Abstract
Four proteins with wall extension activity on grass cell walls were purified from maize (Zea mays) pollen by conventional column chromatography and high-performance liquid chromatography. Each is a basic glycoprotein (isoelectric point = 9.1–9.5) of approximately 28 kD and was identified by immunoblot analysis as an isoform of Zea m 1, the major group 1 allergen of maize pollen and member of the β-expansin family. Four distinctive cDNAs for Zea m 1 were identified by cDNA library screening and by GenBank analysis. One pair (GenBank accession nos. AY104999 and AY104125) was much closer in sequence to well-characterized allergens such as Lol p 1 and Phl p 1 from ryegrass (Lolium perenne) and Phleum pretense, whereas a second pair was much more divergent. The N-terminal sequence and mass spectrometry fingerprint of the most abundant isoform (Zea m 1d) matched that predicted for AY197353, whereas N-terminal sequences of the other isoforms matched or nearly matched AY104999 and AY104125. Highly purified Zea m 1d induced extension of a variety of grass walls but not dicot walls. Wall extension activity of Zea m 1d was biphasic with respect to protein concentration, had a broad pH optimum between 5 and 6, required more than 50 μg mL-1 for high activity, and led to cell wall breakage after only approximately 10% extension. These characteristics differ from those of α-expansins. Some of the distinctive properties of Zea m 1 may not be typical of β-expansins as a class but may relate to the specialized function of this β-expansin in pollen function.
Expansins were first identified a decade ago as mediators of acid-induced plant cell wall extension in cucumber (Cucumis sativus) hypocotyls and oat (Avena sativa) coleoptiles (McQueen-Mason et al., 1992; Li et al., 1993) and since then have been implicated in many aspects of plant growth and development, including cell expansion, leaf organogenesis and phyllotaxy, fruit ripening, root growth, particularly during drought, cell wall disassembly, and penetration of pollen tubes in grass species (for review, see Cosgrove, 2000; Lee et al., 2001; Cosgrove et al., 2002). Currently, two families of expansins, named EXP (α-expansins) and EXPB (β-expansins), have been identified based on protein activity and sequence analysis (Shcherban et al., 1995; Cosgrove et al., 2002; Li et al., 2002). The rice (Oryza sativa) and Arabidopsis genomes also contain a small family of related genes (see http://www.bio.psu.edu/expansins/) that we have named EXPL (expansin-like) and a more distant gene named EXPR (expansin-related). It is unknown whether the proteins encoded by these distantly related genes have expansin's characteristic activity, namely a rapid induction of cell wall extension and an increase in wall stress relaxation.
Although there are now many reports characterizing expansin gene expression, relatively few studies have examined the activity of expansin proteins. It seems that α-expansins are ubiquitous to land plants and loosen cell walls without breakdown of the major wall polysaccharides (for review, see Cosgrove et al., 2002; Li et al., 2002). Similarly, β-expansins are thought to be ubiquitous in land plants but less is known of their mechanism of wall loosening.
The first hint about β-expansin function came when BLAST searches, using α-expansin as query, found approximately 20% identity to a group of proteins known previously as grass group 1 pollen allergens (Shcherban et al., 1995). These allergens were originally identified by immunologists 40 years ago as the main causative agents of hay fever and seasonal asthma induced by grass pollen (Malley et al., 1962; Johnson and Marsh, 1965a, 1965b) and have since been studied mostly by immunologists to define their antigenic epitopes and to understand how they cause human allergy responses. Subsequently, Cosgrove et al. (1997) reported that the group 1 allergen from maize (Zea mays) pollen, called Zea m 1, has wall-loosening activity characteristic of expansins and proposed that these proteins loosen the cell walls of the stigma and style to aid pollen tube penetration. Grass group 1 pollen allergens are now recognized as a subgroup of β-expansins. Although group 1 allergens are expressed specifically in pollen and probably have a unique wall-loosening role in support of pollen function, other β-expansin genes are expressed more widely during the growth and development of vegetative tissues (Lee and Kende, 2001; Schipper et al., 2002; Wu et al., 2001). We sometimes refer to these other β-expansins as the vegetative homologs of group 1 pollen allergens.
The group 1 allergen Zea m 1 was reported to be most effective on grass cell walls (so-called Type II walls), which are distinctive in composition from Type I walls. Type I walls include the cell walls of all dicots and of the non-commelinoid monocot species (Carpita and Gibeaut, 1993; Carpita, 1996). In contrast to Zea m 1 activity, α-expansins from both dicots and grasses are more effective in inducing extension in vitro with dicot (Type I) walls than with grass (Type II) walls (McQueen-Mason et al., 1992; Li et al., 1993; Cho and Kende, 1997). Our tentative interpretation is that α- and β-expansins act on different matrix polysaccharides in the cell wall and that in grasses, the targets of β-expansin action (possibly arabinoxylans) have a more dominant role in wall structure and wall mechanics than they do in dicots and other species with Type I walls. This explanation would account for the selectivity of Zea m 1 toward grass walls and for the presence of β-expansins in dicots. However, this explanation must be considered tentative until more is known about β-expansin action.
Although expansins were originally discovered a decade ago and are now believed to play important roles in regulating plant cell wall extensibility, the exact biochemical mechanism by which these proteins cause plant cell walls to extend is still unclear. Nearly all the details about the biochemical properties and actions of expansin proteins are derived from studies with α-expansins and very little with β-expansins. Recently, Grobe et al. (1999, 2002) proposed that grass pollen group 1 allergens loosen walls via proteolytic action, but this has been refuted by Li and Cosgrove (2001).
To clarify the mechanism of action of β-expansin action, we have developed an efficient purification procedure for Zea m 1, the β-expansin from maize pollen, and we provide a detailed characterization of the biochemical properties and expansin activity of the multiple Zea m 1 isoforms. Although the expansin activity of Zea m 1 was identified previously, neither the properties of this protein nor its activity were characterized in detail, and in some cases, crude pollen extracts, rather than purified proteins, were used for activity analyses (Cosgrove et al., 1997). In this study, we also identify four complete cDNAs for Zea m 1, which previously was known from a partial cDNA (Broadwater et al., 1993), and we report a surprising sequence divergence in Zea m 1 proteins.
RESULTS
Identification of Zea m 1 cDNA Clones
By screening a pollen cDNA library with the partial clone for Zea m 1 (GenBank Accession no. L14271), we identified multiple clones for two distinct Zea m 1 cDNAs, the longest of which were fully sequenced and deposited in GenBank as AY197352 and AY197353. The later clone is a longer version of L14271 and is essentially identical to EXPB1 (Wu et al., 2001), whereas the former clone is a very similar, but distinct, cDNA (see http://www.bio.psu.edu/expansins/ for naming conventions and gene lists). The predicted proteins are made up of 269 and 270 amino acids, have a signal peptide at the amino terminus, and possess the characteristic motifs found for expansins (Cosgrove et al., 2002). They have only approximately 58% protein sequence identity to Lol p 1, the archetype group 1 pollen allergen from ryegrass (Lolium perenne). Although the proteins encoded by the two cDNAs are 98% identical to each other, nucleotide sequence differences in the 5′- and 3′-untranslated regions and in the coding region indicate that these cDNAs represent two distinct genes in the inbred line from which the cDNA library was made. We have designated the gene corresponding to AY197352 as EXPB9 because it is a distinct gene from EXPB1. BLAST analysis of the maize expressed sequence tag (EST) database confirms that EXPB1 and EXPB9 are well represented in pollen and anther cDNA libraries and are not found in cDNA libraries made from tissues that lack pollen. That is, they appear to be pollen specific, as expected for a group 1 allergen.
By BLAST analysis of GenBank, including the maize EST database, we identified two additional maize cDNAs for a second class of Zea m 1 sequences, represented by GenBank AY104125 and AY104999. We have designated the corresponding genes as EXPB10 and EXPB11, respectively. The proteins encoded by these cDNAs are approximately 94% identical to each other and have even greater sequence identity to Lol p 1 (approximately 70%) than do EXPB1 and EXPB9 (see the alignment in Fig. 1). They have approximately 62% sequence identity with EXPB1 and EXPB9. Analysis of the maize EST database indicates that they too are well represented in pollen and anther-specific libraries and are not found in libraries made from other tissues. Moreover, differential screening of our pollen cDNA library also identified 11 clones corresponding to this second class of Zea m 1 (data not shown). Thus, expression all four genes also appears to be pollen specific.
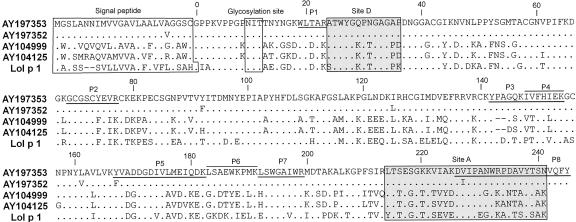
Figure 1.
Protein sequence alignment of Zea m 1 based on four distinctive cDNAs and comparison with Lol p 1 (GenBank accession no. X57678). Predicted proteins are identified by GenBank accession numbers. Dots represent residues that are identical to that of AY197353. Hyphens show gaps to maximize alignment. Boxed annotations include the signal peptide at the N terminus, the conserved glycosylation site near the N terminus of the mature protein, and the recognition sites for monoclonal antibodies for sites A and D. Underlined or above-lined regions indicate the tryptic peptides identified by electrospray ionization (ESI)-mass spectrometry (MS; Supplemental Table S1, http://www.plantphysiol.org).
