, unpublished data). The level of cytokinin is potentially reduced by increased expression of a cytokinin-degrading enzyme, cytokinin oxidase, consistent with previous studies demonstrating that cytokinin oxidase enzyme activity was elevated in response to exogenous cytokinin (Terrine and Laloue, 1980; Palmer and Palni, 1987) and by the induction of a gene encoding a potential cytokinin-conjugating enzyme, a putative glucosyltransferase (At2g16890; Fig. 1A). These feedback responses may explain why we do not observe large secondary transcriptional responses after prolonged exogenous cytokinin treatment. In contrast, expression of the cytokinin receptor, CRE1, is up-regulated over time in response to the hormone, which is similar to the induction of ethylene receptors by ethylene. This slight increase may reflect a potential negative regulator role of CRE1 in cytokinin response because His kinases in other two-component systems can act as both His kinases and phosphatases (Stock et al., 2000).
A second potential pattern that emerges from cytokinin-regulated genes is an effect on redox state. Four peroxidases and two oxidases are down-regulated by cytokinin in multiple treatments, and both a glutathione S-transferase and a glutaredoxin are up-regulated. The down-regulated peroxidase genes are phylogenetically dispersed within the Arabidopsis peroxidase gene family, which includes 73 members (Tognolli et al., 2002). Peroxidases have been implicated in many physiological processes, including hydrogen peroxide detoxification, lignin biosynthesis, and stress responses (Hiraga et al., 2001; Tognolli et al., 2002). IAA oxidase, which catabolizes auxin, is also a peroxidase (Arnison, 1980), and its down-regulation could lead to an alteration in the levels of IAA. This would provide one mechanism for the synergistic effects of these two hormones. Lee (1974) found that treatment of tobacco (Nicotiana tabacum) callus culture with 5 μm cytokinin strikingly reduced the activity of multiple peroxidases, including IAA oxidase activity, although the effect on the IAA oxidase was less pronounced as compared with other peroxidases. Miller (1978, 1979, 1985) noted that cytokinins caused both a rapid promotion and an inhibition of the apparent peroxidations of several compounds, such as coumarate, the effect being dependent on the concentration and timing of application of the hormone. Potentially, auxin levels could be up-regulated to help balance excess cytokinin levels, if the down-regulated peroxidases have any IAA oxidase activity. Interestingly, several auxin-regulated genes, such as IAA3, IAA17, and SAUR AC-1, were elevated by cytokinin treatment.
A number of transcription factors are rapidly up-regulated by cytokinin, including two AP2/ERF family members and a WRKY DNA-binding protein. The two AP2 genes cluster within the same subgroup of the large Arabidopsis ERF/AP2 gene family, and this subgroup as of now has no members with assigned functions (Sakuma et al., 2002). It is possible that they may act in a similar manner to ethylene-responsive ERF genes in closely related clades of the AP2 gene family. In addition, several transcription factors are among the genes that were found to be consistently down-regulated in response to cytokinin treatment (Fig. 1B).
Other genes that we have identified as cytokinin up-regulated reflect previous findings on the effects of cytokinin. Consistent with our identification of a cytokinin up-regulated expansin, cytokinin has been implicated in regulating cell expansion in certain tissues, most notably cotyledons, and a cytokinin up-regulated β-expansin gene has been identified from tobacco (Dowens and Crowell, 1998). Interestingly, there are three protein kinases and a dual-specificity protein phosphatase that are down-regulated by cytokinin and could potentially play a role in cytokinin signal transduction. Three of these genes are more highly down-regulated with extended cytokinin treatment (Fig. 1B), similar to the pattern of another negatively regulated kinase gene identified from tobacco (Schäfer and Schmülling, 2002).
