Abstract
The Arabidopsis genome contains a family of NDR1/HIN1-like (NHL) genes that show homology to the nonrace-specific disease resistance (NDR1) and the tobacco (Nicotiana tabacum) harpin-induced (HIN1) genes. NHL3 is a pathogen-responsive member of this NHL gene family that is potentially involved in defense. In independent transgenic NHL3-overexpressing plant lines, a clear correlation between increased resistance to virulent Pseudomonas syringae pv. tomato DC3000 and enhanced NHL3 transcript levels was seen. These transgenic plants did not show enhanced pathogenesis-related gene expression or reactive oxygen species accumulation. Biochemical and localization experiments were performed to assist elucidation of how NHL3 may confer enhanced disease resistance. Gene constructs expressing amino-terminal c-myc-tagged or carboxyl-terminal hemagglutinin epitope (HA)-tagged NHL3 demonstrated membrane localization in transiently transformed tobacco leaves. Stable Arabidopsis transformants containing the NHL3-HA construct corroborated the findings observed in tobacco. The detected immunoreactive proteins were 10 kD larger than the calculated size and could be partially accounted for by the glycosylation state. However, the expected size was not attained with deglycosylation, suggesting possibly additional posttranslational modification. Detergent treatment, but not chemicals used to strip membrane-associated proteins, could displace the immunoreactive signal from microsomal fractions, showing that NHL3 is tightly membrane associated. Furthermore, immunofluorescence and immunogold labeling, coupled with two-phase partitioning techniques, revealed plasma membrane localization of NHL3-HA. This subcellular localization of NHL3 positions it at an initial contact site to pathogens and may be important in facilitating interception of pathogen-derived signals.
Most plants do not succumb to the many existing potential pathogens, which may be ascribed to the existence of nonhost resistance in plants (Nürnberger and Scheel, 2001). Acquisition of virulence factors has enabled some pathogen strains to overcome this plant species resistance (Van't Slot and Knogge, 2002). In response to those pathogens capable of infection, plants have evolved a repertoire of counter-active defense mechanisms (Nürnberger and Scheel, 2001). One of the best studied defense mechanisms is the genetically defined race-specific resistance that is determined by products of resistance (R) genes that recognize pathogen-specific avirulence (Avr) proteins (Takken and Joosten, 2000).
It was initially proposed that the R proteins act as receptors for pathogen-encoded Avr proteins to initiate a signal activating plant defense responses (Keen, 1990). However, a direct interaction has only been shown for few examples such as the rice (Oryza sativa) Pi-ta protein and its corresponding Avr protein (Jia et al., 2000) or the tomato (Lycopersicon esculentum) Pto and the bacterial avrPto (Tang et al., 1996). In contrast, the product of the tomato R gene, responsible for resistance to Cladosporium fulvum expressing Avr9, cf-9 does not seem to bind Avr9 directly (Luderer et al., 2001). It has been shown recently that epitope-tagged functional cf-9 protein is part of a higher Mr complex, suggesting that R proteins may be part of signaling complexes (Rivas et al., 2002). This may imply that intermediate proteins are required to mediate the interaction of R and Avr proteins. For example, a potential bridging protein is the coprecipitating 75-kD protein found in immune complex with anti-Avr/anti-R antibodies (Leister and Katagiri, 2000). Alternatively, to explain the general lack of direct interaction between R proteins and its cognate Avr proteins, the “guard hypothesis” has been proposed; this predicts that R proteins guard host defense proteins targeted by Avr gene-encoded pathogenicity factors (Van der Biezen and Jones, 1998; Dangl and Jones, 2001). Support for this hypothesis has been recently provided by two papers reporting the guarding of RIN4 by RPS2 (Axtell and Staskawicz, 2003; Mackey et al., 2003). Nevertheless, despite failure to detect direct interactions between R and Avr proteins in many cases, there appears to be a spatial interdependency of these proteins (Bonas and Lahaye, 2001).
NDR1 (nonrace-specific disease resistance) is required for proper function of a subclass of R genes that includes RPM1 (Century et al., 1997; Aarts et al., 1998). Although subcellular localization of NDR1 remains to be demonstrated, it is predicted to be a membrane protein (Century et al., 1997), potentially placing R, Avr, and an R-signaling component at the same subcellular location. Although no obvious biochemical functions could be predicted for the protein, it has recently been proposed that NDR1 could modulate production of reactive oxygen intermediates during resistance (Rusterucci et al., 2001). NDR1 shares some sequence similarities with the tobacco HIN1 (harpin-induced) protein (Century et al., 1997) and approximately 28 putative Arabidopsis proteins (The Arabidopsis Genome Initiative, 2000). The latter were designated as NHL1-28 (for NDR1/HIN1-like) and one to two putative transmembrane domains were predicted for these NHL proteins (Dörmann et al., 2000). These NHLs may not necessarily have similar biochemical properties or signaling roles in plant defense, but might represent protein families sharing common structural motifs because sequence homologies are restricted to short amino acid stretches. We have recently shown that expression for two of these genes, NHL3 and NHL25, is triggered by avirulent but not virulent Pseudomonas syringae pv. tomato strains. In particular, NHL3 expression appears to be suppressed by virulent bacteria and is thus likely to participate in disease resistance (Varet et al., 2002).
Here, we assessed the effect of NHL3 overexpression on the in planta growth of pathogenic bacteria in transgenic plants. To further understand the possible function of NHL3, we localized the epitope-tagged NHL3 proteins in microsomal preparations of tobacco and Arabidopsis leaves. We could show that NHL3 is a glycosylated plasma membrane protein. The NHL3 location and the correlation between NHL3 overexpression and bacterial resistance support the hypothesis that this gene contributes to disease resistance against pathogenic bacteria.
RESULTS
NHL3 Overexpression Correlates with Resistance against Virulent Bacteria
NHL3 has only weak sequence homology (41% similarity/27% identity) to NDR1. Nevertheless, suppression of NHL3 expression by virulent bacteria (Varet et al., 2002) suggests a potential role for this gene in the Arabidopsis disease resistance. To test this hypothesis, we generated transgenic Arabidopsis plants expressing NHL3 cDNA in sense orientation under the control of the strong cauliflower mosaic viral 35S promoter. The resulting transgenic lines showed variable levels of transgene expression and four transgenic lines showing no, intermediate, or high expression were selected for analysis (Fig. 1A, lines S6, S7, S5, and S13). These plants were self-pollinated until the T4 to T5 generation of which expression of the transgene is similar between sibling plants, and the growth of virulent P. syringae pv. tomato DC3000 was determined.