We conclude from these results that the maize genome contains at least four pollen-expressed β-expansin genes that fall within the subgroup known as grass group 1 pollen allergens. This is consistent with Southern-blot analysis (Broadwater et al., 1993), which suggested that maize contains two to four genes for Zea m 1. This conclusion is confirmed by phylogenetic analysis (Fig. 2), which shows that these four sequences fall within the pollen allergen clade of β-expansins. We have divided the group 1 allergens into two classes (A and B), each containing with two maize genes. The maize EST database contains many entries corresponding to these four genes, some with slight sequence variations. Whether these sequence variations represent allelic variations (the libraries were made from different maize lines), sequencing errors, or possibly additional genes for Zea m 1 is not certain at this time.
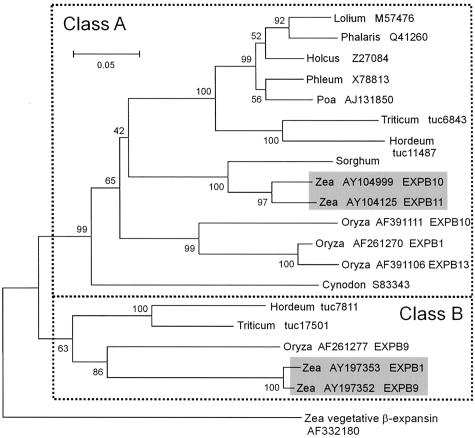
Figure 2.
Phylogenetic tree of protein sequences for Zea m 1 and other grass group 1 pollen allergens. The tree was constructed by neighbor joining and rooted using a maize vegetative β-expansin as the out group; bootstrap values are shown at each node, and GenBank Accession numbers are indicated after the genus name. For maize and rice, the expansin gene designations are also given, e.g. EXPB1. The sorghum sequence is from Knox and Suphioglu (1996b) and is not in GenBank. The Triticum and Hordeum sequences are based on EST assemblies (http://www.plantgdb.org/cgi-bin/PlantGDBblast), and the assembly number is given.
Fractionation of Maize Pollen Protein Yields Four Zea m 1 Isoforms
Maize pollen was extracted in acetate buffer, and the extracted proteins were fractionated by chromatography on carboxymethyl (CM)-Sepharose followed by chromatography on a CM-silica-based HPLC column (Fig. 3; Table I). Following the protocol of Cosgrove et al. (1997), we used heat-inactivated grass coleoptile walls to assay the fractions for rapid induction of wall extension, i.e. detectable within 5 min. About 7% of the total protein and all of the detectable activity were retained on CM-Sepharose. Upon elution with a linear salt gradient, about two-thirds of the eluted proteins had wall extension activity, resulting in approximately 19-fold purification (Table I).
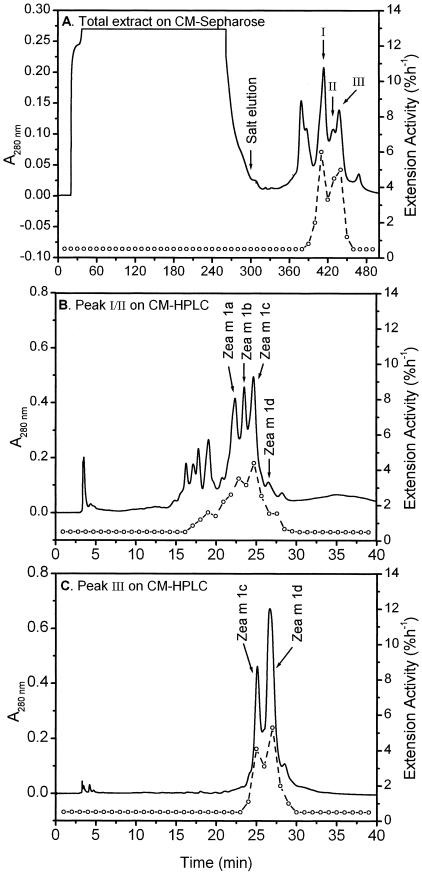
Figure 3.
Purification of Zea m 1 isoforms from maize pollen. A, Fractionation of the maize pollen extract by CM-Sepharose cation exchange chromatography yielded three peaks of activity (I–III). B, Fractionation of peaks I and II from A on a silica-based CM-HPLC column yielded four peaks of activity, which were identified as four isoforms of Zea m 1 by immunoblot analysis. C, Fractionation of peak III from A yielded isoforms Zea m 1c and 1d. Open circles, Wall extension activity of the corresponding fraction.
Table I.
Purification of Zea m 1 from maize pollen
Twenty grams of maize pollen was extracted, centrifuged, and filtered to give the crude extract. This extract was then subjected to conventional chromatography on CM-Sepharose followed by HPLC on CM-silica-based matrix. This purification was repeated two additional times with similar results.
| Purification Step | Protein | Activity | Specific Activity | Purification | Protein Yield |
|---|---|---|---|---|---|
| mg | % h-1 | % h-1mg-1protein | -fold | % | |
| Crude extract | 580.4 | 2,868 | 4.9 | 1 | 100 |
| CM-Sepharose | 35.1 | 3,212 | 91.5 | 19 | 6.0 |
| CM-HPLC | |||||
| Zea m 1a | 3.0 | 410 | 136.7 | 28 | 0.5 |
| Zea m 1b | 3.4 | 429 | 126.2 | 26 | 0.6 |
| Zea m 1c | 5.1 | 663 | 130.0 | 27 | 0.9 |
| Zea m 1d | 8.6 | 1205 | 140.1 | 29 | 1.5 |
For further purification, fractions including peak I/II and peak III were separately loaded onto a CM-silica-based HPLC column (Fig. 3, B and C). Four proteins with wall extension activity were purified to homogeneity as determined by SDS-PAGE (Fig. 4A) and native PAGE (Fig. 4B). As described below, these proteins were identified as isoforms of Zea m 1 and are designated Zea m 1a, Zea m 1b, Zea m 1c, and Zea m 1d, in the order of their elution from the CM-HPLC column. Each consists of a single polypeptide (Fig. 4A) of nearly identical size (Zea m 1a, 28.5 kD; Zea m 1b, 28.4 kD; Zea m 1c, 28.7 kD; and Zea m 1d, 29.0 kD, as estimated by SDS-PAGE).
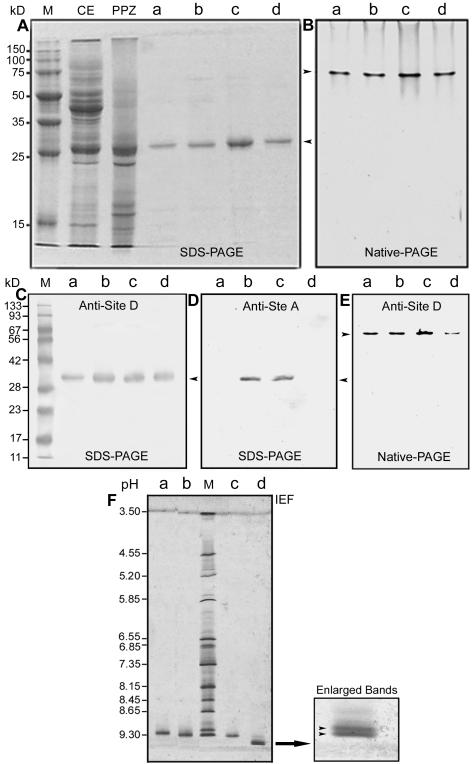
Figure 4.
Electrophoretic and immunoblot analysis of Zea m 1 from the different purification steps. A, Coomassie-stained SDS-PAGE. Lane M, Protein markers; lane CE, crude extract (40 μg); lane PPZ, partially purified Zea m 1 (20 μg) pooled from peaks I to III of the CM-Sepharose column; lanes a-d, Zea m 1a to d (approximately 2 μg lane-1) purified from the CM-HPLC column. B, Coomassie-stained native PAGE. Lanes a to d were loaded with 2 μg of purified Zea m 1a to d, respectively. Arrows indicate the position of Zea m 1 proteins. C, Immunoblot analysis of the Zea m 1a to d separated by SDS-PAGE and probed with McAb antisite D. D, Immunoblot analysis of Zea m 1a to d separated by SDS-PAGE and probed with McAb antisite A. E, Immunoblot analysis of Zea m 1a to d separated by native gel electrophoresis and probed with McAb antisite D. F, Isoelectric focusing (IEF) of Zea m 1a to d. Lane M, pI markers; lanes a to d, purified Zea m 1a to d (approximately 2 μg/lane), respectively. An enlargement of the Zea m 1d band is shown at the right.
The data in Table I indicate that Zea m 1 proteins are abundant in maize pollen, comprising approximately 0.1% of the total pollen mass or approximately 4% of the extracted protein. The relative proportions of Zea m 1a to d isoforms were 15%, 17%, 25%, and 43%, respectively. Because Zea m 1d was the most abundant isoform and was readily purified from maize pollen, we used it for most of the investigations in this paper. We also noted a small peak of activity in an HPLC fraction that lacked Zea m 1 isoforms (at 17–20 min in Fig. 3B); this fraction will be the subject of a future study.
