Abstract
Ribosomal protein S6 (RPS6) is located in the mRNA binding site of the 40S subunit of cytosolic ribosomes. Two maize (Zea mays) rps6 genes were identified that encode polypeptides (30 kD, 11.4 pI) with strong primary amino acid sequence and predicted secondary structure similarity to RPS6 of other eukaryotes. Maize RPS6 was analyzed by the use of two-dimensional gel electrophoresis systems, in vivo labeling with [32P]Pi and immunological detection. Nine RPS6 isoforms were resolved in a two-dimensional basic-urea/sodium dodecyl sulfate-polyacrylamide gel electrophoresis system. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry performed on trypsin-digested isoforms identified four serine (Ser) and one threonine (Thr) residue in the carboxy-terminal region as phosphorylation sites (RRS238KLS241AAAKAS247AAT250S251A-COOH). Heterogeneity in RPS6 phosphorylation was a consequence of the presence of zero to five phosphorylated residues. Phosphorylated isoforms fell into two groups characterized by (a) sequential phosphorylation of Ser-238 and Ser-241 and (b) the absence of phospho-Ser-238 and presence of phospho-Ser-241. The accumulation of hyper-phosphorylated isoforms with phospho-Ser-238 was reduced in response to oxygen deprivation and heat shock, whereas accumulation of these isoforms was elevated by cold stress. Salt and osmotic stress had no reproducible effect on RPS6 phosphorylation. The reduction in hyper-phosphorylated isoforms under oxygen deprivation was blocked by okadaic acid, a Ser/Thr phosphatase inhibitor. By contrast, the recovery of hyper-phosphorylated isoforms upon re-oxygenation was blocked by LY-294002, an inhibitor of phosphatidylinositol 3-kinases. Thus, differential activity of phosphatase(s) and kinase(s) determine complex heterogeneity in RPS6 phosphorylation.
Ribosomal protein S6 (RPS6) assembles onto the 45S rRNA precursor in the nucleolus and is located at the small head region of the cytosolic 40S ribosomal subunit (Nygärd and Nilsson, 1990). RPS6 can be directly cross-linked to mRNA, tRNA, and initiation factors, confirming its location in a region involved in the initiation of translation. RPS6, a 30-kD protein, is the major phosphoprotein of eukaryotic ribosomes. In mammals and fruitfly (Drosophila melanogaster), it is sequentially phosphorylated at five carboxy-terminal Ser residues (Krieg et al., 1988; Fumagalli and Thomas, 2000; Radimerski et al., 2000). RPS6 phosphorylation in the nucleolus occurs during active ribosome biogenesis, whereas RPS6 phosphorylation in the cytoplasm is thought to promote the selective translation of a subset of transcripts, primarily ribosomal protein and elongation factor mRNAs with a 5′-terminal oligopyrimidine tract (5′TOP; Reinhard et al., 1994; Jefferies et al., 1997; Fumagalli and Thomas, 2000). Ribosomes with the highest degree of RPS6 phosphorylation appear to have a selective advantage in mobilization into polysomes (Jefferies and Thomas, 1996; Meyuhas and Hornstein, 2000). RPS6 phosphorylation might not be sufficient to recruit 5′TOP mRNAs, because auxiliary translation factors such as the cellular nucleic acid-binding protein and the La auto-antigen may also be involved (Schlatter and Fussenegger, 2002). In addition, mechanisms independent of S6 phosphorylation like growth or mitogenic stimuli may regulate the translation of 5′TOP mRNAs (Barth-Baus et al., 2002; Stolovich et al., 2002).
The phosphorylation status of mammalian RPS6 is modulated by opposing activities of protein kinases and phosphatases that are activated in response to external stimuli, intracellular signals, and developmental cues. RPS6 phosphorylation requires activation of an S6 kinase, S6K1 or S6K2 (Fumagalli and Thomas, 2000; Kozma and Thomas, 2002). Mitogens, insulin, and abundant amino acids trigger a signal transduction cascade that activates S6K1 by hierarchical phosphorylation events that remodel protein conformation. This pathway includes phosphatidylinositol 3-kinase (PI3K), protein kinase B, atypical protein kinase C, phosphoinositide-dependent kinase 1, and target of rapamycin (TOR) kinase (Kozma and Thomas, 2002). Intracellular concentration of ATP directly controls S6 kinase activity via regulation of TOR activity (Dennis et al., 2001). The activation of S6 kinase is required for completion of the G1 phase and progression to the S phase of the cell cycle and plays a role in the regulation of cell size in mammals and fruitfly (Peterson and Schreiber, 1998; Dennis et al., 1999; Montagne et al., 1999; Pende et al., 2000; Petritsch et al., 2000; Kozma and Thomas, 2002). It is thought that the downstream effector of S6 kinase activation is RPS6 phosphorylation and the consequential effect on the regulation of 5′TOP mRNA translation. The negative regulation of RPS6 phosphorylation is less well understood. In animals, the process is controlled by the Ser/Thr protein phosphatase 2A (PP2A) through regulation of TOR and S6 kinase activity (Millward et al., 1999; Peterson et al., 1999; Westphal et al., 1999; Petritsch et al., 2000; Hartley and Cooper, 2002). A few studies suggest that turnover of phosphates on RPS6 may be regulated. Protein phosphatase 1 (PP1) was identified as the major RPS6 phosphatase in mouse cells (Olivier et al., 1988), and a calyculin-A-sensitive Ser/Thr phosphatase was shown to act downstream of S6 kinase in human hematopoietic cells (Barth-Baus et al., 2002). Thus, the negative regulation of RPS6 phosphorylation may be controlled by phosphatases that act indirectly through regulation of S6 kinase activity or directly by removal of phosphates from the ribosomal protein.
There is evidence that phosphorylation of RPS6 may be regulated in plants during development and in response to changes in the environment. The dephosphorylation of a 30-kD ribosomal protein was observed in response to heat shock in cultured tomato (Lycopersicon esculentum Miller) cells (Scharf and Nover, 1982) and maize (Zea mays) embryos (Beltrán-Peña et al., 2002), as well as oxygen deprivation in maize roots (Bailey-Serres and Freeling, 1990). By contrast, increased phosphorylation of a 30-kD ribosomal protein was observed in maize embryo axes after imbibition and treatment with indole acetic acid (Perez et al., 1990; Beltrán-Peña et al., 2002) and in pumpkin (Cucurbita sp.) cotyledons treated with cytokinin 6-benzylaminopurine (Yakovleva and Kulaeva, 1987). In cultured cells of Arabidopsis, RPS6 accumulated as three phosphorylated and one non-phosphorylated isoform under control conditions and shifted to hypophosphorylated isoforms in response to heat shock (Turck et al., 1998). The expression of Arabidopsis AtS6k2 (Atpk2/ATPK19; At3g08720), an ortholog of mammalian S6K1, in serum-stimulated mammalian cells resulted in an active kinase that could phosphorylate RPS6 of plant and mammalian ribosomes in vitro. As observed for mammalian S6 kinase, activation of AtS6k2 in mammalian cells was inhibited by the PI3K inhibitor wortmannin but not the TOR kinase inhibitor rapamycin. The insensitivity of AtS6k2 to rapamycin was attributed to divergence in the amino-terminal region of the kinase, which is required for rapamycin sensitivity in mammals. Plants possess PI3K and TOR kinases as well as PP2A and PP1 (Smith and Walker, 1996; Drøbak et al., 1999; Menand et al., 2002), however a role for these proteins in the regulation of RPS6 phosphorylation has not been reported.