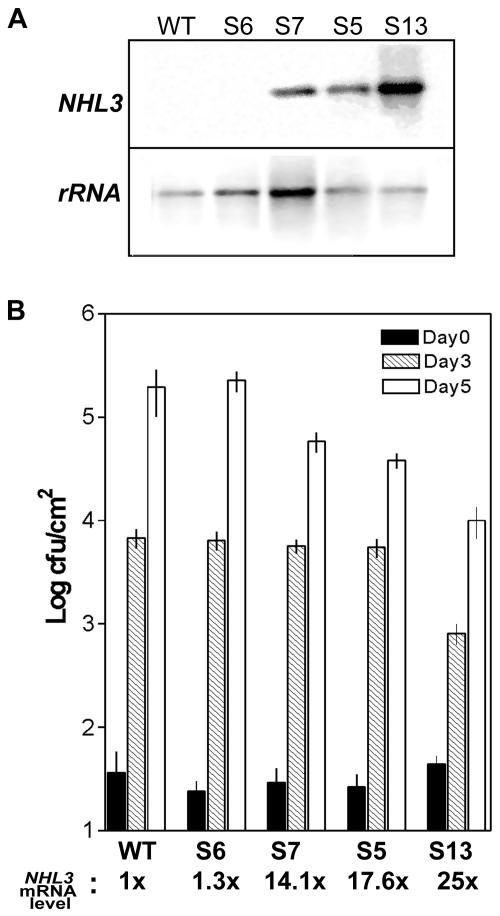
Figure 1.
NHL3 overexpression levels correlate with resistance to virulent bacteria. A, Two micrograms of total RNA prepared from wild-type (WT) or four independent transgenic Arabidopsis lines with NHL3 under the control of the constitutive 35S promoter (S6, S7, S5, and S13) was used for northern blots and was hybridized with the indicated radioactively labeled cDNA probes (rRNA, 26S rDNA). B, Four-week-old plants were inoculated with P. syringae pv. tomato DC3000 at a concentration of 1 × 103 cfu mL–1. Bacterial titers in leaves at the indicated time points were determined. The figure shows the means and ses of a representative experiment. Line S13 consistently showed less bacterial growth in at least four other experiments. Bacterial growth in lines S7 and S5 are intermediate between S13 and the nonexpressing line S6 or wild-type plants. The four transgenic lines show differential expression of the transgene, and the relative mRNA level of NHL3 in the transgenic lines is indicated at the bottom of the figure. The level in the untransformed wild-type (Columbia [Col-0]) plants is set at a reference value of one. An inverse correlation between transgene expression and bacterial growth can be observed.
The transgenic line accumulating high NHL3 mRNA levels (Fig. 1A, line S13) showed significantly reduced bacterial growth 3 and 5 d postinfiltration (Fig. 1B). In contrast, the transgenic line S6, which does not overexpress NHL3, did not differ from the untransformed wild-type plants. The lines S7 and S5, accumulating intermediate levels of NHL3 mRNA, also showed an increased (but less dramatic) resistance to P. syringae pv. tomato DC3000 5 d postinfiltration (Fig. 1B). These observations were confirmed in at least three independent experiments. Notably, when the transgene expression levels were quantified by phosphorimaging of RNA gel blots, a clear correlation between transgene expression and resistance to virulent bacteria could be seen (Fig. 1B). Hence, high expression of NHL3 contributes quantitatively to resistance against P. syringae pv. tomato DC3000. However, when resistance test against two virulent Peronospora parasitica isolates (cv Noks and cv Waco) were performed, no difference in spore germination or sporulation was observed for these two isolates (data not shown). Thus, the enhanced resistance appears to be specific to the tested virulent P. syringae strain.
The overexpressing transgenic lines had no obvious phenotypic or morphological differences from wild-type plants. This distinguishes these lines from enhanced resistance mutants such as constitutive expressor of pathogenesis-related (cpr) genes or mitogen-activated protein kinase 4 (mpk4), which show some developmental defects or are dwarfs (Petersen et al., 2000; Clarke et al., 2001). Defense gene markers such as PR1, PR2, and PDF1.2 were analyzed but no increased expression levels were seen in the transgenic lines (data not shown). Because these genes are known to be responsive to salicylic acid (SA) or jasmonic acid/ethylene, respectively, we infer that levels of these hormones are also not elevated. Similarly, 3,3′-diaminobenzidine staining failed to detect enhanced accumulation of H2O2 in the NHL3 transgenic plants (not shown). Taken together, it appears that NHL3 overexpression does not lead to a general systemic acquired resistance, which is also supported by the lack of resistance to the tested P. parasitica isolates. To assess if the enhanced resistance might be due to a stronger response to virulent bacteria in terms of PR gene expression or hypersensitive reaction (HR) development, we performed RNA gel-blot analysis or trypan-blue staining, respectively, on P. syringae pv. tomato DC3000-infiltrated leaves. However, no significant alterations were observed compared with wild-type plants (data not shown).
Reciprocal experiments to study resistance phenotype through reducing NHL3 expression (antisense gene constructs) or in gene knockouts were also attempted. Seventy putative antisense lines were analyzed, but none showed a convincing reduction of NHL3 mRNA levels after wounding (note that NHL3 is also wound inducible; Varet et al., 2002). Screens for insertion knockouts in the libraries of En-1 transposon (Cologne), dSpm transposon (Norwich), T-DNA tags (Wisconsin), as well as the public T-DNA flanking sequence database (Versailles) were all unsuccessful.
NHL3 Is Predicted to Be a Membrane Protein
Because our analysis above did not provide any clue toward understanding the enhanced resistance brought about by overexpressing NHL3, we next evaluated the localization of the protein. Similar to NDR1 (Century et al., 1997), NHL3 is predicted to be a membrane protein. Computational analysis indicated a putative transmembrane domain between amino acids 52 and 75 (Fig. 2, TM1). A Cys-rich region was predicted as a possible second transmembrane domain (Fig. 2, TM2), albeit with lower probability, and only by a subset of the available membrane prediction programs (http://www.expasy.org). Thus, NHL3 contains one or two potential transmembrane domains. No putative N-terminal cleavable signal sequence was found in NHL3, although this was predicted for NDR1. Furthermore, positively charged amino acids flanking the transmembrane domains are found in NHL3 (Fig. 2). These usually promote cytoplasmic retention during insertion into membranes (Hartmann et al., 1989) and might affect the insertion topology (see “Discussion”).
Figure 2.
Predicted structural features of the NHL3 protein. TM1 is the most likely transmembrane domain, whereas TM2 is a second putative transmembrane domain but predicted with a lower likelihood. Regions flanking transmembrane domain containing positively charged amino acids that favor cytoplasmic exposure are indicated by plus symbols. The predicted N-glycosylation sites are marked with asterisks. The amplified boxes show the amino acid sequences of the indicated domains.