One group of genes (cell cycle and meristem genes) was conspicuously absent from our set of induced genes (Fig. 1A; see Supplemental Table V). The cdc2 protein kinase, CycD3 cyclin, and the STM1 homeobox transcription factor have all been reported previously to be elevated in response to cytokinin (Hemerly et al., 1993; Riou-Khamlichi et al., 1999; Rupp et al., 1999). CycD3 was reported to be induced by cytokinin, based mostly on analysis of suspension cultures. Induction was also observed in whole seedlings, though this was done using liquid-grown seedlings treated for 24 h with zeatin (Riou-Khamlichi et al., 1999), conditions that are sufficiently different from those used for our experiments to potentially account for these contradictory results. Likewise, increases in STM1 expression has been observed only in cytokinin-overproducing plants (Rupp et al., 1999), and cdc2 induction was reported only after 72 h of cytokinin treatment and only in a small subset of root cells (Hemerly et al., 1993). However, we did find several cell cycle genes, including CycD3, that are down-regulated in the cytokinin receptor mutants, suggesting cell cycle genes may be responsive to endogenous cytokinin function. Alternatively, the down-regulation of these cell cycle and meristem genes in wol and cre1-1 may simply reflect the altered tissue composition of the mutants.
Our examination of the temporal expression patterns of genes over our time course of treatments revealed two major patterns. The first is a constant pattern of induction or repression from the first 15-min time point to the 24-h time point, including several of the type-A ARRs, cytokinin oxidase and two AP2s that are constantly induced, and a respiratory burst oxidase and several kinases that are constantly repressed. The second major pattern represented by a small set of genes is one of alteration only after prolonged exposure to cytokinin. This occurs both for induced genes such as a putative disease resistance protein and a glutaredoxin, which are up-regulated after 45 min, and for repressed genes, such as the four peroxidases that do not show down-regulation until after 120 min. While other expression patterns can be identified, they are represented by relatively few genes. Although most rapidly induced genes remain altered at later time points, in many cases, a decrease in the magnitude of the response is observed. This suggests that there is a partial desensitization of the response pathway after prolonged exposure to cytokinin.
We were also able to identify a common upstream sequence motif, GATCTT. The similarity of this motif to the previously identified type-B ARR-binding site suggests that type-B ARR genes may mediate induction of genes in addition to type-A ARRs (Sakai et al., 2000). Further analysis is required to determine the functional significance of this motif in cytokinin regulation.
To better understand the role of the cytokinin receptor CRE1 in mediating cytokinin response, we examined expression profiles of cytokinin treatment in the receptor mutant alleles wol and cre1-1. We found lower baseline expression levels of the type-A ARRs, suggesting that the response to endogenous cytokinin is affected by the loss of this receptor. However, the general trend for gene expression in the presence of cytokinin was a reduction, but not elimination, of the alteration in transcript levels in both receptor mutants. This suggests a redundancy of cytokinin receptor function, probably by the other CHASE domain-containing His kinase homologs, AHK2 and AHK3. There is also another group of genes that show a lack of induction by cytokinin in the receptor mutants or even in some cases a repression in the mutants relative to the wild type. Although some of these genes may be explained by general variation in expression levels, it is possible that for some, the cytokinin signal acts solely through CRE1. Additional analysis of lines disrupted for CRE1 and other cytokinin receptors may shed further light on this.
In conclusion, we have identified through expression profiling a set of cytokinin-regulated genes. Some of these correspond to previously identified cytokinin-regulated genes, and others are novel cytokinin-regulated genes that provide new insight into the mechanism of cytokinin function. More detailed analyses of the function of these novel genes are in progress to better elucidate their role in cytokinin response. Our examination of CRE1 cytokinin receptor mutants suggests that there is redundancy at the level of cytokinin perception, although levels of downstream genes in the absence of exogenous cytokinin are reduced in these receptor mutants. Further analyses of gene expression with full genome arrays and with additional cytokinin mutants should provide additional insight into the function of this hormone.