Identification of Zea m 1 by Immunoblot and Amino Acid Sequence Analyses
The active proteins were identified as group 1 allergens by immunoblot analysis (Fig. 4, C–E) using monoclonal antibody (McAb) against Lol p 1 (antisite D, where the epitope is defined by the Lol p 1 peptide STWYGKPTGAGPK; Hiller et al., 1997; see also the annotation of the sequence alignment in Fig. 1). Lol p 1 is the group 1 allergen of perennial ryegrass pollen and has moderate sequence similarity to Zea m 1. Although the four Zea m 1 isoforms are very similar to each other in their biochemical properties, they differ somewhat in their antigenicity because two of the isoforms (Zea m 1a and 1d) were not recognized by a second McAb against Lol p 1 (antisite A, where the epitope is defined by the peptide YTTEGGTKSEVEDVIPEGWKADTSYSAK; Esch and Klapper, 1989; see also Fig. 1). This antibody did recognize Zea m 1b and Zea m 1c (Fig. 4D). Similar immunoblot results were obtained after protein separation by native PAGE (Fig. 4E) and by IEF (data not shown). In addition, these Zea m 1 isoforms were not recognized by two polyclonal antibodies raised against cucumber α-expansins EXP1 or EXP2; this is consistent with previous observations with a mixture of all Zea m 1 isoforms (Cosgrove et al., 1997).
The N-terminal amino acid sequence for Zea m 1d was determined by automated Edman degradation to be GPPKVPPGO?ITTNYNGKWL, which corresponds to the predicted protein sequence of two of the cDNAs isolated above (GenBank accession nos. AY197352 and AY197353; see Fig. 1), after removal of the signal peptide. The residue at position 9 is Hyp and the residue at position 10 (?) could not be identified because its yield in the sequencing reaction was too low to be detected. However, it is probably a glycosylated Asn because this position conforms to the structural motif “NXT/S” (X represents any amino acid except P) for N-linked glycosylation. Glycosylation interferes with amino acid identification by Edman degradation. A glycosylated Asn in this position is a conserved feature of group 1 allergens (Knox and Suphioglu, 1996b). It is not, however, universally characteristic of β-expansins in general because other members of this family (i.e. the “vegetative homologs”) often have glycosylation sites predicted at other sites (our own observation).
MS of the tryptic peptides of Zea m 1d indicates that it is encoded by AY197353 and not the other cDNAs in Figure 1. Supplemental Table S1 (see supplemental data at http://www.plantphysiol.org) shows that the masses of eight tryptic peptides perfectly matched with those calculated for the tryptic peptides predicted from AY197353. The positions of these peptides are indicated in Figure 1. Some of the peptides detected by ESI-MS remain unidentified (not shown), probably due to the instability of the expected products, posttranslational modifications, or perhaps allelic differences in sequence.
Therefore, we conclude that Zea m 1d corresponds to AY197353. Furthermore, the N terminus of Zea m 1c was found to be GPPKVPPGK?ITATYGKDWL. Assuming the uncertain residue at position 10 to be a glycosylated Asn (see above), this sequence is an identical match with that predicted for AY104125 and is distinct from the other three cDNAs identified in Figure 1. The N-terminal sequence for Zea m 1b was GPPKVPPGK?ITAKYGSDWL, which is an identical match for AY104999. The N-terminal sequence for Zea m 1a was GPPKVPPGK?ITANYGSDWL, which matches that predicted for AY104999, except at position 14, which is an N instead of the predicted K. Because the pollen was collected from a hybrid line different from the inbred source for AY104999, we suspect that Zea m 1a is allelic to AY104999; however, further work is necessary to resolve this point. We also make note that the N terminus in all four proteins matches that expected after removal of the predicted signal peptide (Fig. 1).
All Four Zea m 1 Isoforms Are Basic Glycoproteins
By IEF, we found that Zea m 1a, 1b, and 1c each consisted of a single band of pI 9.12, 9.30, and 9.32, respectively, whereas Zea m 1d split into two bands of pI 9.50 and 9.52 (Fig. 4F). The doublet for Zea m 1d may be due to a charge isoform, minor modification of Zea m 1d, or the presence of a closely related isoform, e.g. perhaps that encoded by AY197352. These results are comparable with the theoretical pI of 8.84 for AY197352/AY197353 and 8.23 for AY104125/AY104999.
All four Zea m 1 isoforms were demonstrated to be glycoproteins by use of the glycan detection kit (Boerhinger Mannheim Co., Indianapolis; data not shown). This is consistent with the “NXT/S” glycosylation motif found near the N terminus of Zea m 1. To confirm the glycosylation, we determined the molecular mass of Zea m 1d by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) and ESI-MS. The values, 27,986 D by MALDI-TOF and 28,066 D by ESI-MS, are approximately 1,200 D higher than the predicted mass of 26,883 D for the mature Zea m 1d protein, encoded by AY197353. Thus, the carbohydrate content of Zea m 1d is estimated to be about 4% and corresponds to that expected for a typical glycan chain decorating a single amino acid residue.
Characteristics of Wall Extension Activity by Zea m 1
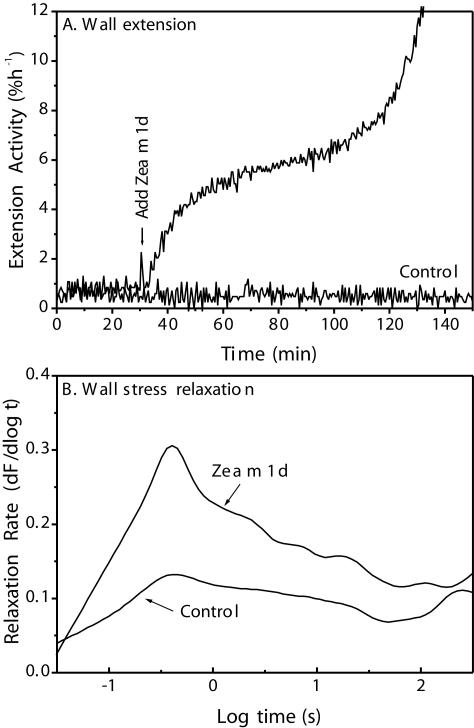
Wall extension assays showed that Zea m 1d rapidly induced extension of heat-inactivated cell walls from wheat (Triticum aestivum) coleoptiles (Fig. 5A). In addition, Zea m 1d enhanced the rate of stress relaxation of wheat coleoptile walls over a broad range of time (approximately 0.1–100 s; Fig. 5B). These properties are characteristic of expansins. Similar results were obtained with the other three Zea m 1 isoforms (data not shown).
Figure 5.
Wall-loosening activities of purified Zea m 1d. A, Induction of wall extension of heat-inactivated wheat coleoptile by Zea m 1d (0.3 mg mL-1). B, Stimulation of wall stress relaxation by 0.10 mg mL-1 Zea m 1d (average of 10 measurements).
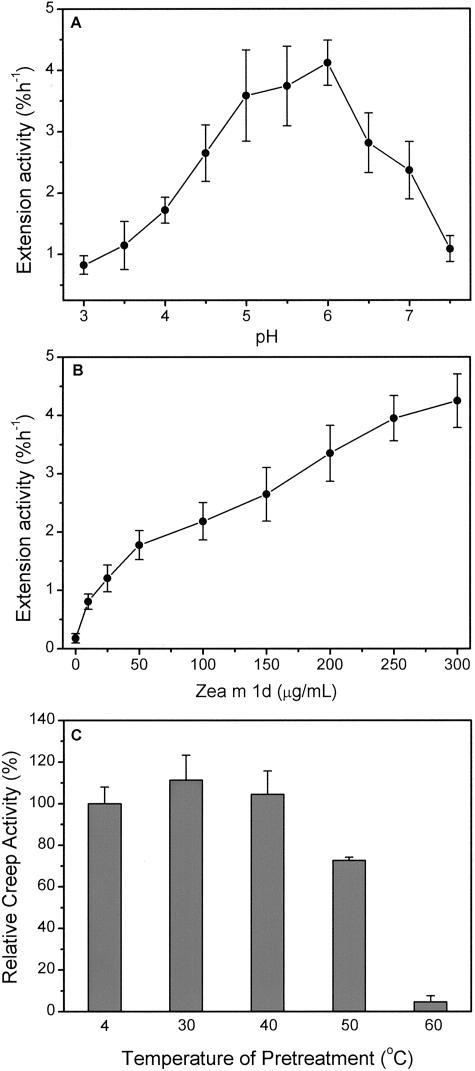
In a buffer of 50 mm 3,3-dimethylglutaric acid (DMGA)-NaOH, Zea m 1d had a broad optimum pH between 5.0 and 6.0 (Fig. 6A). This pH dependence is significantly different from that of α-expansins (see “Discussion”). The buffer used in the assay is an important consideration because we found that wall extension activity of native coleoptile walls (i.e. not inactivated by heat treatment) was significantly affected by the buffering species. When compared with acetate buffer, buffers based on DMGA, citratephosphate, piperazine-N,N′-bis(4-butanesulfonic acid), and piperazine-N,N′-bis(3-propanesulfonic acid) inhibited the native wall extension by 15% to 44%, whereas MES enhanced wall extension by 100%. Although the extension activity using DMGA buffer was 50% to 60% that using acetate buffer, DMGA buffer has the important advantage of adequate buffering capacity over the whole pH range tested here (pH 3.0–7.5).
Figure 6.