Hemagglutinin Epitope (HA)- and c-myc-Tagged NHL3 Are Membrane Localized in Tobacco
An epitope tag strategy was chosen to study NHL3 localization because the specificity of two independent antipeptide antibodies generated for NHL3 could not be demonstrated. Gene constructs were created that expressed NHL3 with a C-terminal HA-epitope tag (NHL3-HA) or an N-terminal c-myc-epitope tag (c-myc-NHL3). Expression of these genes was driven by the dexamethasone (DEX)-inducible promoter (for NHL3-HA) or the constitutive 35S promoter (for c-myc-NHL3). For negative control, the corresponding empty vectors were also included. Leaves of tobacco were transiently transformed with Agrobacterium tumefaciens strains carrying these constructs (see “Materials and Methods” for details). Western-blot analysis of total protein extracts of tobacco leaves infiltrated with A. tumefaciens strains carrying the NHL3-HA construct and treated with DEX revealed three crossreacting bands (Fig. 3A). Only two crossreacting bands were detected in total protein extracts of leaves transformed with A. tumefaciens strains carrying the c-myc-NHL3 construct (Fig. 3B). For both constructs, the detected protein bands were located to the microsomal fraction and no signal was detectable in leaves infiltrated with the respective empty vectors (Fig. 3, A and B). A very weak signal was occasionally visible in the soluble fraction, possibly as a result of the high overexpression levels. The largest immunoreactive band of both constructs is about 37 kD (Fig. 3, A and B), which is approximately 10 kD larger than the predicted Mr of 27 for NHL3.
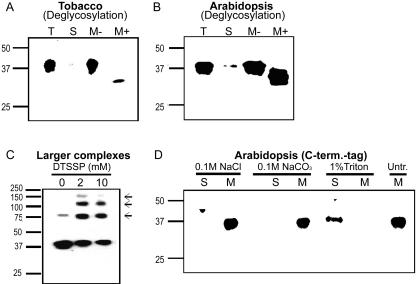
Figure 3.
Epitope-tagged NHL3 is localized to the membrane fractions of transiently transformed tobacco leaves (A and B) or stable transformants of Arabidopsis (C). A and B, Tobacco leaves were inoculated with A. tumefaciens strain GV3101 carrying the respective cloning plasmids (empty vector), the NHL3-HA construct (in A), or the c-myc-NHL3 construct (in B). Total (T) protein extracts were separated into soluble (S) and 100,000g microsomal pellet (M) fractions, and were subjected to western-blot analysis with the appropriate antibody. A, Leaves were sprayed with DEX 24 h postinfiltration and were harvested 24 h later. B, Leaf tissues were harvested 48 h postinfiltration. C, Leaves of transgenic Arabidopsis (T3 generation) harboring the control cloning plasmid (empty vector) or the NHL3-HA construct were harvested 24 h post-DEX treatment and were analyzed as described above for A and B. Three independent transgenic lines of each construct showed identical results and only one representative experiment is shown. Numbers on the left indicate positions of protein size markers in kilodaltons.
HA-Tagged NHL3 Is Membrane Localized in Arabidopsis and Is Glycosylated
To ascertain that the unexpected size of the immunoreactive bands is not due to heterologous expression in tobacco, we conducted similar studies in the homologous system, Arabidopsis. Because A. tumefaciens-mediated transient expression in Arabidopsis was variable in our hands, we stably transformed Arabidopsis (ecotype Col-0) with the gene construct expressing NHL3-HA under the control of the DEX promoter. This construct was selected because it enables an DEX-inducible expression that reflects the temporal expression pattern of NHL3 by avirulent bacteria (A.Varet, unpublished data) and allows studies in the absence of pathogens. Westernblot analysis with the anti-HA antibody showed the presence of protein bands of approximately 37 kD in total and microsomal protein extracts, whereas no signal was detectable in DEX-treated leaves of plants transformed with the control empty vector (Fig. 3C). Notably, the increased size of the protein compared with the predicted size and its microsomal localization are conserved in Arabidopsis and tobacco. This might be indicative of posttranslational modification for NHL3.
Because NHL3 contains four predicted N-glycosylation sites (Fig. 2), we tested if the epitope-tagged proteins are glycosylated in tobacco and Arabidopsis. Solubilized microsomal fractions obtained from transiently transformed tobacco leaves and DEX-treated NHL3-HA Arabidopsis plants were incubated with peptide N-glycosidase F (PNGase F), a glycoamidase that liberates N-linked oligosaccharides from glycoproteins (Tarentino and Plummer, 1987). After PNGase F treatment, smaller bands with a molecular mass of approximately 31 kD were detected in tobacco (Fig. 4A) as well as in Arabidopsis (Fig. 4B), indicating that N-glycosylation accounts at least in part for the larger size of NHL3-HA in SDS-PAGE.
Figure 4.
NHL3-HA in transgenic Arabidopsis is a glycosylated and membrane-associated protein complex. A, Microsomal fractions of tobacco leaves transiently expressing NHL3-HA were incubated for 30 min with buffer (M-) or with PNGase F (M+) and were subjected to western-blot analysis with the anti-HA antibody. B, Microsomal fractions isolated from leaves of DEX-treated transgenic NHL3-HA Arabidopsis plants were treated and analyzed as described in A. C, Higher mobility complexes (arrows) were seen when the protein extracts were treated with the indicated amount of chemical crosslinker, 3,3′-dithiobis(sulfosuccinimidylpropionate) (DTSSP) before western blotting. D, Microsomal fractions isolated from leaves of DEX-treated transgenic NHL3-HA Arabidopsis plants were treated with the indicated substances for 1 h at 4°C. Membranes were then pelleted by ultracentrifugation and the resulting soluble (S) and membrane proteins (M) were analyzed by western blotting.
NHL3-HA May Be Part of Larger Protein Complexes andIs Tightly Membrane Associated
The loading of high protein amounts or extended exposure of western blots revealed the presence of several larger bands in microsomal fractions isolated from transgenic NHL3-HA Arabidopsis (data not shown). Furthermore, deglycosylation procedure also gave rise to a size shift of these larger bands (data not shown). When the protein extracts were treated with the homobifunctional chemical crosslinker, DTSSP, before SDS-PAGE and western blotting, these larger immunoreactive bands appeared, whereas the intensity of the 37-kD band was reduced (Fig. 4C). Four bands were seen in the crosslinking experiments (Fig. 4C), but another two larger bands could be seen when noncrosslinked protein extracts were loaded in the absence of reducing agents (data not shown). Taken together, the sizes of these additional bands suggest that NHL3-HA could form oligomers or be a component of larger protein complexes. These complexes appear to be partially resistant to denaturation during SDS-PAGE.
To further test the prediction that NHL3-HA is a transmembrane protein and is not merely membrane associated, we used various treatments to strip protein subclasses from microsomal fractions isolated from NHL3-HA transgenic Arabidopsis plants. Salt or alkaline treatment, known to be effective in extracting proteins that are peripherally associated with membranes, did not remove the tagged NHL3 protein from the microsomal fraction (Fig. 4D). Only treatment with the nonionic detergent, Triton X-100, could shift the immunoreactive signal to the soluble fraction (Fig. 4D). Thus, NHL3-HA is tightly associated with membranes.