The present study was undertaken to confirm the phosphorylation of RPS6 in root tips of maize and to begin to elucidate the mechanisms that regulate its phosphorylation status. cDNAs that encode RPS6 were characterized, and a polyclonal antiserum against recombinant RPS6 was produced. Immunoblot analysis and in vivo labeling with [32P]Pi confirmed the presence of phosphorylated RPS6 isoforms in ribosomes under normal growth conditions. RPS6 isoforms were resolved in a high-resolution two-dimensional gel system, and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry was used to determine phosphorylation status. We found that several abiotic stimuli affect the phosphorylation of RPS6, apparently via distinct mechanisms. We studied the regulation of RPS6 phosphorylation under control conditions and during recovery from oxygen deprivation by use of the Ser and Thr phosphatase inhibitor okadaic acid (OA) and the PI3K inhibitor LY-294002 (LY). Our results reveal two distinct patterns of maize RPS6 phosphorylation that are dynamically regulated by kinases and phosphatases of variable sensitivity to abiotic stress.
RESULTS
Characterization of Maize Ribosomal Protein S6 cDNAs
A partial maize cDNA encoding ribosomal protein S6 (rps6) was used to isolate two full-coding cDNAs from a root cDNA library. rps6A and rps6B (GenBank accession nos. U92045, 1,039 bp; AF295599, 1,024 bp) were highly similar in their 5′-untranslated (UTR) and coding regions (85% and 94% identity, respectively) and were divergent in their 3′-UTRs (41% identity). Both cDNAs possess a 5′-UTR of 69 nucleotides. Nucleotide differences in the 5′-UTRs were limited to two regions resulting in the presence of three polypyrimidine tracts in rps6A (nucleotides 9–15, 33–38, and 40–45) and one polypyrimidine tract in rps6B (nucleotides 33–38). Both rps6 cDNAs encode a deduced 251-amino acid polypeptide (28.6 kD) of basic charge (pI 11.4; Fig. 1). RPS6A and RPS6B are identical except for polar to nonpolar substitutions of Asn and Ala at residue 150 and Tyr and Ala at residue 204, respectively. Maize RPS6 appears to be encoded by two genes based on Southern-blot analysis of genomic DNA with an rps6A probe (data not shown) and on our identification of two genes by analysis of cDNAs and search of public databases.
Figure 1.
Comparison of the deduced amino acid sequences of RPS6 from plants, fruitfly, rat, and yeast. RPS6 of maize (Zm1: RPS6A, AAB51304; Zm2: RPS6B, AAG02240), Arabidopsis (At1: RPS6B, At5g10360, CAA74381; At2: RPS6A, At4g31700, AAB88298), tobacco (Nicotiana tabacum; Nt, CAA48187), Brewer's yeast (Saccharomyces cerevisiae; Sc, NP_015235), fruitfly (Dm, AAC34306), and Rattus norvegicus (Rn, R3RTS6). The two amino acid differences between maize RPS6A and RPS6B are boxed at positions 150 and 204. Known phosphorylation sites are shown in black (Krieg et al., 1988; Radimerski et al., 2000). Possible phosphorylation sites are shown in gray. A basic-residue triplet (235–237) necessary for initiation of RPS6 phosphorylation in R. norvegicus is boxed. The white, black, and gray bars below the sequences of the protein represent extended, helical, and loop domains, respectively, as determined by the PHDsec algorithm (Rost and Sander, 1994). The thin bar at the carboxy terminus indicates the helical domain as predicted for a rat peptide fragment. Numbering is based on the sequence of maize RPS6A, including gaps. Conserved residues are indicated by a dot, and gaps are indicated with a dash.
The deduced amino acid sequence of RPS6 is highly conserved among eukaryotes with identities ranging from 54.2% between maize and Brewer's yeast, 62.8% between maize and R. norvegicus, and 87.3% between maize and Arabidopsis. Several Ser residues are phosphorylated in the carboxy terminus of RPS6 of rat and fruitfly (Radimerski et al., 2000). Despite limited primary sequence conservation in the carboxy termini of RPS6 of plants and animals, basic residues are prevalent from residues 235 to 237 (numbering refers to Fig. 1), and Ser residues are conserved at position 238 in plants and position 241 in plants and other eukaryotes. Furthermore, the carboxy terminus of RPS6 of R. norvegicus, fruitfly, Arabidopsis, and maize is predicted to be strongly α-helical (residues 189–235), ending with a loop structure (residues 242–252). This prediction is consistent with 1H-NMR spectroscopy of a polypeptide identical to rat RPS6 residues 223 to 253, which confirmed a helical conformation between residues 227 to 243 (numbering refers to Fig. 1; Katahira et al., 1996).
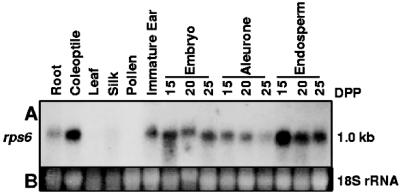
RNA-blot analysis with a probe that would hybridize to RPS6A and RPS6B transcripts revealed that RPS6 mRNA was abundant in organs undergoing rapid cell division (Fig. 2). Coleoptiles, immature ear, embryo, aleurone, and endosperm samples showed high RPS6 transcript levels, whereas root tips and ears just before pollination had lower levels. RPS6 mRNA was abundant in kernel tissues at 15 d post pollination and decreased during aleurone and endosperm maturation. RPS6 transcript levels were low in pollen cells and mature tissues such as leaf and silk. These observations are consistent with the prediction that ribosome biogenesis occurs primarily in mitotically active tissues (Bonham-Smith et al., 1992; Gao et al., 1994; Williams and Sussex, 1995; Bailey-Serres, 1998).
Figure 2.
RPS6 transcript abundance is high in mitotically active organs and tissues of maize. A, Total RNA (20 μg) from various tissue samples (see “Materials and Methods”) was fractionated per lane of an agarose-formaldehyde gel, transferred to a nylon membrane, hybridized with a 32P-labeled rps6A probe, and exposed to x-ray film. B, UV light detection of ethidium bromide-stained 18S rRNA.