MATERIALS AND METHODS
Plant Growth
All plants, except for those in the wol/cre1-1 experiments, were planted at a density of approximately 500 seeds per plate (150 mm) using 0.8% (w/v) top agar (0.8% [w/v] low-melt agarose in 1× Murashige and Skoog salts) on filter paper (150 mm, Whatman no. 3, Whatman, Clifton, NJ) placed on 1% (w/v) agar containing 1× Murashige and Skoog salts + 1% (w/v) Suc buffered to pH 5.7 with MES (Murashige and Skoog media). Seedlings were stratified at 4°C for 4 d, then grown horizontally under constant light (90 μm m–2 s–1) at 22°C for 10 d. The seedlings were then gently scraped off the filter paper into flasks containing 500 mL of liquid Murashige and Skoog media and shaken lightly for 2 h in the light before treatment. At time 0, the indicated concentration of cytokinin (1 or 5 μm BA or 1 μm trans-zeatin) was added, or an equal volume of DMSO was added as a vehicle control to the seedlings, with 0.1% (v/v) DMSO in a final volume of Murashige and Skoog. The treatment of seedlings with cycloheximide was conducted in the same manner as with the cytokinin treatments using either a cytokinin (5 μm BA) or DMSO treatment for 45 min, except that seedlings were treated with 50 μm cycloheximide for 30 min before and during the 45-min cytokinin or DMSO treatment. After the appropriate time in all gene chip treatments, seedlings were harvested and frozen at –80°C until RNA was extracted. For all experiments, RNA from two independent biological replicates was pooled before hybridizing to the chip, except for the wol/cre1-1, for which only one replicate was made. For the time course, there was a gene chip for each of the following: 5 μm BA treatment at 15, 45, and 120 min; two treatments at 480 and 1,440 min; a 1 μm BA treatment at 120 min; and two treatments with 1 μm zeatin at 120 min, as shown in Figure 1A,Figure 1B, along with DMSO control gene chips in parallel with each of the above cytokinin gene chip times. Plants in the wol/cre1-1 experiments were treated in a similar manner as above except that these seedlings were grown vertically on agar plates without filter paper from a line of seeds such that the roots could be harvested easily. After growth at the above conditions for 10 d, roots were harvested and immediately moved to flasks of liquid Murashige and Skoog, where they were treated in a similar manner as the seedlings above. wol- and cre1-treated root samples were normalized to two independent biological wild-type root DMSO-treated gene chip samples. Seedlings used in these experiments treated with 5 μm BA and its DMSO control were of the Wassilewskija ecotype, with 1 μm BA and zeatin treatments, and their DMSO control was of the Columbia ecotype. In the wol/cre1-1 experiment, the Ler ecotype was used as both the wol and the cre1-1 mutants were in the Ler background.
RNA Sample and Microarray Preparation
Total RNA was extracted from frozen plant tissue using TRIzol reagent following the manufacturer's instructions (Introgen, Grand Island, NY).Seven micrograms of total RNA was used to synthesize cDNA. A custom cDNA kit (Life Technologies/Gibco-BRL, Gaithersburg, MD) was used with a T7-(dT)24 primer for this reaction. Biotinylated cRNA was then generated from the cDNA reaction using the BioArray High Yield RNA Transcript Kit (Life Technologies/Gibco-BcL, Gaithersburg, MD). The cRNA was then fragmented in fragmentation buffer (5× fragmentation buffer: 200 mm Tris-acetate [pH 8.1], 500 mm KOAc, and 150 mm MgOAc) at 94°C for 35 min before the chip hybridization. Fifteen micrograms of fragmented cRNA was then added to a hybridization cocktail (0.05 μg μL–1 fragmented cRNA; 50 pm control oligonucleotide B2; BioB, BioC, BioD, and cre1-1 hybridization controls; 0.1 mg mL–1 herring sperm DNA; 0.5 mg mL–1 acetylated bovine serum albumin; 100 mm MES; 1 m NaCl; 20 mm EDTA; and 0.01% [v/v] Tween 20), and 10 μg of cRNA was used for each hybridization. Arrays were hybridized for 16 h at 45°C in a GeneChip Hybridization Oven 640. The arrays were washed and stained with R-phycoerythrin streptavidin in the GeneChip Fluidics Station 400. The arrays were subsequently scanned with the Hewlett-Packard GeneArray Scanner (Hewlett-Packard, Palo Alto, CA). Affymetrix GeneChip Microarray Suite 5.0 software was used for washing, scanning, and basic analysis. Sample quality was assessed by examination of 3′ to 5′ intensity ratios of certain genes.