NHL3-HA Is Localized to Plasma Membrane
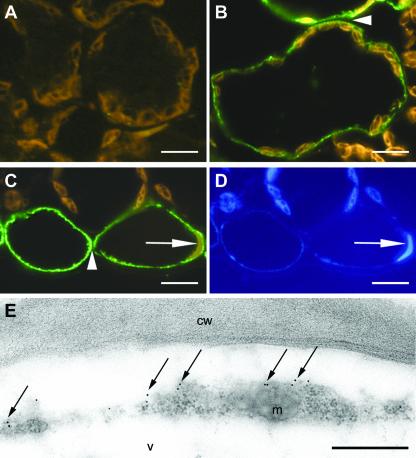
Immunohistochemical experiments were performed to further pinpoint the subcellular localization of NHL3 in the NHL3-HA Arabidopsis plants. No signal was detectable in the sections of plants transformed with the empty control vector (Fig. 5A). In the transgenic NHL3-HA plants, immunodecoration could be clearly detected but was not seen in every cell. This was seen in two different transgenic lines and is unlikely to be a result of inefficient DEX uptake. It could be that some cell-specific silencing might have occurred. Nevertheless, when observed, the immunofluorescent signal was restricted to a thin layer at the periphery of cells (Fig. 5, B and C). When two adjacent cells were labeled, the signals were in two separate lines with the cell wall space unstained (see arrowheads in Fig. 5, B and C). The label was, in most cases, a spotty line indicating regions of high concentration of the epitope. No label was seen in chloroplasts, nuclei, or tonoplasts (Fig. 5, C and D). Little or no signal was seen in the cytoplasm that is expected to encompass organelles such as chloroplasts or nuclei (see Fig. 5, B and C). Hence, despite poor resolution of the thin cytoplasm, it appears that the immunoreactive signal is not found in internal membranes such as endoplasmic reticulum. Immunogold-labeled ultrathin sections for electron microscopy were used to verify this. Even at this high resolution, the cytoplasm in labeled epidermal Arabidopsis cells is still relatively thin compared with the large vacuole (Fig. 5E). Similar results were observed for two independent transgenic lines, and Figure 5E shows a representative section where the detected gold labels were primarily observed at the cell periphery adjacent to the cell wall. Due to a weak plasmolysis, the plasma membrane is detached from the cell wall (Fig. 5E), which further highlights the absence of signals in the cell wall or extracellular matrix. In some cases, the labels were detected in membrane protrusions toward the cell wall, indicating plasma membrane localization. Controls performed by immunodecoration of sections of leaves harboring the control empty vector did not exhibit labeling (data not shown).
Figure 5.
Immunolabeling localizes NHL3-HA to the periphery of the cell. Cross-sections of leaves from DEX-treated transgenic plants harboring the empty pTA7002 vector control (A) or NHL3-HA construct (B–E) were probed with an anti-HA antibody followed by an Alexa488-coupled (Molecular Probes, Eugene, OR) secondary antibody giving green signals (A–D) or by a secondary antibody conjugated with colloidal gold (E). In leaf sections of plants transformed with the empty vector control, only brownish autofluorescence is visible (A), whereas mesophyll (B) and epidermal cells (C) of plants expressing NHL3-HA show label. In both cases, cell walls do not exhibit label (arrow heads in B and C). Moreover, the label is restricted to the peripheral side of the cytoplasm as visible near the nucleus (arrows in C and D), which is visualized by the concomitant 4,6-diamidino-2-phenylindole (DAPI) staining (D). Immunogold labeling of ultrathin sections exhibits the majority of label at the periphery of the cytoplasm adjacent to the cell wall (arrows in E). cw, cell wall; v, vacuole; m, mitochondrion. Bars = 10 μm in A through D; bar = 0.5 μm in E.
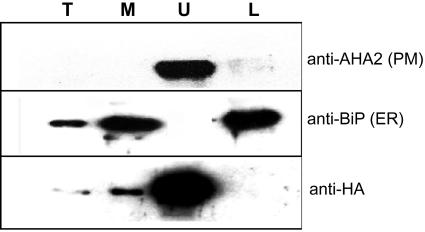
To strengthen the microscopy data, microsomes prepared from DEX-treated transgenic NHL3-HA plants were subjected to aqueous two-phase partitioning. The efficiency of the partitioning process was confirmed with antibodies for the plasma membrane marker, proton-ATPase AHA2 (Palmgren et al., 1991) and endoplasmic reticulum lumenal binding protein (BiP) (Fig. 6). The NHL3-HA protein was exclusively found in the upper phase where plasma membrane proteins are enriched (Fig. 6).
Figure 6.
NHL3-HA is highly enriched in the upper phase of two-phase partitioning fraction. Two-phase partitioning was used to separate plasma membrane and internal membranes. Five micrograms of proteins from microsomal (M), upper phase (U), and lower phase (L) preparations was loaded with 2 μL of total (T) extracts (approximately 30 mg of ground tissue boiled in 200 μL of SDS buffer) and was subjected to western blotting. Antibodies specific for amino acids 6 through 51 of AHA2, a plasma membrane-localized proton ATPase, and against the plant-specific endoplasmic reticulum lumenal binding protein BiP were used to show the efficiency of the fractionation process. Identical results were obtained in two different transgenic lines, and only a representative experiment is shown.
DISCUSSION
NHL3 transcripts were previously shown to accumulate during several incompatible interactions, upon in planta expression of AvrRpt2, and also very rapidly but transiently after mechanical wounding (Varet et al., 2002). Because the former two treatments cause HR, one might speculate that the onset and progressive development of HR results in cellular damage that subsequently leads to a sustained, wound-like induction of NHL3 expression. In such a scenario, NHL3 expression would be a secondary effect that has no consequence for pathogen defense. However, NHL3 expression is also strongly induced by a nonpathogen of Arabidopsis, P. syringae pv. phaseolicola and a Type III secretion mutant of P. syringae pv. tomato DC3000 (Varet et al., 2002). Arabidopsis leaves infiltrated with either one of these strains do not show any HR or necrosis, thus ruling out the secondary wounding as a causal induction of NHL3 expression. We have previously suggested that virulent P. syringae pv. tomato DC3000 probably suppresses NHL3 expression through a translocated Type III effector protein (Varet et al., 2002). This led us to propose that NHL3 is possibly important in the defense response against P. syringae pv. tomato DC3000. The NHL gene family shows similarities to the tobacco HIN1 gene (Dörmann et al., 2000), and because HIN1 is often used as a marker for the HR (Gopalan et al., 1996), NHL3 expression could affect the HR and thus contribute to resistance during pathogen attack. To address this possibility, we generated NHL3-overexpressing transgenic lines, but did not observe increase in HR-like necrosis under long-day or short-day conditions (data not shown). However, we could demonstrate a subtle but distinct increased resistance to virulent P. syringae pv. tomato DC3000 (Fig. 1B). Overexpression of NHL2, which is most homologous to NHL3 among the NHLs, was shown to lead to light-dependent speck-like lesion development, but not to increased resistance (Dörmann et al., 2000). This indicates that NHL2 and NHL3 might be functionally distinct.