Identification of RPS6 Isoforms by in Vivo Phosphorylation and Immunological Detection
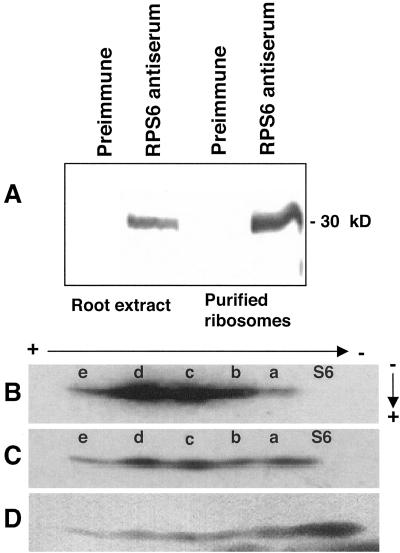
To identify RPS6, a polyclonal antiserum was prepared against recombinant maize RPS6 produced in Escherichia coli. This antiserum recognized, with high specificity, a 30-kD polypeptide of a crude cell extract and purified ribosomes from maize root tips (Fig. 3A). To examine the phosphorylated isoforms of maize RPS6, ribosomes were isolated from root tips of seedlings grown in air after in vivo labeling with [32P]Pi and fractionated on a two-dimensional gel electrophoresis system optimized for resolution of basic ribosomal proteins of mammals (Krieg et al., 1988). The first dimension was a basic-urea tube gel in which acidic-charged proteins migrate more slowly than basic-charged proteins. The second dimension gel was an acidic-urea slab gel in which fractionation was based on apparent molecular mass. After two-dimensional gel electrophoresis, the proteins were transferred to nitrocellulose for autoradiographic detection and immunological identification with the RPS6 antiserum. Autoradiography revealed a 30-kD protein resolved as five phosphorylated isoforms (Fig. 3B, isoforms S6a–S6e). The increasing autoradiographic detection of the progressively more acidic isoforms S6a, S6b, S6c, and S6d is consistent with an increase number of phosphorylated residues. The low autoradiographic signal of isoform S6e indicates that it was less abundant than the other isoforms. Immunodetection of RPS6 on the same blot confirmed the identity of the phosphorylated proteins. The most basic isoform (S6) was not detected by in vivo phosphorylation (Fig. 3B) but was visible on the protein blot (Fig. 3C), indicating that it is non-phosphorylated. Isoforms S6 and S6a overlapped partially in this gel but were fully resolved when the first dimension was run longer (as shown in Figs. 4 and 5). All six RPS6 isoforms and ribosomal proteins were detected following staining of the nitrocellulose filter with colloidal gold (Fig. 3D). Comparison of the immunodetection and colloidal gold staining of RPS6 suggests that phosphorylation status may have an effect on the recognition of the protein by the polyclonal antiserum. These results indicate that a non-phosphorylated and multiple phosphorylated isoforms of RPS6 accumulate in root tips of maize seedlings.
Figure 3.
Identification of isoforms of maize RPS6 by in vivo labeling with [32P]Pi and immunodetection. A, Immunological identification of RPS6 with an antiserum prepared against recombinant maize RPS6. Total root protein (25 μg) and purified ribosomes (4.5 μg) from root tips (apical 1 cm) of 4-d-old seedlings were fractionated by 12% (w/v) SDS-PAGE, transferred to nitrocellulose, and incubated with rabbit preimmune serum (1:1,250 dilution) or polyclonal RPS6 antiserum (1:1,250). The migration of molecular mass markers is indicated to the right. B to D, Basic/acidic-urea two-dimensional gel electrophoresis of [32P]Pi-labeled maize ribosomal proteins. Seedlings were in vivo labeled with [32P]Pi for 3 h under aerobic conditions. Ribosomes were isolated from root tips (apical 1 cm) of 4-d-old seedlings, and rRNA was removed by extraction with acetic acid. Ribosomal proteins were fractionated over a specialized basic/acidic-urea two-dimensional gel system and were transferred to nitrocellulose. The direction and polarity of electrophoresis is shown with arrows and charge symbols. The isoform designated S6 is non-phosphorylated. Isoforms designated with the letters a to e are phosphorylated. B, Radiolabeled proteins detected by autoradiography. C, RPS6 detected by incubation of the nitrocellulose membrane with RPS6 antiserum (1:1,250 dilution) as in A. D, Detection of ribosomal proteins by colloidal gold staining of the nitrocellulose membrane.
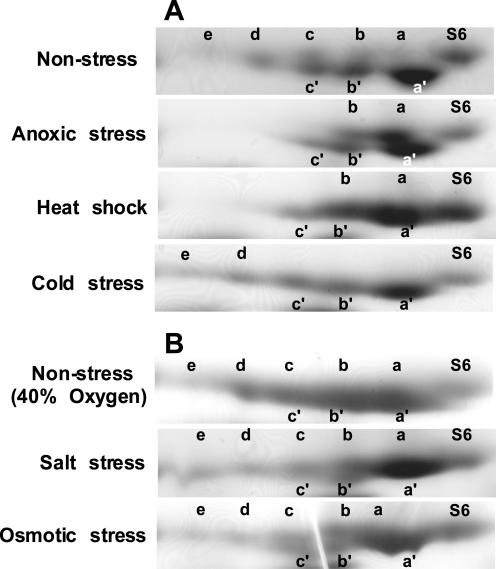
Figure 4.
Basic-urea/Laemmli SDS-PAGE two-dimensional gel electrophoresis of ribosomal proteins from root tips following abiotic stresses. Ribosomes were isolated from root tips (apical 1 cm) of 4-d-old seedlings, rRNA was removed by extraction with glacial acetic acid, and ribosomal proteins were separated using a basic-urea/Laemmli SDS-PAGE two-dimensional gel electrophoresis system and visualized by staining with Coomassie Blue. The region of each gel with RPS6 is shown. The direction and polarity of electrophoresis is as described for Figure 3. Isoform S6 is non-phosphorylated. Isoforms identified with the letters a to e migrated slowly, and isoforms designated a′ to c′ migrated rapidly in the Laemmli SDS-PAGE dimension. A, Seedlings were grown in air (Non-stress), submerged and sparged with argon for 6 h to deprive of oxygen (Anoxic stress), transferred to 42°C for 2 h (Heat shock), or transferred to 8°C for 8 h (Cold stress). B, Seedlings were submerged and sparged with 40% (v/v) O2 for 6 h (Non-stress, 40% Oxygen), submerged in 0.1 m NaCl and sparged with 40% (v/v) O2 for 6 h (Salt stress), or submerged in 0.9 m mannitol sparged with 40% (v/v) O2 for 6 h (Osmotic stress).
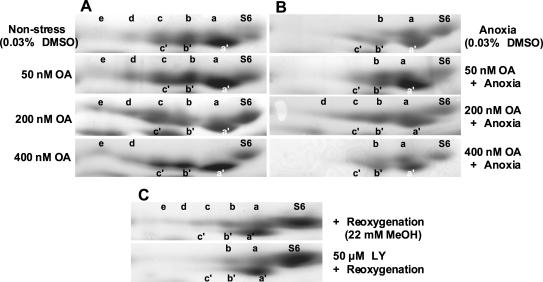
Figure 5.