Data Analysis
Affymetrix gene chip data files were imported into GeneSpring 5.0 software and normalized as recommended by the GeneSpring manual for Affymetrix gene chips. Using 50% median normalization, each treatment chip was specifically normalized to a control chip treated with DMSO for a similar time to the actual treatment. Once the data were normalized within GeneSpring, there were analyzed as a ratio to signal in a variety of ways. Data for all experimental treatments are from a single gene chip compared with its respective DMSO time point control. Data for tables and figures were restricted such that values were considered only if genes had positive expression levels for both treated and control microarrays and if the raw value for a gene on a treated microarray was above a raw expression level of 500 or 175 in the DMSO or treated sample when examining down- and up-regulated genes, respectively. A raw level cutoff of 500 was assigned to all gene chip experiments conducted on seedlings with 5 μm BA or the corresponding DMSO control, whereas a raw cutoff of 175 was assigned to all other gene chips experiments. These raw level cutoffs were assigned because expression levels of genes below these cutoff thresholds frequently showed variability greater than 2-fold and often had Affymetrix flag calls of Absent (data not shown).
Statistical analyses were performed on Affymetrix gene chip treatments normalized as described above and examined as a log ratio to have a normally distributed population for parametric analysis. Welch's t tests were performed using the Statistical Group Comparison tool in GeneSpring 5.0 software using the GeneSpring Cross-Gene Error model variances. Comparisons were made between treatments of cytokinin or DMSO either for individual time point treatments or across all time points after a Welch's ANOVA determined that there was no significant effect of the time treatment of samples. A P value cutoff of 0.05 was selected for all tests. In addition, the Benjamini and Hochberg False Discovery Rate Multiple Testing Correction option was applied to the examination of the cytokinin up-regulated genes to eliminate against false positives in the larger number of significant genes identified there.
The search for potential cis-acting regulatory sequence motifs in the cytokinin up-regulated genes was conducted using the Find Potential Regulatory Sequence tool in GeneSpring 5.0 software. We searched using Find New Sequences to look for oligonucleotides 5 to 13 bp in length that were enriched in the open reading frames from 0 to 1,000 bp upstream of the top 17 cytokinin up-regulated genes from Figure 1A compared with other genes in the Arabidopsis genome. The sequences GATCTT, GATCTTA, AGATCTTA, AAGATC, and AAGATCTT were identified as having single P values below 0.05, observed rates 2 to 3 times the expected rate for those sequences. The only sequences also identified using the above criteria were AAAAAGA and AAAGAAAA, which are likely to simply reflect the A/T-rich value of the non-coding region in the Arabidopsis genome.
Supplementary Material
Acknowledgments
We would like to thank Mike Vernon and the University of North Carolina Affymetrix chip facility for their help in labeling and reading the Arabidopsis Affymetrix GeneChips. We also thank the members of the Kieber lab for their helpful comments and suggestions.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.021436.
This work was funded by the National Science Foundation (grant no. DBI–0077503) and by the National Institutes of Health (grant no. GM4421–01).
The online version of this article contains Web-only data. The supplemental material is available at http://www.plantphysiol.org.