Dependence of wall extension activity of Zea m 1d on pH, concentration, and temperature pretreatment. A, Activity as a function of pH. Zea m 1d was applied at 0.3 mg mL-1 to heat-inactivated walls from wheat coleoptiles in 50 mm DMGA adjusted to different pH values. B, Activity as a function of Zea m 1d concentration, assayed in 50 mm sodium acetate (pH 4.5). Data represent means ± se of the induced extension rate, expressed as percentage increase in length per hour (mean of at least five replicates). C, Thermostability of Zea m 1d. Purified Zea m 1d was pre-incubated for 4 h at different temperatures and then assayed (at 0.3 mg mL-1) for wall extension activity with heat-inactivated wheat coleoptile wall in 50 mm sodium acetate (pH 4.5), with 5 mm dithiothreitol (DTT); activity is expressed as the percentage of control which was Zea m 1d incubated for the same time at 4°C. Data reported are the averages + SE of at least four measurements.
Wall extension activity increased with increasing concentrations of Zea m 1d in a biphasic manner (Fig. 6B). A steep increase in activity between 0 and 100 μg mL-1 was followed by a more gradual increase to concentrations of at least 300 μg mL-1. This concentration dependence is more complicated than that found for α-expansins (McQueen-Mason et al., 1992; Li et al., 1993), which show saturating activity at approximately 30 μg mL-1.
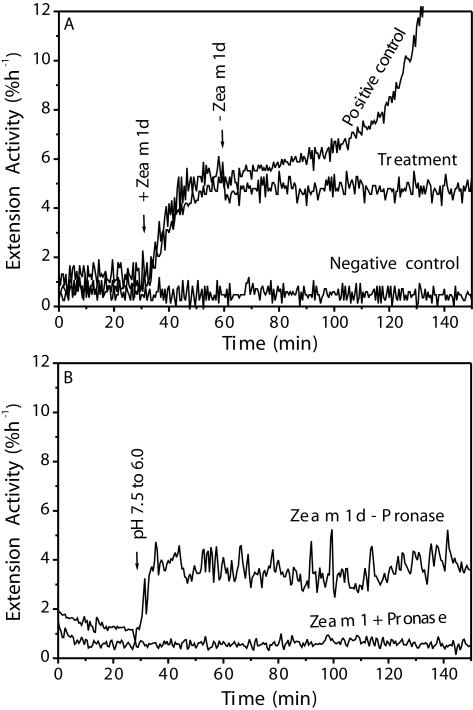
Zea m 1d appeared to bind tightly to the coleoptile wall because wall extension activity was not removed by exchanging the protein solution with fresh buffer lacking Zea m 1d (Fig. 7A). The persistence of wall extension activity also might be explained by an irreversible modification of the wall by Zea m 1d, such that the wall remained extensible even after expansin removal. To evaluate this possibility, we first incubated coleoptile walls with Zea m 1d, then treated the walls with Pronase, a powerful wide-spectrum protease preparation. Protease treatment completely abolished Zea m 1d-induced wall extension activity, whereas the control walls were still extensible (Fig. 7B). Taken together, these results indicate that Zea m 1d is not readily washed off of coleoptile walls and that the bound Zea m 1d continues to catalyze wall extension even after buffer exchange to remove the unbound Zea m 1d.
Figure 7.
Residual wall-loosening activity after removal of unbound Zea m 1 and after protease treatment. A, After 30 min of extension of the heat-inactivated wall in 50 mm sodium acetate (pH 4.5), the incubation solution was changed for one containing 0.3 mg mL-1 Zea m 1d, and the wall was extended for a further 30 min. The solution in the extensometer cuvette was exchanged 5× with buffer without Zea m 1d (“Treatment”). The negative control contained no Zea m 1d, whereas the positive control continued extending in 0.3 mg mL-1 Zea m 1d. B, Destruction of wall extension activity by Pronase. Heat-inactivated wheat coleoptiles were pre-incubated for 1 h with 0.1 mg mL-1 of Zea m 1d in 50 mm sodium acetate, pH 4.5. After three washes with 50 mm MES, 1 mm EDTA, and 5 mm DTT (pH 6.0), they were then further pretreated for 1 h at room temperature with 2 mg mL-1 of Pronase in the washing buffer. The walls were then assayed for residual Zea m 1 activity. At the time indicated by the arrow, the buffer was switched from pH 7.5 buffer to pH 6.0 buffer (50 mm DMGA in both cases). All of the above experiments were performed at least five times with similar results.
Another point to be noted is that walls treated continuously with pure Zea m 1d did not maintain a stable creep rate indefinitely, but the rate began to increase after approximately 10% extension, and this increase soon led to breakage (e.g. starting at 100–120 min in the positive control of Fig. 7A). Wall breakage was delayed or did not occur in the walls where the free Zea m 1 was removed by buffer exchange; this result suggests that breakage was caused by additional loosening action of the Zea m 1d in solution. This behavior differs from that of α-expansins, which induce prolonged extension before breakage. It also did not occur in crude pollen extracts, which suggests the presence of agents in the pollen extract that modify Zea m 1d extension activity.
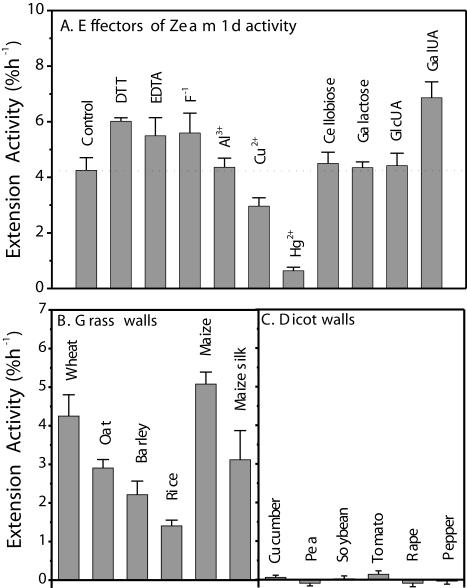
For further comparison with α-expansin properties (McQueen-Mason et al., 1992), we assessed the effect of a number of substances on Zea m 1d activity (Fig. 8A). At 10 mm, the thiol-reducing reagent DTT, the divalent ion chelator EDTA, and fluoride ion increased Zea m 1d activity by 42%, 29%, and 32%, respectively. At 2 mm, Al3+ did not affect the Zea m 1d activity, although it prevented walls from breaking at the later stage of the extension assay. Cu2+ inhibited the extension activity by 30%, and the sulfhydryl group-modifying reagent Hg2+ (2 mm) almost completely abolished Zea m 1 activity, suggesting involvement of a sulfhydryl group in maintaining Zea m 1 in an active conformation. GalUA, at 100 mm, enhanced Zea m 1d activity by 62%, whereas GlcUA, Gal, and cellobiose had little effect at the same concentration.
Figure 8.
Dependence of Zea m 1 activity on various agents and on source of cell wall. A, Effects of thiol reagent, chelator, inorganic ion, sugar, and sugar acids on Zea m 1 activity. Heat-inactivated wheat coleoptiles were initially extended in 50 mm sodium acetate (pH 4.5). After 30 min, the bathing solution was replaced by the same buffer containing 0.3 mg mL-1 Zea m 1d and effectors: 10 mm DTT, EDTA, F-1, 2 mm Al3+, Cu2+, Hg2+, or 100 mm Gal, cellobiose, and GlcUA, and GalUA. Data represent means + se of the induced extension rate, expressed as percentage increase in length per hour (n = 5–8). B and C, Substrate preference of Zea m 1d. Heat-inactivated walls of Type II (B) and Type I (C) were assayed using Zea m 1d (0.3 mg mL-1). Wall extension activity is expressed as the increase in extension rate (percentage per hour). Negative data arise in some Type I walls because of nonzero extension rates before addition of Zea m 1d. Data are means + se of at least five measurements.
Zea m 1 Acts Selectively on Grass Cell Walls
Purified Zea m 1d induced extension of coleoptile cell walls from various grass species and walls from maize silks (Fig. 8B). In contrast, dicot cell walls did not extend in response to Zea m 1d (Fig. 8C). Similar results were obtained with the other three Zea m 1 isoforms (data not shown). This result is in agreement with the previous observation of Cosgrove et al. (1997) obtained with crude maize pollen extracts and shows that grass walls are much more responsive to all four Zea m1 isoforms than dicot (or Type I) walls.
Zea m 1 Is Resistant to Denaturation by Methanol Boiling and Heat Treatments
In earlier studies, both the native wall extension activity (Cosgrove, 1989; Keller and Cosgrove, 1995; Wu et al., 1996) and purified α-expansin activity (McQueen-Mason et al., 1992) were shown to survive methanol boiling, a treatment that is commonly used to eliminate enzymatic activity before viscoelastic assays of isolated cell walls (Cosgrove, 1993). We found that Zea m 1 was likewise resistant to methanol boiling. When maize pollen was boiled for 5 min in 100% methanol, approximately 80% of the extension activity was retained in pollen extracts, as compared with that from untreated pollen. Furthermore, when lyophilized Zea m 1d was suspended in pure methanol, boiled for 5 min, and resuspended in 50 mm sodium acetate and 5 mm DTT (pH 4.5), about 85% of the Zea m 1d extension activity was recovered, as compared with protein not boiled in methanol. Thus, resistance to denaturation by hot methanol seems to be a property characteristic of both α- and β-expansins.