Alternatively, a different outcome could be caused by the different starting bacterial inoculum used. In contrast to 2 × 106 cfu mL–1 used by Dörmann et al. (2000), we used a lower titer of 103 cfu mL–1, which probably better reveals a weak resistance effect and, therefore, is more sensitive. For instance, it has been reported that the cpr/npr1 double mutants are susceptible to P. syringae pv. maculicola ES4326 at a greater inoculum, but are resistant at a lower dose of the same bacteria, indicating that resistance can be overcome by higher titer in certain cases (Clarke et al., 2000). Interestingly, the resistance in our NHL3 transgenic plants correlates with the accumulated steadystate NHL3 mRNA levels and suggests a dose-dependent effect of NHL3 gene expression toward enhanced resistance to P. syringae pv. tomato DC3000, with the line S13 that accumulates the highest NHL3 transcript levels supporting the least bacterial growth (Fig. 1B). Such transgene expression level-dependent increased resistance has also been reported for transgenic expression of NPR1 in Arabidopsis (Cao et al., 1998). Although it is theoretically possible to use the DEX-inducible NHL3-HA lines to verify a controlled dose-dependent transgene expression effect, this was not done because the DEX system has been associated with aberrant defense-related effects in some transgenic lines (Kang et al., 1999). Unfortunately, despite extensive screenings, attempts to isolate insertion knockouts of NHL3 failed and antisense NHL3 plants did not show the desired suppression of NHL3 mRNA levels (data not shown), such that the converse “loss-of-function” experiments could not be performed.
Although enhanced resistance was seen in the overexpressing lines, no obvious phenotypic or morphological alterations were observed. The mechanism of the enhanced resistance to P. syringae pv. tomato DC3000 is not a result of increased HR or constitutive defense activation as no elevated PR (PR1, PR2, and PDF1.2) gene expression was observed. Furthermore, the lack of increased resistance to two strains of virulent P. parasitica (data not shown) also argues against pleiotrophic effects or constitutive systemic acquired resistance. Hence, the overexpression of NHL3 leads to a resistance mechanism that is distinct from the pathways in enhanced resistance mutants such as cpr1, cpr5, cpr6, or mpk4, which are smaller in stature and show constitutive PR gene expression (Clarke et al., 2000, 2001; Petersen et al., 2000). NDR1 has been proposed to link reactive oxygen species to SA production (Shapiro and Zhang, 2001). Although NHL3 is a member of the NDR1/HIN1-like gene family, we failed to detect higher SA or reactive oxygen species in the NHL3-overexpressing plants. Because NHL3 only exhibits weak sequence homology to NDR1, it is possible that the two proteins are functionally different. The lack of PR gene expression in the NHL3-overexpressing lines does not allow us to speculate on the possible pathway(s) in which the enhanced resistance operates, but it is noteworthy that enhanced disease resistance can occur in the absence of constitutive PR gene expression as is documented for the edr1 mutant (Frye and Innes, 1998; Frye et al., 2001). Previous expression analysis did not show increased NHL3 expression in response to avirulent P. parasitica (Varet et al., 2002); the resistance mediated by NHL3 overexpression might thus be limited to P. syringae and could also be a direct antibacterial effect of the gene product.
The lack of suitable antibodies does not permit us to follow up on previous mRNA expression analysis to determine if the native NHL3 protein also accumulates after pathogen infection. It is also conceivable that the observed transcript accumulation (Varet et al., 2002) is required to maintain NHL3 protein level as a result of higher protein turnover of proteins during infection. Degradation of membrane proteins, such as the peripheral membrane R protein, RPM1, in a similar time frame with the HR has been previously reported (Boyes et al., 1998). Thus, it is interesting to note that NHL3 is also predicted to be membrane localized (Fig. 2).
Knowledge of NHL3 localization at the subcellular level will assist in the elucidation of its biochemical function and mode of action in conferring enhanced disease resistance when overexpressed. With the aid of two different epitope tags, we confirmed the computer-based prediction of membrane localization using agroinfiltrated tobacco leaves (Figs. 2 and 3). The presence of an epitope tag on either termini did not affect membrane localization of the NHL3 protein (Fig. 3, A and B). Moreover, the detection of the c-myc-NHL3 fusion protein indicates the absence of a cleavable N-terminal signal peptide. Conversely, the detection of the C-terminal HA-epitope tag suggests no processing of the C terminus, which occurs in certain proteins such as glycosyl-phosphatidyl-inositol-anchored membrane proteins. This membrane localization, at least for NHL3-HA, is not an artifact of heterologous expression in tobacco because it could be verified in transgenic Arabidopsis (Fig. 3C). If transgene expression of the HA-tagged protein in both plant species had led to an incorrect localization, high levels of immunoreactive signals would have additionally been found in soluble fractions. However, the NHL3-HA fusion protein was always found at higher levels in the microsomal fractions. Thus, we conclude that the tagged NHL3 is membrane localized.
Deglycosylation experiments demonstrated that NHL3 is a glycosylated membrane protein and glycosylation accounts partially for the larger size observed in SDS-PAGE (Fig. 4, A and B). Thus, the detection of two to three immunoreactive bands could be a result of incomplete or different levels of glycosylation. Surprisingly, treatment with PNGase F did not restore the mobility of the immunoreactive protein to that of the calculated NHL3 molecular mass. Because PNGase F does not hydrolyze N-glycans if they carry α-1–3-linked core Fuc residues (Tarentino and Plummer, 1987), one or more of the four predicted glycosylation sites might contain such glycans. Alternatively, the unexpected mobility in SDS-PAGE despite deglycosylation could be due to other posttranslational modifications or aberrant mobility of the naked protein in SDS-PAGE.
Using different reagents, we could further show that NHL3 is not peripherally bound but is tightly membrane associated (Fig. 4D). Consistent with this finding, all analysis programs predicted NHL3 to contain a transmembrane stretch between amino acids 52 and 75, and therefore suggested it to be a type III membrane protein (Fig. 2, TM1). Type III membrane proteins lack cleavable signal peptide, have an amino-terminal hydrophilic ectodomain (i.e. extracellular or lumenal depending on localization), a single hydrophobic transmembrane stretch, and a carboxyl-terminal cytoplasmic domain. Predictions of protein topology are based on the charge difference rule where positive charges flanking the transmembrane domain promote cytoplasmic retention and negative charges promote translocation into the endoplasmic reticulum lumen (Hartmann et al., 1989; Singer, 1990; Parks and Lamb, 1991). The charge partition near the most likely transmembrane domain of NHL3 (amino acids 52–75, see Fig. 2) supports this prediction. However, all the predicted N-glycosylation sites would thus be localized on the cytoplasmic side with this topology. Therefore, the C terminus would not have passed through the endoplasmic reticulum and cannot be glycosylated. Because our deglycosylation experiments suggested that at least one or more of these sites are used, NHL3 is probably not a type III membrane protein. Alternatively, the SOSUI program (Hirokawa et al., 2000) predicts NHL3 as a protein with two transmembrane domains (see Fig. 2). In this case, the N- and C-terminal ends are ectodomains (extracellular) with only a small loop in the cytoplasmic face between the two transmembrane helices. This prediction is the most likely NHL3 topology because it is then in agreement with the use of the N-glycosylation sites present on the C-terminal region.