References
- Arnison PG (1980) A redox model of the mechanism of action of indole acetic acid (auxin) and other plant growth regulators. Speculations Sci Technol 3: 5–15 [Google Scholar]
- Binns AN (1994) Cytokinin accumulation and action: biochemical, genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol 45: 173–196 [Google Scholar]
- Brandstatter I, Kieber JJ (1998) Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 10: 1009–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell DN, Amasino RM (1994). Cytokinins and plant gene regulation. In DWS Mok, MC Mok, eds, Cytokinins: Chemistry, Activity and Function. CRC Press, Boca Raton, FL, pp 233–242
- Crowell DN, Kadlecek AT, John MC, Amasino RM (1990) Cytokinin-induced mRNAs in cultured soybean cells. Proc Natl Acad Sci USA 87: 8815–8819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agostino I, Deruère J, Kieber JJ (2000) Characterization of the response of the Arabidopsis ARR gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowens BP, Crowell DN (1998) Cytokinin regulates the expression of a soybean β-expansin gene by a post-transcriptional mechanism. Plant Mol Biol 37: 437–444 [DOI] [PubMed] [Google Scholar]
- Haberer G, Kieber JJ (2001) Cytokinins: new insights into a classic phytohormone. Plant Physiol 128: 354–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly AS, Ferreira P, de Almeida Engler J, Van Montagu M, Engler G, Inze D (1993) cdc2a expression in Arabidopsis is linked with competence for cell division. Plant Cell 5: 1711–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S, Sasaki K, Ito H, Ohashi Y, Matsui H (2001) A large family of class III plant peroxidases. Plant Cell Physiol 42: 462–468 [DOI] [PubMed] [Google Scholar]
- Houba-Hérin N, Pethe C, d'Alayer J, Laloue M (1999) Cytokinin oxidase from Zea mays: purification, cDNA cloning and expression in moss protoplasts. Plant J 17: 615–626 [DOI] [PubMed] [Google Scholar]
- Hutchison CE, Kieber JJ (2002) Cytokinin signaling in Arabidopsis. Plant Cell 14: S47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J (2001) Two-component circuitry in Arabidopsis signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Imamura A, Hanaki N, Umeda H, Nakamura A, Suzuki T, Ueguchi C, Mizuno T (1998) Response regulators implicated in His-to-Asp phosphotransfer signaling in Arabidopsis. Proc Natl Acad Sci USA 95: 2691–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Kakimoto T (2001) Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate:ATP/ADP iospentenyltransferases. Plant Cell Physiol 42: 677–685 [DOI] [PubMed] [Google Scholar]
- Kiba T, Taniguchi M, Imamura A, Ueguchi C, Mizuno T, Sugiyama T (1999) Differential expression of genes for response regulators in response to cytokinins and nitrate in Arabidopsis thaliana. Plant Cell Physiol 40: 767–771 [DOI] [PubMed] [Google Scholar]
- Kiba T, Yamada H, Mizuno T (2002) Characterization of the ARR15 and ARR16 response regulators with special reference to the cytokinin signaling pathway mediated by the AHK4 histidine kinase in roots of Arabidopsis thaliana. Plant Cell Physiol 43: 1059–1066 [DOI] [PubMed] [Google Scholar]
- Koshiba T, Ballas N, Wong L-M, Theologis A (1995) Transcriptional regulation of PS-IAA4/5 and PS-IAA6 early gene expression by indoleacetic acid and protein synthesis inhibitors in pea (Pisum sativum). J Mol Biol 253: 396–413 [DOI] [PubMed] [Google Scholar]
- Lee TT (1974) Cytokinin control in subcellular localization of indoleacetic acid oxidase and peroxidase. Phytochemistry 13: 2445–2453 [Google Scholar]
- Liang X, Abel S, Keller JA, Shen NF, Theologis A (1992) The 1-aminocyclopropane-1-carboxylate synthase gene family of Arabidopsis thaliana. Proc Natl Acad Sci USA 89: 11046–11050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrmann J, Harter K (2002) Plant two-component signaling systems and the role of response regulators. Plant Physiol 128: 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrmann J, Buchholz G, Keitel C, Sweere C, Kircher S, Bäurle I, Kudla J, Harter K (1999) Differentially-expressed and nuclear-localized response regulator-like proteins from Arabidopsis thaliana with transcription factor properties. J Plant Biol 1: 495–506 [Google Scholar]