To assess its sensitivity to heat denaturation in aqueous solutions, Zea m 1d was pre-incubated for 4 h in 50 mm sodium acetate and 5 mm DTT (pH 4.5) at different temperatures, then assayed for wall extension activity (Fig. 6C). Zea m 1d retained full activity at 30°C and 40°C, 72% of its activity at 50°C, and none of its activity at 60°C. Moreover, Zea m 1d maintained full activity after incubation at 31°C for 48 h or after storage at -20°C for more than 2 years. Similarly, about 91% of the Zea m 1d activity was recovered after it was lyophilized to a dried powder and stored at 4°C for 6 months. Thus, native Zea m 1d is very stable and does not exhibit the instability of recombinant Phl p 1, the group 1 allergen of Phleum pretense (Grobe et al., 1999).
DISCUSSION
In this study, we developed a simple and efficient protocol to purify Zea m 1, a pollen β-expansin (group 1 allergen) of maize, and we characterized the physical properties and wall rheological activity of the purified isoforms. This is an advance over the previous report of Zea m 1 wall-loosening activity (Cosgrove et al., 1997), which used either whole pollen extracts or partially purified fractions in which the different Zea m 1 isoforms were not separated. We resolved four Zea m 1 isoforms, each of which induced rapid wall extension, enhanced wall stress relaxation, and was selective for grass cell walls. A similar selectivity of activity was reported previously for whole pollen extracts from maize (Cosgrove et al., 1997) but not tested for purified Zea m 1.
Although our results confirmed the wall extension activity of purified Zea m 1 isoforms and showed that they have the same specific activity (Table I), we found significant and unexpected activity differences compared with α-expansins, most notably in the pH dependence of wall extension, the amount of protein needed to induce wall extension, and the tendency to cause cell wall breakage after a brief period of extension. These differences in properties suggest that the activity of Zea m 1 (and by extrapolation group 1 allergens from other grass species) differs from that of other α- and β-expansins believed to mediate acid-induced wall extension in vegetative tissues. One might consider that the premature wall breakage found in our study is due to some peculiarity of the grass wall that becomes weakened upon heat inactivation. However, this is not the case because breakage is less evident when such walls are treated with α-expansins or with whole pollen extracts (Cosgrove et al., 1997). The latter observation also suggests that additional factors in the pollen extracts may modify the action of Zea m 1 on cell walls. Furthermore, wall breakage at an early stage of extension is not typical of acid-induced extension of native walls from grasses (Rayle and Cleland, 1972; Cleland et al., 1987; Cosgrove and Li, 1993; Li et al., 1993; Cho and Kende, 1997). Thus, the wall breakage hints at an additional activity in Zea m 1 that may not be characteristic of all expansins or of acid growth in vegetative tissues. The biphasic character of the Zea m 1 concentration dependence likewise hints at an additional activity not found with α-expansins, which have a simple, monophasic, saturating concentration dependence (McQueen-Mason et al., 1992; Li et al., 1993, 1998). Finally, compared with α-expansins, Zea m 1 must be supplied at relatively high concentration to induce wall extension.
These unexpected characteristics of Zea m 1 activity may relate to the specialized role of these proteins in pollen function (Cosgrove et al., 1997), as opposed to control of cell enlargement. Consistent with this idea, phylogenetic analysis indicates that grass pollen allergens are distinctive in protein sequence from the majority of β-expansins expressed in vegetative tissues of rice and maize. These differences in sequence and protein characteristics may indicate functional specialization related to the unusual role of these grass pollen-specific expansins.
Also suggestive of a distinctive role, we note that Zea m 1 has a broad pH optimum (between 5 and 6) for wall extension activity, whereas pH optima for α-expansins and native acid-induced wall extension are 4.5 or lower (Cosgrove, 1989; McQueen-Mason et al., 1992; Li et al., 1993, 1998). Although the wall pH of maize silks has not been measured directly, it is likely to be in the range of pH 5 to 6, as reported in many other plant tissues (Vesper, 1985; Felle, 1998; Peters et al., 1998; Soga et al., 2000; Fasano et al., 2001). This means that Zea m 1 will likely function with maximal activity at the normal pH found at its site of action and that modest changes in silk wall pH will have little effect on wall loosening by Zea m 1. Thus, Zea m 1 does not appear to be adapted as an agent of “acid growth.”
In addition to its pH dependence and its tendency to cause wall breakage, Zea m 1 also differs from α-expansin in its selectivity of grass walls over dicot walls, its abundance, its glycosylation, and its solubility. The α-expansin proteins that have been studied to date are relatively rare, non-glycosylated proteins (for review, see Cosgrove et al., 2002), whereas Zea m 1 and group 1 allergens from other grass species are glycosylated proteins abundantly expressed in pollen. Unlike α-expansins, which readily precipitate at concentrations <100 μg mL-1 (D.M. Durachko and D.J. Cosgrove, unpublished data), we found Zea m 1 to be highly soluble and readily concentrated to more than 30 mg mL-1. This high solubility probably is due to the high number of charged residues in Zea m 1, a property that is common to other group 1 allergens. Like α-expansins, Zea m 1 tolerates boiling methanol, is stimulated by thiol reductants and chelators of divalent cations, and is inhibited by Cu2+ and Hg2+; unlike α-expansins, it is not inhibited by Al3+.
At present, it is not possible to conclude that the distinctive properties of Zea m 1 are also characteristic of β-expansins in general. This is so because β-expansin proteins from vegetative tissues have not been characterized with regard to their wall extension activities and because the group 1 allergens are distinctive in sequence. A β-expansin (CIM1) from soybean (Glycine max) cell cultures has been characterized partially (Downes et al., 2001) but not with respect to its potential wall extension properties. It is apparent from sequence analysis that α-expansins typically lack motifs for N-linked glycosylation, whereas β-expansins characteristically possess such motifs and have been shown to be glycosylated in the few examples studied (Marsh, 1975; Howlett and Clarke, 1981; Cottam et al., 1986; Petersen et al., 1995; Knox and Suphioglu, 1996b; Downes et al., 2001). However, we doubt that β-expansins from vegetative tissues cause early cell wall breakage or have a pH optimum >5, as found for Zea m 1, because acid-induced extension of native walls (not heat inactivated) from grass coleoptiles does not exhibit these properties, and it is likely that β-expansins are major catalysts of acid-induced extension in grass walls (Cosgrove and Li, 1993; Li et al., 1993; Cho and Kende, 1997; Cosgrove et al., 1997).
Although the work reported here was not undertaken to address directly the question of the possible proteolytic activity of grass group 1 allergens (Grobe et al., 1999, 2002; Li and Cosgrove, 2001), some of our observations bear on this question. We found Zea m 1 to be very stable during extraction, purification, and storage, to the extent that addition of protease inhibitors was not required during these steps. The addition of 5 mm DTT stabilized activity and prevented formation of dimers, trimers, etc. through disulfide bonds (data not shown). Our observations are consistent with the notable stability of most natural and recombinant group 1 allergens. For example, about 100 mg of Lol p 1 was purified from ryegrass pollen by multiple steps including time-consuming conventional chromatography without use of protease inhibitors (Johnson and Marsh, 1965b; Marsh et al., 1966). Suck et al. (1999) purified milligrams of natural Phl p1 by two chromatographic steps without addition of protease inhibitors except EDTA (a metallo-protease inhibitor).
In contrast, Grobe et al. (1999) concluded that recombinant Phl p1 was a protease because when expressed in Pichia pastoris, it was highly unstable and was rapidly degraded; they reported it to have some characteristics of a Ser protease, although their sequence analysis suggested it to be a Cys protease. Undermining these results, the same group (Poppelmann et al., 2002) later reported that expression of Phl p 1 in P. pastoris induced a highly active Ser protease. Our experience with Zea m 1 purification confirms our recent conclusion that grass group 1 allergens do not possess proteolytic activity (Li and Cosgrove, 2001), and we suspect that the instability of recombinant Phl p1 reported by Grobe et al. (1999) was due to protease contamination.
Downes et al. (2001) studied the proteolytic processing of CIM1, a vegetative β-expansin expressed in soybean cell cultures. The protein that accumulated in the culture medium was proteolytically cleaved in discrete steps, including removal of a short peptide containing a predicted glycosylation site at the N terminus. We did not find evidence for such proteolytic cleavage in any of the four Zea m 1 isoforms. Because Zea m 1 isoforms were active in causing wall extension, proteolytic processing is not necessary for expansin activity (see also Li and Cosgrove, 2001). The significance of proteolytic processing of CIM1 in soybean cultures is unknown but probably is the first stage in CIM1 degradation by extracellular proteases.
Zea m 1 Isoforms
The presence of multiple isoforms of Zea m 1 is typical of the situation found in other grass pollens. Multiple forms (or isoallergens) of grass group 1 pollen allergens were first described by Johnson and Marsh (1965a, 1965b) for Lol p 1 from ryegrass pollen. Similarly, four isoforms of Poa p 1 from Poa pratensis were observed by two-dimensional gel electrophoresis and immunoblotting (Ekramoddoullah, 1990) and six isoforms of Phl p 1 from P. pretense (Petersen et al., 1993, 1995).