Immunohistochemistry coupled with two-phase partitioning experiments further allowed us to unequivocally pinpoint the subcellular localization of the tagged NHL3 to the plasma membrane (Figs. 5 and 6). Intriguingly, many R/Avr gene products are also localized to plant membranes and, in particular, plasma membranes. RPM1, which confers resistance to P. syringae expressing avrRpm1 or avrB (Grant et al., 1995), has been shown to be plasma membrane associated (Boyes et al., 1998). AvrRpm1 and AvrB have myristoylation sites that determine their membrane localization and are crucial to their Avr activity (Nimchuk et al., 2000). The tomato Pto R protein similarly possesses myristoylation motifs and its corresponding P. syringae Avr protein AvrPto is plasma membrane localized (Shan et al., 2000). Like NHL3, cf-9 is a glycosylated plasma membrane protein (Piedras et al., 2000), and moreover, this localization is required for its function (Van der Hoorn et al., 2001). The barley (Hordeum vulgare) mlo gene, which confers broad spectrum disease resistance against the pathogenic powdery mildew fungus (Büschges et al., 1997), encodes a membrane protein with seven transmembrane-spanning helices (Devoto et al., 1999). Hence, membrane and, in particular, plasma membrane appear to be important sites where pathogen signal recognition and/or generation of downstream defense signals occur.
Interestingly, the use of large amounts of protein in western blotting led to the detection of larger complexes in membrane fractions. These appear to be very stable interactions that can partially resist denaturation during SDS-PAGE. The chemical crosslinking experiments (Fig. 4C) further demonstrated that these complexes are not due to trapping within membrane vesicle, but to true proximity of the interacting proteins (note that DTSSP has a spacer arm of only 12Å). The mobility of these crosslinked bands corresponds to that of dimers, trimers, and larger oligomers. Thus, NHL3 may interact with itself and/or with other proteins. In view of the enhanced resistance upon overexpressing NHL3 (Fig. 1), it is tempting to speculate that it may possibly interact with other signaling proteins required for resistance. As proposed by Grant and Mansfield (1999), such a signal linker protein complex in the membrane may thus participate in intercepting pathogen-derived signals and transducing this into effective resistance reactions to pathogens.
In summary, we have provided evidence that overexpression of NHL3, which has sequence similarities to NDR1, can confer enhanced resistance to virulent P. syringae pv. tomato DC3000 and that it encodes a glycosylated plasma membrane protein. In conjunction with our previous studies showing possible suppression of NHL3 expression by virulent bacteria (Varet et al., 2002), the current data on plasma membrane localization and the predicted membrane topology will be instrumental in future studies to understand the mechanism by which NHL3 contributes to plant defense responses to bacteria.
MATERIALS AND METHODS
Plants and Growth Conditions
Tobacco (Nicotiana benthamiana) plants were grown in the greenhouse at 22°C in a 14-h light/10-h dark cycle. Six- to 8-week-old plants were used for infiltration experiments. Arabidopsis ecotype Col-0 plants were grown in a phytochamber (Heraeus Voetsch, Balingen, Germany) at 22°C under short-day conditions (8-h light/16-h dark cycle) for infection experiments or under long-day conditions (16-h light/8-h dark cycle) for seed set in a potting mixture consisting of soil:sand (2:1).
Bacterial and Oomycete Inoculation
The bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (Staskawicz et al., 1987) was used. Bacteria were grown at 28°C in medium (King et al., 1954) containing 50 μg mL–1 rifampicin. Plants were syringe infiltrated with bacterial suspensions of 103 cfu mL–1 in 10 mm MgCl2 as described previously (Kiedrowski et al., 1992). Bacterial growth within leaf tissues was monitored 3 and 5 d postinfiltration. Three leaf punches (6 mm in diameter) were homogenized in 200 μL of 10 mm MgCl2 with the aid of plastic disposable pestles. A dilution series in 10 mm MgCl2 was plated on King's B agar plates containing rifampicin and cycloheximide and the bacterial population was counted 2 d later. Leaf material was also harvested from transgenic plants for RNA extraction and RNA-blot analysis (Maniatis et al., 1982). Peronospora parasitica infections were performed with the strains cv Noks and cv Waco as described in McDowell et al. (2000).
Vector Construction, Plant Transformation, and Other Molecular Techniques
An HA epitope was added to NHL3 by cloning its cDNA without the translational stop codon into the XhoI and HindIII sites of the pKHS-0 vector (a gift from Eric Marois and Ulla Bonas, Institute of Genetics, Martin-Luther-University, Halle/Saale, Germany). The NHL3-HA construct was then excised as an XhoI/SpeI DNA fragment and was cloned into the DEX-inducible expression vector pTA7002 (Aoyama and Chua, 1997). The last two amino acids of NHL3 were removed as a result of the cloning. The empty vector and NHL3-HA construct were transformed into Agrobacterium tumefaciens strain GV3101. Oligonucleotide sequences coding for an N-terminal c-myc tag were annealed and inserted into the pRT100 vector between the XhoI and NcoI site. NHL3 cDNA was inserted between the NcoI and the XbaI sites of this modified vector. Empty vector and c-myc-NHL3 were subcloned into the pCB302 binary vector (Xiang et al., 1999) using the HindIII site and were transformed into A. tumefaciens strain GV3101. All constructs were transformed into Arabidopsis Col-0 plants by the floral dip method (Clough and Bent, 1998). RNA extraction and northern-blot analysis were performed as described previously (Varet et al., 2002).
A. tumefaciens Transient Expression Assays
The transient expression assays in tobacco were performed as described in Nimchuk et al. (2000) by infiltrating a bacterial suspension at an OD600 of one. For the NHL3-HA construct, plants were sprayed with 20 μm DEX (Sigma, Taufkirchen, Germany) 24 h after inoculation to induce expression and were harvested 24 h later. For the c-myc-NHL3 construct, leaf discs were harvested 48 h after inoculation.
Plant Membrane Fractionation, Immunoblot Analysis, and Chemical Crosslinking
For tobacco and Arabidopsis, 15 leaf discs of each sample were extracted in 350 μL of buffer and were fractionated as described (Nimchuk et al., 2000). Ultracentrifugation was performed at an acceleration of 100,000g in the absence of CaCl2, and the resulting pellet was resuspended in 350 μL of Tris-EDTA, pH 8.0. For each fraction, 10 μL was loaded onto a 12% (w/v) SDS-PAGE gel.