Basic-urea/Laemmli SDS-PAGE two-dimensional gel electrophoresis analysis of RPS6 from root tips following treatment with phosphatase and kinase inhibitors under non-stress, anoxia, and re-oxygenation conditions. The direction and polarity of electrophoresis is as described for Figure 3. Ribosomes were isolated from root tips (apical 1 cm) of 4-d-old seedlings and were analyzed by basic-urea/Laemmli SDS-PAGE two-dimensional gel electrophoresis as described for Figure 4. A, Seedlings treated for 6 h in air with the apical 1 cm of the primary root tip immersed in induction buffer (IB) with 0.03% (w/v) dimethylsulfoxide (DMSO) and 0, 50, 200, or 400 nm OA. B, Seedlings treated for 6 h under argon with the apical 1 cm of the primary root tip immersed in IB with 0.03% (w/v) DMSO and OA. C, Seedlings deprived of oxygen for 6 h by submergence in IB under argon and re-oxygenated in a humidified chamber open to the air for 2 h with the apical 1 cm of the primary root tip immersed in IB with 22 mm methanol and 0 or 50 μm LY.
Mass Spectrometric Identification of Phosphorylated RPS6 Isoforms
The resolution of RPS6 isoforms was further improved by use of the basic-urea gel in the first dimension and a 12% (w/v) SDS-polyacrylamide gel in the second dimension. Nine RPS6 isoforms were detected in ribosomes from root tips of seedlings grown under control conditions (Fig. 4A, Non-stress). This system revealed additional complexity in the proteins designated S6a, S6b, and S6c in Figure 3 (compare Figs. 3, B–D, with 4 and 5) due to increased resolution of proteins in the SDS-PAGE dimension. The more abundant and more rapidly migrating forms were designated S6a′, S6b′, and S6c′, whereas the more slowly migrating and less abundant isoforms were designated S6a, S6b, and S6c.
MALDI-TOF mass spectrometry was used to analyze these proteins after in-gel digestion with trypsin. All nine proteins were positively identified as RPS6 based on detection of peptides within 80 or 200 ppm, where ppm equals ([observed mass (D) - theoretical mass]/theoretical mass), of the predicted mass values of trypsin-digested maize RPS6A and RPS6B (data not shown). Peptide matches covered up to 62.9% of RPS6. Due to the limited sequence variation between RPS6A and RPS6B, only a few peptides could be assigned to a specific gene product. Peptides specific for RPS6A were detected in the digests of S6a, S6b, S6e, S6a′, and S6c′, and peptides specific for RPS6B were detected in the digests of S6e and S6b′ (data not shown). These results indicate that products of rps6A and rps6B are present in ribosomes of root tips, and the migration of the S6 isoforms is not due to differences in the mobility of the gene products.
To confirm the phosphorylation status of the RPS6 isoforms and to identify the phosphorylation sites of RPS6, peptides were generated by trypsin-digestion in the presence of barium hydroxide. Addition of a phosphate group increases the mass of Ser or Thr by 80 D, however the concomitant decrease in the charge of the residue reduces the frequency of ionization in MALDI-TOF analysis. Barium hydroxide treatment of phospho-Ser and phospho-Thr residues results in β-elimination of H3PO4 (98 D), giving rise to dehydro-Ala and dehydroamino-2-butyric acid, respectively. MALDI-TOF analysis of trypsin-digested and barium hydroxide-treated peptides provided evidence that the Ser and Thr residues within the carboxy-terminal region are sites of phosphorylation (Table I). There was no evidence of phosphorylation at other sites in RPS6 (data not shown). Tryptic peptides of isoform S6 showed no evidence of phosphorylation, consistent with the failure of [32P]Pi to label this isoform, whereas mass values of peptides from S6a, S6b, S6c, S6d, and S6e corresponded to a mono-, di-, tri-, tetra-, and pentaphosphorylation of the carboxy region, respectively. The phosphorylation of these proteins appeared to be sequential for the first two Ser residues (S238KLS241), however the order of phosphorylation of residues in the peptide fragment AS247AAT250S251A-COOH could not be determined. Three isoforms, S6c, S6a′, and S6b′, appeared to be triphosphorylated. S6c mass values corresponded to peptides with phosphorylated Ser-238, Ser-241, and one site in the peptide AS247AAT250S251A-COOH. In S6a′ and S6b′, Ser-238 was not phosphorylated, based on the detection and abundance of a peptide matching the mass of non-phosphorylated RRS238K. This peptide was also detected in the MALDI-TOF analysis of S6a′ without barium hydroxide treatment (data not shown). Both of these isoforms showed phosphorylation of Ser-241 and two residues in the peptide AS247AAT250S251A-COOH. S6c′ also lacked phosphorylation of Ser-238 but was tetra-phosphorylated in the peptide LS241AAAKAS247AAT250S251A-COOH. These results indicate that the degree of RPS6 phosphorylation determined migration in the basic-urea gel and the phosphorylation of Ser-238 determined migration in the SDS-polyacrylamide gel dimension.
Table I.
Identification of carboxy-terminal phosphorylation sites of S6 isoforms by MALDI-TOF mass determination analysis of Ba(OH)2-treated trypsin-digested peptides
Peptide mass and sequence is provided for observed peptides with a mass value ± 1.0 D of the theoretical mass.
| S6 Isoform | Experimental Replicates | Peptide Mass (D) | Peptide Sequencea (No. of Phosphates in Peptide) | No. of Phosphates in Carboxy Terminus | Phosphorylation Status of Carboxy-Terminal Region RRS238KLS241AAAKAS247AAT250S251A-COOHa,b,c |
|---|---|---|---|---|---|
| S6 | 5 | 559.585 | LSAAAK(0) | 0 | SKLSAAAKASAATSA-COOH |
| 577.521 | ASAATSA(0) | ||||
| 774.496 | SKLSAAAK(0) | ||||
| 1,118.87 | LSAAAKASAATSA(0) | ||||
| 1,334.02 | SKLSAAAKASAATSA(0) | ||||
| S6a | 1 | 528.332 | RRSK(1) | 1 | SKLSAAAKASAATSA-COOH |
| 577.568 | ASAATSA(0) | ||||
| 1,315.77 | SKLSAAAKASAATSA(1) | ||||
| S6b | 3 | 528.806 | RRSK(1) | 2 | SKLSAAAKASAATSA-COOH |
| 542.808 | LSAAAK(1) | ||||
| 738.668 | SKLSAAAK(2) | ||||
| 1,298.04 | SKLSAAAKASAATSA(2) | ||||
| S6c | 3 | 527.247 | RRSK(1) | 3 | SKLSAAAKASAATSA-COOH |
| 543.117 | LSAAAK(1) | ||||
| 561.137 | ASAATSA(1) | ||||
| 739.342 | SKLSAAAK(2) | ||||
| 1,083.610 | LSAAAKASAATSA(2) | ||||
| S6d | 4 | 528.315 | RRSK(1) | 4 | SKLSAAAKASAATSA-COOH |
| 542.134 | ASAATSA(2) | ||||