There are at least two possible reasons for such isoform multiplicity, namely: (a) multiple genes or alleles encoding somewhat different forms of the protein, and (b) variable posttranslations modifications, including glycosylation, hydroxylation of Pro residues, disulfide bond formation, Met oxidation, etc. (Petersen et al., 1993, 1995, 1997; Knox and Suphioglu, 1996b; Hiller et al., 1997). In maize, we found two divergent classes of Zea m 1 cDNAs with quite different protein sequence (only approximately 62% identical to each other). This sequence divergence is larger than what has been noted previously. Each of these two classes is encoded by at least two maize genes, giving a total of at least four genes encoding Zea m 1 isoforms. A similar situation seems to hold for rice, which likewise has both divergent classes of group 1 allergens (Fig. 2), with three members in Class A and one member in Class B. BLAST searches of the rice EST database (data not shown) indicates that these four rice genes are expressed in panicles at the flowering stage, i.e. when mature pollen is present; thus, they are presumptive group 1 allergens. EST analysis likewise indicates both classes are also found in wheat and barley (Hordeum sativum; Fig. 2). Other grass species lack Class B representatives in GenBank. It is possible that these other grass species also contain Class B group 1 pollen allergens, but they simply have not been identified yet due to the limited sequence analysis in these species; alternatively, Class B group 1 allergens may be present only in a subset of grasses. It is also notable that although the group 1 allergens are members of the β-expansin gene family, phylogenetic analysis always shows them to be clustered and separated from the main group of β-expansins. This divergence in sequence and specificity of expression may point to a significant divergence in biochemical function that is specific to pollen function.
Despite their sequence divergence, the four isoforms of Zea m 1 that we purified and characterized in this study show rather similar biochemical characteristics, as has been found for other grass pollen group 1 isoallergens (Knox and Suphioglu, 1996a, 1996b). The presence of multiple group 1 isoforms in many different grasses points to a selection pressure maintaining this trait. We suspect that the multiple isoforms may have distinctive developmental or biochemical functions that have not yet been discerned from in vitro analyses.
MATERIALS AND METHODS
Chemicals
Mouse monoclonal antibodies (antisites A and D) raised against Lol p 1 were kindly provided by Dr. David G. Klapper (Department of Microbiology and Immunology, University of North Carolina School of Medicine, Chapel Hill). Ammonium persulfate (electrophoresis reagent), Coomassie Brilliant Blue R-250, Ponceau S, cytochrome C, and DMGA were purchased from Sigma-Aldrich Corp. (St. Louis). Piperazine-N,N′-bis(4-butanesulfonic acid) and piperazine-N,N′-bis(3-propanesulfonic acid) were purchased from GFS Chemicals, Inc. (Columbus, OH). Methanol (HPLC grade) was purchased from Mallinckrodt Baker, Inc. (Phillipsberg, NJ). Sequencing grade modified porcine trypsin was purchased from Promega (Madison, WI). All other chemicals used for electrophoresis were obtained from Research Organics, Inc. (Cleveland).
Plant Materials
Maize (Zea mays) pollen and silks were collected in August 1998 and 1999 from maize plants grown in State College, PA, and stored at -80°C. Seeds of wheat (Triticum aestivum L. cv Pennmore), oat (Avena sativa L. cv Olge), barley (Hordeum sativum L. cv Barsoy), rice (Oryza sativa L. cv Nipponbare), and maize cv FR1064 × LH185 were germinated in moist Metro-Mix 360 growing medium (Scotts-Sierra Horticultural Products Co., Marysville, OH) at 27°C to 29°C in complete darkness for 3 to 5 d. Seeds of cucumber (Cucumis sativus L. cv Burpee Pickler), pea (Pisum sativum L., cv Alaska), soybean (Glycine max L. Merr. cv Williams 82), tomato (Lycopersicon esculentum cv Rutgers), oilseed rape (Brassica napus cv Qinyu No. 2), and pepper (Capsicum annuum L. cv Long Red Cayenne) were sown on wet germination paper (Kimpak K-22, Seedburo Equipment Co., Chicago) at 27°C to 29°C in darkness for 4 to 6 d. Coleoptiles from etiolated grass seedlings were cut, gently abraded by rubbing them between two fingers coated with a slurry of well washed carborundum (320 grit, Fisher Scientific Inc., Fair Lawn, NJ), separated from primary leaves, and then stored at -20°C before use. Dicot hypocotyls were quickly excised from etiolated seedlings under room light and directly frozen at -20°C for wall extension assays.
cDNA Isolation
Poly(A+) RNA was isolated from young starch-filled pollen from maize inbred line K21, and a cDNA library was constructed in the Lambda ZAP cloning vector (Stratagene Corp., La Jolla, CA) with EcoRI/NotI adapters (Invitrogen Corp., Carlsbad, CA). The library was screened by hybridization with a partial Zea m 1 cDNA (GenBank Accession no. L14271). Eleven clones were isolated and upon sequencing were found to correspond to two distinctive cDNAs identified here as GenBank AY197353 (seven clones) and GenBank AY197352 (four clones). One additional clone (pSF23) was identified that closely matched AY197353, except for two small insertions in the 3′-untranslated region.
Eleven cDNAs corresponding to GenBank AY104125 and AY104999 were also identified by differential screening of the pollen library. Duplicate filters were hybridized utilizing both a subtracted probe (made by hybridization of P32-labeled pollen cDNA with excess biotinylated endosperm mRNA, reaction with streptavidin, followed by phenol extraction) and an unsubtracted pollen P32-cDNA probe; in the latter case, signals were compared with those on a duplicate filter hybridized with endosperm P32-cDNA.
Purification of β-Expansin Proteins
Approximately 20 g of maize pollen was extracted in 80 mL of 50 mm sodium acetate (pH 4.5) for 1 h at 4°C. The extract was centrifuged at 15,000g at 4°C and then loaded onto a CM-Sepharose Fast Flow (Pharmacia Biotech AB, Uppsala) column (15 × 300 mm) equilibrated in 20 mm sodium acetate (pH 4.5). The column was washed with the same buffer until the A280 returned to baseline and then eluted at 2 mL min-1 with a linear gradient of NaCl (0–500 mm in 2.5 h) followed by 1 h in 500 mm NaCl in the same buffer. Fractions were collected at 4 mL per tube.
The fractions from CM-Sepharose column chromatography were desalted and concentrated by ultrafiltration (Ultrafree-15 centrifugal filter device-Biomax 10K NMWL membrane; Millipore Co., Bedford, MA). Active fractions were pooled and filtered (Ultrafree-MC 0.45-μm centrifugal filter, Millipore Co.) and then loaded onto a silica-based CM-HPLC column (4.6 × 250 mm, Synchropak CM300/6.5 μm, ISCO Inc., Lincoln, NE). Proteins were eluted at 1 mL min-1 with a linear gradient of 0 to 550 mm NaCl and 20 mm sodium acetate (pH 4.5) for 40 min. The fractions were collected by peak mode and desalted as above before storing at -20°C. Proteins were quantified colorimetrically with the Coomassie Plus Protein Assay Reagent (Pierce, Rockford, IL) according to the manufacturer's instructions.
Wall Extension and Stress Relaxation Assays
Wall extension activity was measured with a constant load extensometer (Cosgrove, 1989) using grass coleoptile walls as a sensitive substrate for β-expansin activity (Cosgrove et al., 1997). In brief, sample tissues (except grass coleoptile) prepared as above were quickly abraded with carborundum to disrupt the cuticle, submerged in boiling distilled water for 15 s, and secured between two clamps (with 5 mm between the clamps) under constant tension. To compensate for the varying thickness of the cell wall specimens, a 5-g weight was used to keep the silk walls under constant tension, whereas a 20-g weight was used for the coleoptile and hypocotyl walls. Protein fractions in 50 mm sodium acetate (pH 4.5) were added to the extensometer cuvettes (volume = 0.20 mL) after the walls were initially bathed in the same buffer for about 30 min. Protein fractions were assayed using wheat coleoptile walls unless otherwise indicated because they were easy to prepare and had low breakage and a lower baseline extension rate.
For stress relaxation assays, wheat coleoptiles, prepared as described above, were clamped (5 mm between the jaws) in a custom-made tensile tester (Cosgrove, 1989). Heat-inactivated walls were pretreated for 10 min in either 50 mm sodium acetate (pH 4.5) or the same buffer containing 0.1 mg mL-1 of each Zea m 1 isoform, then stored on ice before stress relaxation measurement. Each tissue segment was extended at a rate of 170 mm min-1 until a stress of 20 g was attained and thereafter held at a constant strain. Stress was recorded over 5 min by a computer with a minimum sampling rate of 2 ms, gradually increasing to 2 s. The relaxation spectrum was calculated as the derivative of the stress with respect to log (time).
Electrophoresis
Proteins were separated by discontinuous SDS-PAGE in a minigel apparatus (Protean II, Bio-Rad Laboratories, Hercules, CA) using 3.4% (w/v) stacking polyacrylamide gel and 12% (w/v) separation gel according to the method of Laemmli (1970). Samples were first heated to 95°C for 5 min in sample buffer with or without 100 mm DTT. Minigels containing proteins were stained with 0.1% (w/v) Coomassie Brilliant Blue R-250 in a solution containing 10% (v/v) acetic acid and 30% (v/v) methanol. After destaining, gels were photographed with a Kodak Digital Science DC40 camera (Eastman Kodak Co., Rochester, NY), and the Mr of each Zea m 1 isoform was estimated with Kodak 1d image analysis software. The protein marker for SDS-PAGE was from Novagen Inc. (catalog no. 69149, Madison, WI).