Chemical crosslinking with DTSSP (Pierce, Rockford, IL) was performed by adding the indicated amount of the crosslinker to microsomes prepared in phosphate-buffered saline (PBS). Reactions were incubated at room temperature for 30 min before SDS-PAGE and western-blot analysis.
Two-phase partitioning experiments were performed as described by Larsson et al. (1987). Membrane proteins in the upper and lower phase were pelleted by ultracentrifugation as described above, and the protein content was determined by bicinchoninic acid method (Pierce) with bovine serum albumin (BSA) as reference.
For immunodetection, blots were probed with a monoclonal antibody to the HA epitope (Roche Applied Science, Mannheim, Germany) at a dilution of 1:1,000 or a monoclonal antibody to the c-myc epitope (Sigma) at a dilution of 1:500. The mouse monoclonal anti-BiP antibody (Stressgen Biotechnologies, Victoria, British Columbia, Canada) was used at 2 μg mL–1 dilution, and the rabbit polyclonal anti-AHA2 (gift from Michael Palmgren's laboratory) was used at 1: 3,000 dilution. Peroxidase-conjugated anti-mouse immunoglobulin (Ig) G at 1:10,000 dilution (Sigma) or anti-rabbit IgG at 1:3,000 dilution (Bio-Rad, Munich) was used as a secondary antibody.
Deglycosylation and Detergent Treatments
Microsomes prepared from plants expressing NHL3-HA were treated with denaturation buffer and PNGase F from the deglycosylation kit (Roche Applied Science, Mannheim, Germany) for 30 min at 37°C. The proteins were subjected to SDS-PAGE and western blotting with the anti-HA antibody. To strip subclasses of proteins from membranes, aliquots of microsomal fractions were incubated in NaCl or NaCO3 solutions at a final concentration of 0.1 m, or in the presence of 1% (v/v) Triton X-100 for 1 h at 4°C. After incubation, samples were centrifuged (100,000g for 1.5 h), the supernatant was retained, and the pellet was resuspended in Tris-EDTA, pH 8.0. The proteins were subjected to SDS-PAGE and western blotting with the anti-HA antibody.
Immunohistochemical Techniques and Microscopy
Three-week-old Arabidopsis plants were sprayed with 20 μm DEX. After 24 h, small pieces of leaves were fixed with 4% (w/v) paraformaldehyde/0.1% (v/v) Triton X-100 in PBS (135 mm NaCl, 3 mm KCl, 1.5 mm KH2PO4, and 8 mm Na2HPO4), dehydrated by a graded series of ethanol, and embedded in polyethylene glycol for fluorescence microscopy as described (Hause et al., 1996). Sections of 2-μm thickness were immunolabeled with the mouse monoclonal antibody to the HA epitope (diluted 1:500 in PBS containing 2% [w/v] acetylated BSA and 1 mg mL–1 goat IgG). Subsequently, an anti-mouse-IgG antibody conjugated with Alexa488 (Molecular Probes) was used according to the supplier's instructions. Immunodecorated sections were stained with 0.1 μg mL–1 DAPI (Sigma) for 15 min and were mounted in Citifluor/glycerol. The fluorescence of immunolabeled NHL3-HA and of DAPI-stained nuclei was visualized with an epifluorescence microscope (Axioskop; Zeiss, Jena, Germany) using the appropriate filter combinations. Micrographs were taken by a CCD camera (Sony, Tokyo) and were processed through the Photoshop program (Adobe Systems, Seattle).
For electron microscopy, ethanol of dehydrated specimens was substituted by LR-White (Polysciences, Warrington, PA). Immunolabeling of ultrathin sections was carried out with the mouse monoclonal antibody to the HA epitope (diluted 1:500 in PBS containing 1% [w/v] acetylated BSA and 0.1% [v/v] Tween 20) and a goat anti-mouse IgG conjugated with 10 nm colloidal gold (Sigma). After immunolabeling, sections were poststained with uranyl acetate and lead citrate. Sections were visualized with a electron microscope (TEM 900; Ziess).
Acknowledgments
We thank Guido van den Ackerveken (Utrecht University, Utrecht, The Netherlands) for performing the resistance test with Peronospora parasitica, and Michael Palmgren for the gift of anti-AHA2 antibodies (Royal Veterinary and Agricultural University, Copenhagen, Denmark). Many thanks also to Rebecca Boston (North Carolina State University, Raleigh, NC) and Estelle Hrabak (University of New Hampshire, Durham, NH) for advice on antibodies against membrane markers. We also thank Thomas Lahaye, Norbert Nass (Martin-Luther University, Halle/Saale, Germany), and Boris Szurek (Unité de Recherche en Génomique Végétale, Evry, France) for critical comments on the manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.020438.
This work was supported in part by “Fonds der chemischen Industrie.”
References
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 95: 10306–10311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Chua NH (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ (2003) Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377 [DOI] [PubMed] [Google Scholar]
- Bonas U, Lahaye T (2001) Plant disease resistance triggered by pathogen-derived molecules: refined models of specific recognition. Curr Opin Microbiol 5: 44–50 [DOI] [PubMed] [Google Scholar]
- Boyes DC, Jaesung N, Dangl JL (1998) The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci USA 95: 15849–15854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, Van Daelen R, Van der Lee T, Diergaarde P, Groenendijk J et al. (1997) The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88: 695–705 [DOI] [PubMed] [Google Scholar]
- Cao H, Li X, Dong X (1998) Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc Natl Acad Sci USA 95: 6531–6536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century KS, Shapiro AD, Repetti PP, Dahlbeck D, Holub E, Staskawicz BJ (1997) NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278: 1963–1965 [DOI] [PubMed] [Google Scholar]
- Clarke JD, Aarts N, Feys BJ, Dong X, Parker JE (2001) Constitutive disease resistance requires EDS1 in the Arabidopsis mutants cpr1 and cpr6 and is partially EDS1-dependent in cpr5. Plant J 26: 409–420 [DOI] [PubMed] [Google Scholar]
- Clarke JD, Volko SM, Ledford H, Ausubel FM, Dong X (2000) Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Devoto A, Piffanelli P, Nilson I-M, Wallin E, Panstruga R, Von Heijne G, Schulze-Lefert P (1999) Topology, subcellular localization and sequence diversity of the Mlo family in plants. J Biol Chem 274: 34993–35004 [DOI] [PubMed] [Google Scholar]
- Dörmann P, Gopalan S, He SY, Benning C (2000) A gene family in Arabidopsis thaliana with sequence similarity to NDR1 and HIN1. Plant Physiol Biochem 38: 789–796 [Google Scholar]
- Frye CA, Innes RW (1998) An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye CA, Tang D, Innes RW (2001) Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc Natl Acad Sci USA 98: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalan S, Wei W, He SY (1996) hrp gene-dependent induction of hin1: a plant gene activated rapidly by both harpins and the avrPto gene-mediated signal. Plant J 10: 591–600 [DOI] [PubMed] [Google Scholar]