| 542.134 | LSAAAK(1) | ||||
| 739.102 | SKLSAAAK(2) | ||||
| 1,066.31 | LSAAAKASAATSA(3) | ||||
| S6e | 6 | 528.651 | RRSK(1) | 5 | SKLSAAAKASAATSA-COOH |
| 1,245.53 | SKLSAAAKASAATSA(5) | ||||
| S6a′ | 7 | 546.136 | RRSK(0) | 3 | SKLSAAAKASAATSA-COOH |
| 542.362 | LSAAAK(1) | ||||
| 542.362 | ASAATSA(2) | ||||
| 913.404 | RSKLSAAAK(1) | ||||
| 1,279.93 | SKLSAAAKASAATSA(3) | ||||
| S6b′ | 2 | 547.123 | RRSK(0) | 3 | SKLSAAAKASAATSA-COOH |
| 542.277 | LSAAAK(1) | ||||
| 542.277 | ASAATSA(2) | ||||
| 757.147 | SKLSAAAK(1) | ||||
| 1,279.87 | SKLSAAAKASAATSA(3) | ||||
| 1,066.06 | LSAAAKASAATSA(3) | ||||
| S6c′ | 3 | 546.942 | RRSK(0) | 4 | SKLSAAAKASAATSA-COOH |
| 541.839 | LSAAAK(1) | ||||
| 1,048.46 | LSAAAKASAATSA(4) |
Bold type indicates phosphorylated residue
White box indicates phosphorylation at one of three possible residues
Shaded box indicates phosphorylation at two of three possible residues
Altered RPS6 Phosphorylation in Response to Abiotic Stresses
The two-dimensional basic-urea/SDS-PAGE was used to examine the effect of abiotic stresses on the phosphorylation status of RPS6 in root tip ribosomes. Figure 4 compares the isoforms of RPS6 from seedlings grown in air (non-stress) and following oxygen deprivation (anoxia), heat shock, osmotic, salt, and cold stress. Consistent with the previous observation that 24 h of oxygen deprivation blocked in vivo phosphorylation of a 30-kD ribosomal protein (Bailey-Serres and Freeling, 1990), the level of hyperphosphorylated isoforms of RPS6 was dramatically reduced in response to 6 h of anoxia (Fig. 4A). Oxygen deprivation promoted the accumulation of S6, S6a, and S6b, concomitant with a reduction in S6c, S6d, and S6e. Heat shock had a similar negative effect on levels of S6c, S6d, and S6e. Incubation of seedlings at 8°C for 6 h resulted in a significant reduction in S6a, S6b, and S6c; an increase in S6d and S6e; and maintenance of S6, S6a′, S6b′, and S6c′. By contrast, transfer of seedlings to 0.1 m sodium chloride or 0.9 m mannitol solutions aerated with 40% (v/v) O2 for 6 h had no reproducible effect on RPS6 phosphorylation, as compared with the 40% (v/v) O2 control (Fig. 4B). These results indicate that anoxia stress and heat shock reduced the levels of the highly phosphorylated RPS6 isoforms, whereas cold stress promoted the accumulation of these isoforms.
Regulation of RPS6 Phosphorylation in Response to and Recovery from Oxygen Deprivation
To explore the regulation of RPS6 phosphorylation we analyzed the effect of several pharmacologicals on the response to and recovery from oxygen deprivation. The effect of OA, a Ser and Thr protein phosphatase inhibitor, on RPS6 phosphorylation was examined (Fig. 5A). The inhibitory effect of OA is concentration dependent; PP2A is inhibited by 10- to 100-fold lower concentrations of OA than PP1 (Smith and Walker, 1996). Under non-stress conditions, treatment of root tips with 50 nm OA for 6 h resulted in a slight but reproducible depletion in S6a and an increase in abundance of S6b, S6c, and S6d. An effect of OA was more evident after treatment for 6 h with 200 nm OA. In this case, the levels of the hyperphosphorylated isoforms S6c, S6d, and S6e were increased, and the level of the hypophosphorylated isoform S6a was decreased. Levels of S6c′ also appeared to increase as a consequence of 200 nm OA. Treatment with 400 nm OA resulted in isoform levels similar to that observed following cold stress, where only S6, S6a′, S6b′, S6c′, S6d, and S6e accumulated.
To address whether an OA-sensitive phosphatase is involved in the reduction in RPS6 phosphorylation observed during anoxia, seedlings were deprived of oxygen for 6 h in the presence of this inhibitor (Fig. 5B). The decrease in the di-, tri-, and tetraphosphorylated forms, S6b, S6c, and S6d, was partially inhibited by 200 nm OA. The accumulation of isoforms S6b, S6c, and S6d in this sample was confirmed by MALDI-TOF analysis (data not shown). There was no effect on RPS6 phosphorylation when anoxia treatment was performed in the presence of 400 nm OA, despite the clear effect of this level of OA on aerobic roots. In addition, the abundance of S6a′, S6b′, and S6c′ was not dramatically altered by OA treatment under anoxia. Together, these results support the conclusion that OA-sensitive protein phosphatase(s) regulate levels of hyper-phosphorylated RPS6 isoforms under control growth conditions and contribute to the reduction in the hyperphosphorylated RPS6 isoforms in response to oxygen deprivation.
To determine whether the alteration in RPS6 phosphorylation in response to anoxia is reversible, we investigated the phosphorylation status of the protein in seedlings deprived of oxygen for 6 h and then returned to air for 2 h (Fig. 5C). Re-oxygenation resulted in an increase in the phosphorylated isoforms S6b, S6c, S6d, and S6e and a decrease in S6b′ and S6c′. This recovery of sequential phosphorylation of RPS6 could be due to the activation of a kinase, such as S6 kinase. To address whether recovery phosphorylation of RPS6 is impaired by an inhibitor of PI3K-mediated S6 kinase activation in animals, re-oxygenation was performed in the presence of 50 μm LY, a synthetic PI3K inhibitor that is more stable than fungal wortmannin (Vlahost et al., 1994; Downing et al., 1996). The presence of LY during re-oxygenation reduced the recovery of RPS6 phosphorylation, as indicated by the absence of S6c, S6d, and S6e. The treatment of seedlings with LY for 2 h under non-stress conditions did not have a reproducible effect on RPS6 phosphorylation (data not shown).
DISCUSSION
Maize RPS6 Is Phosphorylated at Six Carboxy-Terminal Residues
Maize RPS6 is encoded by at least two highly conserved genes with abundant transcripts in tissues and organs with high rates of cell division. The primary amino acid sequence of the carboxy-terminal region of maize RPS6 is divergent from that of other eukaryotes. Nonetheless, this region possesses potential phosphorylation sites (KRRS238KLS241AAAKAS247AAT250S251A-COOH) based on the conservation of physical features required for S6 kinase recognition in animals, notably Ser residues following three to four basic residues at a junction between a helical and unstructured region (Ferrari et al., 1992; Fig. 1).