Native PAGE was modified from Panyim and Chalkley (1969) and performed on a Bio-Rad Mini Protean II system. The gel contained 10% (w/v) acrylamide, 0.27% (w/v) bisacrylamide, 50 mm acetic acid (pH 4.5), and 2.5 m urea. Samples were loaded in 50 mm sodium acetate (pH 4.5) and 10% (w/v) Suc and run in 50 mm sodium acetate (pH 4.5) at 120 V for 2 h. Bromphenol blue was added to sample at a final concentration of 0.01% (w/v) to improve visibility of proteins during loading samples. The electrodes were reversed at the power supply so that the positively charged Zea m 1 protein could migrate to the bottom of the gel (the negative cathode). The gels were stained as described for SDS-PAGE gels. Cytochrome C was used as a front dye for indicating the progress of electrophoresis.
For IEF, Ampholine PAGplate (pH 3.5–9.5; T = 5%, C = 3%) and Broad pI Calibration Kit (pH 3–10) were from Pharmacia Biotech AB. The gel was mounted onto a water-cooled (15°C) 2117 Multiphor II electrophoresis unit (Pharmacia Biotech AB), and 2 μg of each Zea m 1 isoform and 20 μL of pI standards were focused for 1.5 h at 1,500 V, 50 mA, and 30 W. Immediately after IEF, the gel was fixed, washed, and visualized with Coomassie Brilliant Blue R-250. The migrations of the Zea m 1 bands and pI standards were documented with a Kodak DC40 digital camera, and the pI of each Zea m 1 isoform was determined by the Kodak analysis software.
Immunoblot Analysis
For SDS-PAGE and native PAGE gels, proteins were electrophoretically transferred on a semidry blot apparatus (Trans-Blot SD Semi-Dry Electrophoretic Transfer Cell, Bio-Rad Laboratories) to a Protran BA nitrocellulose membrane (Schleicher & Schull, Keene, NH). Transfers were carried out in a solution of 192 mm Gly, 25 mm Tris, and 20% (v/v) methanol at 1.2 mA cm2 for 1 h. Proteins in IEF gels were transferred by capillarity onto the same membrane for 1.5 h. After transfer, membranes were stained with Ponceau S solution for protein detection. For immunodetection of Zea m 1 proteins, the membranes were blocked with 10% (v/v) horse serum in phosphate-buffered saline containing 0.05% (v/v) Tween 20 and 5 mm sodium azide, incubated for 2 h with the same solution containing mouse monoclonal antibodies against Lol p 1 (antisite A, 1:150 [v/v] dilution; antisite D, 1:200,000 [v/v] dilution), washed twice with phosphate-buffered saline containing 0.05% (v/v) Tween 20 and 5 mm sodium azide and with Tris-buffered saline containing 0.05% (v/v) Tween 20 and 5 mm sodium azide, and then incubated for 1 h with goat anti-rabbit IgG (heavy and light chains)-conjugated alkaline phosphatase conjugate (dilution of 1:1,000 [v/v]; Sigma-Aldrich Corp.). The protein-containing membranes were washed and then developed with 0.1 mg mL-1 5-bromo-4-chloro-3-indolyl phosphate and 0.2 mg mL-1 nitroblue tetrazolium (Sigma-Aldrich Corp.) in the substrate buffer (100 mm Tris/HCl [pH 9.5], 100 mm NaCl, and 5 mm MgCl2). The prestained protein ladder was purchased from Life Technologies/Gibco-BRL (catalog no. 10748-010, Rockville, MD).
Carbohydrate Detection
The carbohydrate moiety of Zea m 1 was detected by the glycan detection kit (Boerhinger Mannheim Co.) according to the manufacturer's description. In brief, approximate 1 μg of each Zea m 1 isoform was dissolved in 0.1 m sodium acetate buffer (pH 5.5). The vicinal hydroxyl groups of Zea m 1 carbohydrate moieties were oxidized to aldehydes in 30 mm sodium meta-periodate for 20 min in the dark at room temperature and labeled with digoxigenin-succinyl-epsilon-amidocaproic acid hydrazide for 1 h at room temperature. The labeled Zea m 1 isoforms were then separated on SDS-PAGE, transferred to a nitrocellulose membrane, and visualized as described above for western blotting, except that alkaline phosphatase-labeled anti-digoxigenin antibody was used.
MS
The highly purified Zea m 1d was analyzed at the Penn State MS Center. MALDI-TOF mass spectra were obtained on a Voyager-DE STR MALDI-TOF (PerSeptive Biosystems, Foster City, CA), whereas ESI-MS was carried out on Mariner Electrospray-TOF workstation (PerSeptive Biosystems, Framingham, MA), which was coupled with a model 1100 HPLC (Hewlett-Packard Co., Palo Alto, CA). For MALDI-TOF analysis, 1 μL of the protein sample containing Zea m 1d was dissolved into water:methanol:acetic acid/(49:50:1 [v/v]) at approximately 2 pmol μL-1 and then mixed with 1 μL of 10 μg mL-1 sinapinic acid in acetonitrile:water:trifluoroacetic acid (70:29:0.1 [v/v]) as a matrix solution. One microliter of this mixture was deposited on the target plate and dried to form uniform crystals. Spectra were accumulated from 76 laser shots (nitrogen laser = 337 nm). For ESI-MS analysis, the Zea m 1d Protein or its tryptic peptides were automatically loaded into the HPLC, linearly eluted using water:acetonitrile:formic acid solvent system (pH 2.5) at 50 μL min-1 from a microbore column (BetaBasic C18, 1 × 50 mm, Keystone Scientific Co., Bellefonte, PA) and directly introduced into ESI-MS system. The mass scale was calibrated with sodium trifluoroacetate cluster ions before sample analysis.
For tryptic peptide analysis, 25 μL of Zea m 1d solution, containing 7.5 μg of Zea m 1d, was dried under vacuum. After the protein was redissolved in 50 mm ammonium bicarbonate (pH 8.5), 1 μg of sequencing-grade modified trypsin was added, and the mixture was incubated at 37°C for 15 h. The proteolytic fragments were then stored at -80°C for ESI-MS analysis. Peptide mass data were collected and then analyzed with web-based software provided by the ExPASy Molecular Biology Server (http://www.expasy.org).
N-Terminal Amino Acid Sequence Analysis
Samples of HPLC purified Zea m 1a to d were subjected to N-terminal amino acid sequence analysis at the Macromolecular Core Facility of College of Medicine (The Pennsylvania State University, Hershey). Approximately 1 nmol of Zea m 1d was dissolved in a minimal amount of neat trifluoroacetic acid and spotted onto a polyvinylidene difluoride membrane for sequencing by automated Edman degradation on a pulsed-liquid-phase microsequencer (model 477A Protein Sequencer, Perkin-Elmer/Applied Biosystems, Foster City, CA) with an online 120A HPLC for analyzing the phenylthiohydantoin amino acid derivatives.
Phylogenetic Analysis
Mature protein sequences (without the signal peptide) were aligned using the slow-accurate method of ClustalW within the Megalign program (DNASTAR, Inc., Madison, WI). Method parameters were: gap penalty of 10 and gap length penalty of 0.10. Phylogenetic trees were constructed by the neighbor-joining method with MEGA2 version 2.1 software (Sudhir Kumar, Koichiro Tamura, Ingrid B. Jakobsen, and Masatoshi Nei, Arizona State University, Tempe), using p distances, pair-wise deletion, and 1,000 replications.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
The authors are grateful to Dr. David G. Klapper (Department of Microbiology and Immunology, University of North Carolina School of Medicine, Chapel Hill) for the gifts of monoclonal antibodies against Lol p 1, Ms. Anne Stanley (Macromolecular Core Facility, College of Medicine, the Pennsylvania State University, Hershey) for N-terminal amino acid sequence analysis of Zea m 1, Dr. Mark W. Shieh (Pennsylvania State University, University Park, PA) for sequencing of Zea m 1 clones, and Dr. Teh-hui Kao (Department of Biochemistry and Molecular Biology, the Pennsylvania State University, University Park) for use of the Multiphor II electrophoresis unit. We also thank Daniel M. Durachko, Melva Perich, Mara Guttmann, Mark D. Spiro, Mark W. Shieh, Sheng Yuan, Tanya Shcherban, and Yajun Wu (Pennsylvania State University, University Park) for help with maize pollen collection.
This work was supported by the U.S. Department of Energy (grant no. DE–FG02–84ER13179) and by the National Institutes of Health (grant no. GM60397).
The online version of this article contains Web-only data. The supplemental material is available at http://www.plantphysiol.org.