- Grant M, Mansfield J (1999) Early events in host-pathogen interactions. Curr Opin Plant Biol 2: 312–319 [DOI] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269: 843–846 [DOI] [PubMed] [Google Scholar]
- Hartmann E, Rapoport TA, Lodish HF (1989) Predicting the orientation of eukaryotic membrane-spanning proteins. Proc Natl Acad Sci USA 86: 5786–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause B, Demus U, Teichmann C, Parthier B, Wasternack C (1996) Developmental and tissue-specific expression of JIP-23, a jasmonate-inducible protein of barley. Plant Cell Physiol 37: 641–649 [DOI] [PubMed] [Google Scholar]
- Hirokawa T, Uechi J, Sasamoto H, Suwa M, Mitaku S (2000) A triangle lattice model that predicts transmembrane helix configuration using a polar jigsaw puzzle. Protein Eng 13: 771–778 [DOI] [PubMed] [Google Scholar]
- Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J 19: 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HG, Fang Y, Singh KB (1999) A glucocorticoid-inducible transcription system causes severe growth defects in Arabidopsis and induces defense-related genes. Plant J 20: 127–133 [DOI] [PubMed] [Google Scholar]
- Keen NT (1990) Gene-for-gene complementarity in plant-pathogen interactions. Annu Rev Genet 24: 447–463 [DOI] [PubMed] [Google Scholar]
- Kiedrowski S, Kawalleck P, Hahlbrock K, Somssich IE, Dangl JL (1992) Rapid activation of a novel plant defense gene is strictly dependent on the Arabidopsis RPM1 disease resistance locus. EMBO J 11: 4677–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EO, Ward NK, Raney DE (1954) Two simple media for the demonstration of pyrocyanin and fluorescein. J Lab Clin Med 44: 301–307 [PubMed] [Google Scholar]
- Larsson C, Widell S, Kjellbom P (1987) Preparation of high-purity plasma membranes. Methods Enzymol 148: 558–569 [Google Scholar]
- Leister RT, Katagiri F (2000) A resistance gene product of the nucleotide binding site: leucine rich repeats class can form a complex with bacterial avirulence proteins in vivo. Plant J 22: 345–354 [DOI] [PubMed] [Google Scholar]
- Luderer R, Rivas S, Nürnberger T, Mattei B, van den Hooven HW, Van der Hoorn RAL, Romeis T, Wehrfritz JM, Blume B, Nennstiel D et al. (2001) No evidence for binding between resistance gene product Cf-9 of tomato and avirulence gene product AVR9 of Cladosporium fulvum. Mol Plant-Microbe Interact 14: 867–876 [DOI] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389 [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch E, Sambrook J (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- McDowell JM, Cuzick A, Can C, Beynon J, Dangl JL, Holub EB (2000) Downy mildew (Peronospora parasitica) resistance genes in Arabidopsis vary in functional requirements for NDR1, EDS1, NPR1 and salicylic acid accumulation. Plant J 22: 523–529 [DOI] [PubMed] [Google Scholar]
- Nimchuk Z, Marois E, Kjemtrup S, Leister RT, Katagiri F, Dangl JL (2000) Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 101: 353–363 [DOI] [PubMed] [Google Scholar]
- Nürnberger T, Scheel D (2001) Signal transmission in the plant immune response. Trends Plant Sci 6: 372–379 [DOI] [PubMed] [Google Scholar]
- Palmgren MG, Sommarin M, Serrano R, Larsson C (1991) Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H+-ATPase. J Biol Chem 266: 20470–20475 [PubMed] [Google Scholar]
- Parks GD, Lamb RA (1991) Topology of eukaryotic type II membrane proteins: importance of N-terminal positively charged residues flanking the hydrophobic domain. Cell 64: 777–787 [DOI] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE et al. (2000) Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Piedras P, Rivas S, Droge S, Hillmer S, Jones JDG (2000) Functional, c-myc-tagged Cf-9 resistance gene products are plasma-membrane localized and glycosylated. Plant J 21: 529–536 [DOI] [PubMed] [Google Scholar]
- Rivas S, Romeis T, Jones JDG (2002) The Cf-9 disease resistance protein is present in an approximately 420-kilodalton heteromultimeric membrane-associated complex at one molecule per complex. Plant Cell 14: 689–702 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rusterucci C, Aviv DH, Holt BF III, Dangl JL, Parker JE (2001) The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13: 2211–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, Thara VK, Martin GB, Zhou J-M, Tang X (2000) The Pseudomonas AvrPto protein is differentially recognized by tomato and tobacco and is localized to the plant plasma membrane. Plant Cell 12: 2323–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro AD, Zhang C (2001) The role of NDR1 in avirulence gene-directed signaling and control of programmed cell death in Arabidopsis. Plant Physiol 127: 1089–1101 [PMC free article] [PubMed] [Google Scholar]
- Singer SJ (1990) The structure and insertion of integral proteins in membranes. Annu Rev Cell Biol 6: 247–296 [DOI] [PubMed] [Google Scholar]
- Staskawicz BJ, Dahlbeck D, Keen N, Napoli C (1987) Molecular characterization of cloned avirulence genes from race0 and race1 of Pseudomonas syringae pv. glycinea. J Bacteriol 169: 5789–5794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken FLW, Joosten MHAJ (2000) Plant resistance genes: their structure, function and evolution. Eur J Plant Pathol 106: 699–713 [Google Scholar]
- Tang X, Frederick RD, Zhou J, Halterman DA, Jia Y, Martin GB (1996) Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274: 2060–2063 [DOI] [PubMed] [Google Scholar]
- Tarentino AL, Plummer TH Jr (1987) Peptide-N4-(N-acetyl-β-glucosaminyl) asparagine amidase and endo-β-N-acetylglucosaminidase from Flavobacterium meningosepticum. Methods Enzymol 138: 770–778 [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Van der Biezen EA, Jones JDG (1998) Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci 23: 454–456 [DOI] [PubMed] [Google Scholar]
- Van der Hoorn RAL, Van der Ploeg A, de Wit PJ, Joosten MH (2001) The C-terminal dilysine motif for targeting to the endoplasmic reticulum is not required for Cf-9 function. Mol Plant-Microbe Interact 14: 412–415 [DOI] [PubMed] [Google Scholar]
- Van't Slot KAE, Knogge W (2002) A dual role of microbial pathogen-derived effector proteins in plant disease and resistance. Crit Rev Plant Sci 21: 229–271 [Google Scholar]
- Varet A, Parker J, Tornero P, Nass N, Nürnberger T, Dangl JL, Scheel D, Lee J (2002) NHL25 and NHL3, two NDR1/HIN1-like genes in Arabidopsis thaliana with potential role(s) in plant defense. Mol Plant-Microbe Interact 15: 608–616 [DOI] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ (1999) A mini binary vector series for plant transformation. Plant Mol Biol 40: 711–717 [DOI] [PubMed] [Google Scholar]