Ribosomes from root tips of maize seedlings grown under control conditions posses nine isoforms of RPS6, as determined by use of a specialized two-dimensional gel electrophoresis system and confirmed by MALDI-TOF mass spectrometry. The accumulation of phosphorylated isoforms was confirmed by in vivo labeling with [32P]Pi. The phosphorylation status of each isoform was determined by analysis of tryptic peptides after the elimination of H3PO4 from the phosphorylated residues by barium hydroxide modification. The differences in electrophoretic migration of the nine isoforms corresponded to variations in Ser and Thr phosphorylation. RPS6 accumulated in non-phosphorylated (S6) and mono- (S6a), di- (S6b), tri- (S6c), tetra- (S6d) and penta- (S6e) phosphorylated isoforms. These isoforms appear to result from sequential phosphorylation at Ser-238 and Ser-241, followed by the phosphorylation of Ser-247, Thr-250, and Ser-251. The order of phosphorylation in the tryptic fragment AS247AAT250S251A-COOH could not be determined. The isoforms S6a′, S6b′, and S6c′ migrated more rapidly in the SDS-PAGE dimension. The distinct mobility of the three triphosphorylated isoforms, S6c, S6a′, and S6b′, suggests that heterogeneity in phosphorylation sites affects the structure of the carboxy terminus. A possible explanation is that, first, the phosphorylation of the first two Ser residues of the carboxy-terminal region (RRS238KLS241) in S6c could disrupt the helical coil region modeled from residues 227 to 243 (Fig. 1). Second, a more condensed conformation of the carboxy-terminal region, due to the absence of phosphorylation at Ser-238, could result in more rapid migration in the SDS-PAGE dimension as observed for S6a′ and S6b′. Finally, variation in the two phosphorylated residues in the AS247AAT250S251A-COOH region of S6a′ and S6b′ could result in a distinction in mobility in the basic-urea dimension. In summary, RPS6 isoforms can be divided into two groups characterized by, (a) sequential phosphorylation of Ser-238 and Ser-241 followed by phosphorylation of more carboxy sites (S6a, S6b, S6c, S6d, and S6e), and (b) no phosphorylation at Ser-238 but phosphorylation at Ser-241 and more carboxy sites (S6a′, S6b′, and S6c′). These distinctions could arise from multiple mechanisms of phosphorylation and/or dephosphorylation.
Regulation of Sequential Phosphorylation of RPS6 Ser-238 and Ser-241 by Opposing Actions of Kinases and Phosphatases
The phosphorylation of RPS6 was altered in response to oxygen deprivation, heat shock, and cold stress but not salt or osmotic stress (Fig. 4). Oxygen deprivation and heat shock promoted the accumulation of hypophosphorylated isoforms, S6a and S6b, whereas cold stress resulted in a loss of these isoforms. Insight into the regulation of the response to abiotic stress was obtained by treatment of seedlings with the Ser/Thr phosphatase inhibitor OA (Fig. 5, A and B). OA inhibits PP2A at 100-fold lower concentrations than PP1 in plants (Smith and Walker, 1996; Cohen, 1997; Millward et al., 1999), however the efficiency of the inhibitor on specific phosphatases was not monitored in our experiments. OA treatment under non-stress conditions promoted the accumulation of hyper-phosphorylated forms of RPS6, indicating that maximum phosphorylation was negatively regulated by protein phosphatase(s) in the absence of abiotic stress. Treatments with 50 and 200 nm OA during oxygen deprivation reduced the loss of the hyper-phosphorylated isoforms, suggesting that an increase in phosphatase activity was responsible for the loss of these forms during oxygen limitation. High concentrations of OA under non-stress conditions and cold stress reduced levels of the partially phosphorylated isoforms S6a, S6b, and S6c, confirming that hyper-phosphorylation of RPS6 is the consequence of inhibition of a Ser/Thr phosphatase. This observation is consistent with the finding that cold stress reduced PP2A phosphatase activity in alfalfa (Medicago sativa) seedlings (Monroy et al., 1998). By contrast, moderate but not high concentrations of OA inhibited the loss of hyper-phosphorylated RPS6 during oxygen deprivation. The limited effect of 400 nm OA under low oxygen conditions could indicate complexity in the OA-sensitive protein phosphatases involved in the regulation of RPS6 phosphorylation. Phosphatases could inactivate S6 kinase and upstream kinases, as observed in mammals where PP2A is found in a complex with S6 kinase and TOR kinase (Novak-Hofer and Thomas, 1985; Ballou et al., 1988; Millward et al., 1999; Westphal et al., 1999). On the other hand, a phosphatase could act directly on the ribosomal protein, as suggested by the description of a phosphatase that acts downstream of S6 kinase in hematopoietic cells (Barth-Baus et al., 2002). Our results indicate that OA-sensitive phosphatase(s) are involved in the regulation of RPS6 phosphorylation under non-stress, cold stress, heat stress, and oxygen deprivation conditions, however additional studies are required to identify the phosphatases and to elucidate their distinct roles.
To investigate the regulation of kinases in the phosphorylation of RPS6, we examined the recovery of hyper-phosphorylated isoforms upon re-oxygenation of seedlings (Fig. 5C). LY, an effective inhibitor of PI3K signaling and S6 kinase activation in animals (Kozma and Thomas, 2002), was shown to inhibit the recovery of hyper-phosphorylated S6c, S6d, and S6e. LY and the fungal metabolite wortmannin inhibit PI3K and PI4K activities in plants (Matsuoka et al., 1995; Jung et al., 2002). The inhibition of S6c, S6d, and S6e accumulation following re-oxygenation by LY suggests that RPS6 phosphorylation may be added to the plethora of roles for phosphatidylinositol signaling in plants (Drøbak et al., 1999; Stevenson et al., 2000). Inhibition of recovery of RPS6 phosphorylation by LY is consistent with the finding that the ortholog of mammalian S6K, Arabidopsis AtS6k2, was functionally activated by a PI3K pathway in mammalian cells (Turck et al., 1998). These results suggest that the sequential phosphorylation of Ser-238 and Ser-241 is regulated by a phosphatidylinositol kinase(s)-signaling pathway.
Relationship between RPS6 Isoforms with Differential Ser-238 Phosphorylation
In contrast to isoforms S6a to S6e, isoforms S6a′, S6b′, and S6c′ were characterized by the absence of phosphorylation at Ser-238 and the presence of phosphorylation at Ser-241. Both S6a′ and S6b′ were triphosphorylated, indicating heterogeneity in the phosphorylation of modified sites within the distal carboxy terminus, AS247AAT250S251A-COOH. Isoforms S6a′, S6b′, and S6c′ could be produced by (a) a phosphatase that dephosphorylates Ser-238 and more carboxy sites, with dephosphorylation of Ser-241 occurring last, (b) poor fidelity in phosphorylation of Ser-238 by the plant S6 kinase, or (c) the activity of a distinct S6 kinase that skips Ser-238 and phosphorylates Ser-241 and more carboxy sites. In general, S6a′ was more abundant than S6b′ and S6c′. The limited effect of OA on the abundance of these isoforms suggests that they were not produced by dephosphorylation of the hyper-phosphorylated forms S6d and S6e. A relationship between S6a′ and the non-phosphorylated isoform S6 was revealed in re-oxygenated seedlings. During the recovery from oxygen deprivation, levels of S6b, S6c, S6d, and S6e recovered, except when the PI3K inhibitor LY was present. Whether or not LY was present, levels of S6b′ and S6c′ were reduced following re-oxygenation. The loss of isoforms lacking phospho-Ser-238 indicates that ribosomes with this form of RPS6 may be recycled into the substrate of the kinase that phosphorylates in a sequential manner.