References
- Broadwater AH, Rubinstein AL, Chay CH, Klapper DG, Bedinger PA (1993) Zea mI, the maize homolog of the allergen-encoding Lol pI gene of rye grass. Gene 131: 227-230 [DOI] [PubMed] [Google Scholar]
- Carpita NC (1996) Structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol 47: 445-476 [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1-30 [DOI] [PubMed] [Google Scholar]
- Cho HT, Kende H (1997) Expansins and internodal growth of deepwater rice. Plant Physiol 113: 1145-1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland RE, Cosgrove DJ, Tepfer M (1987) Long-term acid-induced wall extension in an in-vitro system. Planta 170: 379-385 [PubMed] [Google Scholar]
- Cosgrove DJ (1989) Characterization of long-term extension of isolated cell walls from growing cucumber hypocotyls. Planta 177: 121-130 [PubMed] [Google Scholar]
- Cosgrove DJ (1993) Wall extensibility: its nature, measurement and relationship to plant cell growth. New Phytol 124: 1-23 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (2000) Loosening of plant cell walls by expansins. Nature 407: 321-326 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Bedinger PA, Durachko DM (1997) Group I allergens of grass pollen as cell wall-loosening agents. Proc Natl Acad Sci USA 94: 6559-6564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ, Li Z-C (1993) Role of expansin in cell enlargement of oat coleoptiles: analysis of developmental gradients and photocontrol. Plant Physiol 103: 1321-1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ, Li L-C, Cho H-T, Hoffmann-Benning S, Moore RC, Blecker D (2002) The growing world of expansins. Plant Cell Physiol 43: 1436-1444 [DOI] [PubMed] [Google Scholar]
- Cottam GP, Moran DM, Standring R (1986) Physicochemical and immunochemical characterization of allergenic proteins from rye-grass (Lolium perenne) pollen prepared by a rapid and efficient purification method. Biochem J 234: 305-310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes BP, Steinbaker CR, Crowell DN (2001) Expression and processing of a hormonally regulated beta-expansin from soybean. Plant Physiol 126: 244-252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekramoddoullah AKM (1990) Two dimensional gel electrophoretic analysis of Kentucky bluegrass and rye grass pollen allergen. Int Arch Allergy Appl Immunol 93: 371-377 [DOI] [PubMed] [Google Scholar]
- Esch RE, Klapper DG (1989) Isolation and characterization of a major cross-reactive grass group I allergenic determinant. Mol Immunol 26: 557-561 [DOI] [PubMed] [Google Scholar]
- Fasano JM, Swanson SJ, Blancaflor EB, Dowd PE, Kao TH, Gilroy S (2001) Changes in root cap pH are required for the gravity response of the Arabidopsis root. Plant Cell 13: 907-921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felle HH (1998) The apoplastic pH of the Zea mays root cortex as measured with pH-sensitive microelectrodes: aspects of regulation. J Exp Bot 49: 987-995 [Google Scholar]
- Grobe K, Becker W-M, Schlaak M, Petersen A (1999) Grass group I allergens (β-expansins) are novel, papain-related proteinases. Eur J Biochem 263: 33-40 [DOI] [PubMed] [Google Scholar]
- Grobe K, Poppelmann M, Becker W-M, Petersen A (2002) Properties of group I allergens from grass pollen and their relation to cathepsin B, a member of the C1 family of cysteine proteinases. Eur J Biochem 269: 2083-2092 [DOI] [PubMed] [Google Scholar]
- Hiller KH, Esch RE, Klapper DG (1997) Mapping of an allergenically important determinant of grass group I allergens. J Allergy Clin Immunol 100: 335-340 [DOI] [PubMed] [Google Scholar]
- Howlett BJ, Clarke AE (1981) Isolation and partial characterization of two antigenic glycoproteins from rye-grass (Lolium perenne) pollen. Biochem J 197: 695-706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P, Marsh DG (1965a) “Isoallergens” from rye grass pollen. Nature 206: 935-937 [DOI] [PubMed] [Google Scholar]
- Johnson P, Marsh DG (1965b) The isolation and characterization of allergens from the pollen of rye grass (Lolium perenne). Eur Poly J 1: 63-77 [Google Scholar]
- Keller E, Cosgrove DJ (1995) Expansins in growing tomato leaves. Plant J 8: 795-802 [DOI] [PubMed] [Google Scholar]
- Knox RB, Suphioglu C (1996a) Environmental and molecular biology of pollen allergens. Trends Plant Sci 1: 156-164 [Google Scholar]
- Knox RB, Suphioglu C (1996b) Pollen allergens: development and function. Sex Plant Reprod 9: 318-323 [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680-685 [DOI] [PubMed] [Google Scholar]
- Lee Y, Choi D, Kende H (2001) Expansins: ever-expanding numbers and functions. Curr Opin Plant Biol 4: 527-532 [DOI] [PubMed] [Google Scholar]
- Lee Y, Kende H (2001) Expression of beta-expansins is correlated with internodal elongation in deepwater rice. Plant Physiol 127: 645-654 [PMC free article] [PubMed] [Google Scholar]
- Li L-C, Cosgrove DJ (2001) Grass group I pollen allergens (β-expansins) lack proteinase activity and do not cause wall loosening via proteolysis. Eur J Biochem 268: 4217-4226 [DOI] [PubMed] [Google Scholar]
- Li L-C, Wang X-C, Jing J-H (1998) The existence of expansin and its properties in the hypocotyls of soybean seedlings. Acta Bot Sin 40: 627-634 [Google Scholar]
- Li Y, Darley CP, Ongaro V, Fleming A, Schipper O, Baldauf SL, McQueen-Mason SJ (2002) Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiol 128: 854-864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z-C, Durachko DM, Cosgrove DJ (1993) An oat coleoptile wall protein that induces wall extension in vitro and that is antigenically related to a similar protein from cucumber hypocotyls. Planta 191: 349-356 [Google Scholar]
- Malley A, Reed CE, Lietze A (1962) Isolation of the allergen from timothy pollen. J Allergy 33: 84-93 (123) [DOI] [PubMed] [Google Scholar]
- Marsh DG (1975) Allergens and the genetics of allergy. In Sela M, ed, The Antigens, Vol 3. Academic Press, New York, pp 271-359 [Google Scholar]
- Marsh DG, Milner FH, Johnson P (1966) The allergenic activity and stability of purified allergens from the pollen of common rye grass (Lolium perenne). Int Arch Allergy 29: 521-535 [DOI] [PubMed] [Google Scholar]
- McQueen-Mason SJ, Durachko DM, Cosgrove DJ (1992) Two endogenous proteins that induce cell wall extension in plants. Plant Cell 4: 1425-1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panyim S, Chalkley R (1969) High resolution acrylamide gel electrophoresis of histones. Arch Biochem Biophys 130: 337-346 [DOI] [PubMed] [Google Scholar]
- Peters WS, Lüthen H, Böttger M, Felle H (1998) The temporal correlation of changes in apoplast pH and growth rate in maize coleoptile segments. Aust J Plant Physiol 25: 21-25 [Google Scholar]
- Petersen A, Becker W-M, Moll H, Blumke M, Schlaak M (1995) Studies on the carbohydrate moieties of the timothy grass pollen allergen Phl p I. Electrophoresis 16: 869-875 [DOI] [PubMed] [Google Scholar]
- Petersen A, Becker W-M, Schlaak M (1993) Characterization of grass group I allergen in timothy grass pollen. J Allergy Clin Immunol 92: 789-796 [DOI] [PubMed] [Google Scholar]
- Petersen A, Grobe K, Lindner B, Schlaak M, Becker W-M (1997) Comparison of natural and recombinant isoforms of grass pollen allergens. Electrophoresis 18: 819-825 [DOI] [PubMed] [Google Scholar]
- Poppelmann M, Becker WM, Petersen A (2002) Combination of zymography and immunodetection to analyze proteins in complex culture supernatants. Electrophoresis 23: 993-997 [DOI] [PubMed] [Google Scholar]
- Rayle DL, Cleland R (1972) The in vitro acid-growth response: relation to in vivo growth responses and auxin action. Planta 104: 282-296 [DOI] [PubMed] [Google Scholar]
- Schipper O, Schaefer D, Reski R, Fleming A (2002) Expansins in the bryophyte Physcomitrella patens. Plant Mol Biol 50: 789-802 [DOI] [PubMed] [Google Scholar]
- Shcherban TY, Shi J, Durachko DM, Guiltinan MJ, McQueen-Mason SJ, Shieh M, Cosgrove DJ (1995) Molecular cloning and sequence analysis of expansins-a highly conserved, multigene family of proteins that mediate cell wall extension in plants. Proc Natl Acad Sci USA 92: 9245-9249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soga K, Wakabayashi K, Hoson T, Kamisaka S (2000) Hypergravity-induced increase in the apoplastic pH and its possible involvement in suppression of β-glucan breakdown in maize seedlings. Aust J Plant Physiol 27: 967-972 [PubMed] [Google Scholar]
- Suck R, Hagen S, Cromwell O, Fiebig H (1999) Rapid and efficient purification of Phleum pratense major allergens Phl p 1 and group Phl p 2/3 using a two-step procedure. J Immunol Methods 229: 73-80 [DOI] [PubMed] [Google Scholar]
- Vesper MJ (1985) Use of a pH-response curve for growth to predict apparent wall pH in elongating segments of maize coleoptiles and sunflower hypocotyls. Planta 166: 96-104 [DOI] [PubMed] [Google Scholar]
- Wu Y-J, Meeley RB, Cosgrove DJ (2001) Analysis and expression of the α-expansin and β-expansin gene families in maize. Plant Physiol 126: 222-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y-J, Sharp RE, Durachko DM, Cosgrove DJ (1996) Growth maintenance of the maize primary root at low water potentials involves increases in cell-wall extension properties, expansin activity, and wall susceptibility to expansins. Plant Physiol 111: 765-772 [DOI] [PMC free article] [PubMed] [Google Scholar]