- Mähönen AP, Bonke M, Kauppinen L, Riikonon M, Benfey P, Helariutta Y (2000) A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev 14: 2938–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RC, Mok C, Mok DWS (1999a) A gene encoding the cytokinin enzyme zeatin O-xylosyltransferase of Phaseolus vulgaris. Plant Physiol 120: 553–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RC, Mok MC, Habben JE, Mok DWS (2001) A maize cytokinin gene encoding an O-glucosyltransferase specific to cis-zeatin. Proc Natl Acad Sci USA 98: 5922–5926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RC, Mok MC, Mok DWS (1999b) Isolation of a cytokinin gene, ZOG1, encoding zeatin O-glucosyltransferase from Phaseolus lunatus. Proc Natl Acad Sci USA 96: 284–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Mizuno T (2000) Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol 41: 1002–1012 [DOI] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JAH (2002) Cell cycle regulated gene expression in Arabidopsis. J Biol Chem 277: 41987–42002 [DOI] [PubMed] [Google Scholar]
- Miller C (1979) Cytokinin inhibition of respiration by cells and mitochondria of soybean, Glycine max (L) Merrill. Planta 146: 503–511 [DOI] [PubMed] [Google Scholar]
- Miller CO (1978) Cytokinin modification of metabolism of p-coumaric acid by a cell suspension of soybean (Glycine max (L.) Merrill). Planta 140: 193–199 [DOI] [PubMed] [Google Scholar]
- Miller CO (1985) Possible regulatory roles of cytokinins: NADH oxidation by peroxidase and a copper interaction. Plant Physiol 79: 908–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok DW, Mok MC (2001) Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol 89: 89–118 [DOI] [PubMed] [Google Scholar]
- Mok DWS, Mok MC (1994) Cytokinins: Chemistry, Activity and Function. CRC Press, Boca Raton, FL
- Morris RO, Bilyeu KD, Laskey JG, Cheikh NN (1999) Isolation of a gene encoding a glycosylated cytokinin oxidase from maize. Biochem Biophys Res Commun 255: 328–333 [DOI] [PubMed] [Google Scholar]
- Palmer M, Palni LMS (1987) Substrate effects on cytokinin metabolism in soybean callus tissue. J Plant Physiol 126: 365–371 [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JA (1999) Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Rupp H-M, Frank M, Werner T, Strnad M, Schmülling T (1999) Increased steady state mRNA levels of the STM and KNATI homeobox genes in cytokinin overproducing Arabidopsis thaliana indicate a role for cytokinins in the shoot apical meristem. Plant J 18: 557–563 [DOI] [PubMed] [Google Scholar]
- Sakai H, Aoyama T, Oka A (2000) Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J 24: 703–711 [DOI] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A (2001) Arabidopsis ARR1 is a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521 [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Suzuki M, Takei K, Deji A, Taniguchi M, Sugiyama T (1998) A response-regulator homologue possibly involved in nitrogen signal transduction mediated by cytokinin in maize. Plant J 14: 337–344 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Schäfer S, Schmülling T (2002) The CRK1 receptor-like kinase gene of tobacco is negatively regulated by cytokinin. Plant Mol Biol 50: 155–165 [DOI] [PubMed] [Google Scholar]
- Scheres B, DiLaurenzio L, Willemsen V, Hauser M-T, Janmaat K, Weisbeek P, Benfey PN (1995) Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the embryonic axis. Development 121: 53–62 [Google Scholar]
- Stock A, Robinson V, Goudreau P (2000) Two-component signal transduction. Annu Rev Biochem 69: 183–215 [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Takei K, Sakakibara H, Taniguchi M, Sugiyama T (2001) Nitrogen-dependant accumulation of cytokinins in root and the translocation to leaf: implication of cytokinin species that induces gene expression of maize response regulator. Plant Cell Physiol 42: 85–93 [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Kiba T, Sakakibara H, Ueguchi C, Mizuno T, Sugiyama T (1998) Expression of Arabidopsis response regulator homologs is induced by cytokinins and nitrate. FEBS Lett 429: 259–262 [DOI] [PubMed] [Google Scholar]
- Terrine C, Laloue M (1980) Kinetics of N6-(D2-isopentenyl)adenosine degradation in tobacco cells: evidence of regulatory mechanisms under control of cytokinins. Plant Physiol 65: 1090–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognolli M, Penel C, Greppin H, Simon P (2002) Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 288: 129–138 [DOI] [PubMed] [Google Scholar]
- Ueguchi C, Sato S, Kato T, Tabata S (2001) The AHK4 gene involved in the cytokinin-signaling pathway as a direct receptor molecule in Arabidopsis thaliana. Plant Cell Physiol 42: 751–755 [DOI] [PubMed] [Google Scholar]
- Urao T, Yakubov B, Satoh R, Yamaguchi-Shinozaki K, Seki M, Hirayama T, Shinozaki K (2000) A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11: 1743–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H, Suzuki T, Terada K, Takei K, Ishikawa K, Miwa K, Yamashino T, Mizuno T (2001) The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol 41: 1017–1023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.