Kinases and phosphatases dynamically regulate two modes of RPS6 phosphorylation in root tips of maize, characterized by (a) the sequential phosphorylation of Ser-238 and Ser-241 and (b) the absence of Ser-238 phosphorylation and presence of Ser-241 phosphorylation. The demonstration that RPS6 phosphorylation is altered by OA and LY, although not unequivocal, indicates that phosphatase(s) and phosphatidylinositol-regulated kinase(s) are involved in the regulation of RPS6 phosphorylation. The identification of RPS6 phosphorylation sites and characterization of the complexity in RPS6 phosphorylation provides the foundation for an analysis of the functional significance of RPS6 phosphorylation. It should now be possible to determine whether the modulation of RPS6 phosphorylation contributes to the regulation of mRNA translation, such as differential translation of ribosomal protein mRNAs observed in response to auxin treatment or dehydration stress (Beltrán-Peña et al., 2002; R. Kawaguchi, T. Girke, E. Bray, and J. Bailey-Serres, unpublished data).
MATERIALS AND METHODS
Plant Material and Growth Conditions
Maize (Zea mays) seeds from the public inbred line B73 (gift of Pioneer-Hi-Bred International, Johnston, IA) were imbibed for 8 h andgerminated in the dark for 4 to 5 d at room temperature (23°C–25°C). For in vivo labeling of phosphates, 3 μL of 10 mCi mL-1 [32P]orthophosphate (2000 Ci mmol-1; PerkinElmer Life Sciences, Boston) was applied onto the tips of 4-d-old seedling roots for 3 h in Eppendorf tubes at room temperature. Root tips were harvested by freezing seedlings on a metal plate over dry ice, removal of the apical 1 cm, transfer to liquid nitrogen, and storage at -80°C.
Abiotic stress treatments included anoxic stress, heat shock, osmotic stress, salt stress, and cold stress. For oxygen deprivation, 50 g of seedlings was submerged in 3 L of IB (0.5 mm Tris-HCl, pH 8.0, and 7.5 μg mL-1 chloramphenicol) and sparged with 99.995% (v/v) argon to gradually establish anoxia within 6 h as described (Fennoy and Bailey-Serres, 1995; Manjunath et al., 1998). Alternatively, the apical 2 cm of the primary root tip was submerged in 800 μL of IB in an Eppendorf tube and was gassed with 99.995% (v/v) argon in a humidified chamber. Both methods of treatment resulted in similar effects on RPS6 phosphorylation (see “Results”). For heat shock, dark-grown seedlings were transferred to a 42°C growth chamber in the dark for 2 h; for cold stress, seedlings were incubated at 8°C in the dark for 6 h (Manjunath and Sachs, 1997); for osmotic stress, seedlings were submerged in IB containing 0.9 m mannitol and sparged with 40% (v/v) O2 for 6 h; and for salt stress, seedlings were submerged in IB containing 0.1 m NaCl and sparged with 40% (v/v) O2 for 6 h. Air grown seedlings were used as the control for anoxia, heat shock, and cold stress. Seedlings submerged in IB and sparged with 40% (v/v) O2 for 6 h were used as the control for salt and osmotic stress. All abiotic stress treatments were replicated at least three times.
Chemical solutions were applied to the apical 1 cm of root tips of seedlings in an Eppendorf tube. OA sodium salt (Calbiochem, La Jolla, CA) dissolved in DMSO with a final solvent concentration of 0.03% was used at 0, 50, 200, and 400 nm. Samples were held in a humidified chamber open to the air (non-stress) or under 99.995% (v/v) argon (anoxia) for 6 h. For control treatments, IB was supplemented with the solvent (0.03% [w/v] DMSO). For re-oxygenation after 6 h of oxygen deprivation, seedlings were transferred to a humidified chamber open to the air for 2 h with the apical 1 cm of the root tip in IB containing 0 or 50 μm LY HCl (RBI, Natick, MA) dissolved in methanol with a final solvent concentration of 22 mm. All chemical treatments were replicated at least three times.
Plant tissues used for RNA extraction were obtained as follows: Coleoptiles were collected from 5-d-old seedlings; leaves of ear husks, silks, and pollen were collected from field-grown plants at anthesis and kernel tissues; embryo (scutellum and embryonic axis), aleurone (aleurone and attached pericarp), and endosperm were collected at 15, 20, and 25 d post pollination. These samples were harvested directly into liquid nitrogen and stored at -80°C.
cDNA Library Screening, DNA Sequencing, and RNA-Blot Analysis
Full-coding cDNAs (GenBank accession nos. U92045 [rps6-1, renamed rps6A] and AF295599 [rps6-2, renamed rps6B]) were isolated from a library prepared from 6-h anoxic roots (3-d-old seedlings) of the maize inbred B73 (kindly provided by Dr. M.M. Sachs, U.S. Department of Agriculture/Agricultural Research Service, University of Illinois, Urbana, IL) as described (Manjunath et al., 1999). A partial maize rps6 cDNA clone (5C01A12; kindly provided by Dr. T. Helentjaris, Maize cDNA Project) was used as the initial hybridization probe at moderate stringency (hybridization overnight in 7% [w/v] SDS and 50 mm NaPO4, pH 7.0, at 60°C; then washed 20 min with 2× SSC and 0.1% [w/v] SDS at 55°C). cDNAs were sequenced on both strands by cycle sequencing (PerkinElmer Instruments, Norwalk, CT) and analyzed by an ABI 373a Stretch/377 DNA sequencer (Applied Biosystems, Foster City, CA) at the DNA Sequencing Core Laboratory (University of Florida, Gainesville, FL). Sequence identity scores were determined using a global pair wise alignment tool (Myers and Miller, 1988). Structural conformation of deduced proteins were predicted by use of the PHDsec algorithm (Rost and Sander, 1994).
Total RNA was extracted from sample tissues by use of a guanidinium method (Glisin et al., 1974). Total RNA (20 μg per lane) was electrophoresed on a 2.2 m formaldehyde 1.3% (w/v) agarose gel, stained with ethidium bromide, photographed, transferred to a nylon membrane, and probed with a 32P-labeled SacI-BamHI fragment containing the complete coding sequence of rps6A (GenBank accession no. U92045) at high stringency as described (Manjunath et al., 1998).
Overexpression of Recombinant RPS6 in Escherichia coli, Purification, and Polyclonal Antiserum Production
The complete coding sequence of maize rps6A was amplified by PCR (30 cycles: 2 min, 96°C, 1.5 min, 42°C; and 2 min 72°C) using a custom primer that hybridized at the 5′ end of the coding sequence (5′-GGGGGGCGCCATGAAGTTTCAACATCGCG-3′) and a universal M13 primer that hybridized to the vector at the 3′ end of the cDNA. The product was cloned into the EheI and BamHI restriction sites of pPRO-EX (Invitrogen, Carlsbad, CA). The resulting construct was sequenced and used to overexpress recombinant RPS6 with a 25-residue amino-terminal extension including a His6-tag and tobacco etch virus (TEV) protease cleavage site. Protein was overexpressed in E. coli DH5α cells by induction with 2 mm isopropylthio-β-galactoside and growth at 37°C for 3 h. Cells were lysed in buffer B (8 m urea, 0.1 m sodium phosphate, 0.01 m Tris-HCl, pH 8.0, and 20 mm imidazole) for 2 h at room temperature. Cell debris was removed by centrifugation at 10,000g for 15 min. Nickel-nitrilotriacetic acid agarose beads (Qiagen, Chatsworth, CA), equilibrated in buffer B, were added to the supernatant and stirred at room temperature for 1 h. The slurry was packed into a 1-mL column, washed five times in buffer B and five times in buffer C (8 m urea, 0.1 m sodium phosphate, 0.01 m Tris-HCl, pH 6.3, and 20 mm imidazole), and eluted using buffer C containing 250 mm imidazole. Purified protein was concentrated by centrifugation using a Centricon-3 tube (Millipore, Bedford, MA). Polyclonal antiserum was produced in rabbits by injection of 1 mg of recombinant His6-TEV-tagged RPS6 in Freund's adjuvant (Robert Sargeant, Ramona, CA).
Extraction of Ribosomal Proteins
Ribosome isolation, ribosomal protein extraction from rRNA, and two-dimensional PAGE were as described previously with the following modifications (Siegmann and Thomas, 1987). Root tissue (1–2 g fresh weight) was ground in liquid nitrogen with a mortar and pestle, homogenized with a Dounce homogenizer in ribosome extraction buffer (200 mm Tris-HCl, pH 7.5, 200 mm KCl, 25 mm EGTA, 36 mm MgCl2, 1 mm sodium molybdate, 1 mm dithiothreitol [DTT], 50 μg mL-1 cyclohexamide, 50 μg mL-1 chloramphenicol, 80 mm β-glycerophosphate, 1% [v/v] Triton X-100, 1% [v/v] Brij 35, 1% [v/v] Tween-40, and 1% [v/v] NP-40), and incubated on ice for 20 min. Sodium molybdate and β-glycerophosphate were included as nonspecific protein phosphatase inhibitors. Cell debris was removed by centrifugation at 10,000g for 15 min at 4°C. The supernatant was layered over a Suc cushion (1.3 m Suc, 400 mm Tris-HCl, pH 7.5, 200 mm KCl, 5 mm EGTA, 36 mm MgCl2, 1 mm sodium molybdate, 1 mm DTT, 50 μg mL-1 cyclohexamide, 50 μg mL-1 chloramphenicol, and 80 mm β-glycerophosphate), and ribosomes were pelleted by centrifugation 18 h at 149,000g and resuspended in Staehlin A buffer (20 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 1 mm sodium molybdate, and 1 mm DTT) by rotation of the sample for 1 h at 4°C. Ribosome quantity was determined by measuring A260 (A260/11.1 is proportional to micrograms per microliter of ribosomes). For each gel sample, 0.5 mg of ribosomes was diluted in 0.1 volume of 1 m MgCl2 and 2 volumes of freshly opened glacial acetic acid (Sigma-Aldrich, St. Louis), vortexed for 1 h at 4°C, and centrifuged 10 min at 4°C, at 14,000g to remove rRNA. Ribosomal proteins were precipitated by the addition of 5 volumes of acetone and brief centrifugation at 14,000g, washed with agitation with two changes of acetone over 24 h and twice with ethanol for 60 min, and then dried in air.
Two-Dimensional PAGE
The ribosomal protein pellet was resuspended in 15 μL of fresh sample buffer (8 m urea, 40 mm Tris-HCl, pH 8.6, 2.3 mm EDTA Na4, 0.06% [v/v] TEMED, 52 mm boric acid, and 5% [v/v] β-mercaptoethanol [BME]) at room temperature for 20 min before electrophoresis. Separation in the first dimension was in a specialized basic-urea polyacrylamide electrophoresis system (6 m urea, 8% [w/v] acrylamide, 0.3% [w/v] bis-acrylamide, 21 mm EDTA Na4, 520 mm boric acid, 200 mm Tris-HCl, pH 8.6, 0.1% [w/v] ammonium persulfate, and 0.1% [v/v] TEMED) in fresh running buffer (6.5 mm EDTA Na4, 156 mm boric acid, and 120 mm Tris-HCl, pH 8.6; Krieg et al., 1988). Electrophoresis was performed in an isoelectric focusing gel apparatus using glass tubes (130 × 3.5 mm; Hoefer Scientific, Richmond, CA) for 26 h at 143 V, with migration toward the cathode. Separation in the second dimension was in an acidic-urea polyacrylamide gel (Krieg et al., 1988) or a Laemmli SDS-polyacrylamide gel (12% [w/v] acrylamide, 0.3% [w/v] bis-acrylamide, and 0.1% [w/v] SDS; Pino et al., 1995). After the extrusion from the glass tubes, first-dimension gels were incubated in the equilibration buffer for 15 min (0.05 m Tris-HCl, pH 6.8, 5% [v/v] BME, 10% [v/v] glycerol, 2% [w/v] SDS, and 0.1% [w/v] bromphenol blue). Tube gels were trimmed to fit the mini-gel apparatus by removal of 5 cm from the top and 1 cm from the bottom of the gel and embedded to the top of the second-dimension gel with agarose (0.05 m Tris-HCl, pH 6.8, 5% [v/v] BME, 10% [v/v] glycerol, 2% [w/v] SDS, and 1% [w/v] agarose). The second-dimension electrophoresis was performed using Mini-PROTEAN II cell (Bio-Rad Laboratories, Hercules, CA) with resolving gel size of 800 × 500 × 1.75 mm and stacking gel of 800 × 200 × 1.75 mm. Electrophoresis was performed for 90 min at 180 V until a prestained carbonic anhydrase (30 kD; Amersham Biosciences, Buckinghamshire, UK) reached the bottom of the gel. Gels were stained with Coomassie Blue R250 for 24 h, destained, and scanned for image analysis (Epson Perfection 1200U Scanner, Epson America, Torrance, CA).
Immunological Detection of RPS6
Proteins were fractionated by one-dimensional Laemmli SDS-PAGE or two-dimensional basic/acidic-urea gel electrophoresis and were transferred to nitrocellulose according to standard procedures (Manjunath et al., 1999). For immunological detection of RPS6, membranes were blocked for 1 h in PBST (1 mm Na2HPO4, 0.14 mm KH2PO4, 13.7 mm NaCl, 0.27 mm KCl, and 0.1% [v/v] Tween 20) that contained 5% (w/v) non-fat dry milk powder and were incubated with a 1:1,250 dilution of polyclonal RPS6 antiserum in PBST for 1 h. Blots were washed twice in PBST with 1% (w/v) non-fat dry milk powder for 5 min and were incubated in a 1:25,000 dilution of rabbit IgG horseradish peroxidase conjugate (Bio-Rad Laboratories) for 1 h. Blots were washed again three times in PBST and the antibody-antigen interaction was detected by chemiluminescence using the ECL reagent (Amersham Biosciences) After immunological identification, nitrocellulose membranes were stained with Colloidal Gold Staining Reagent (Bio-Rad Laboratories).
Mass Spectrometric Analysis of RPS6
For mass spectrometry, proteins were cut-out of Coomassie Blue-stained gels with a sterile scalpel, de-stained by three 15 min washes in 50% (v/v) acetonitrile (ACN), 25 mm ammonium bicarbonate (pH 8.0), and one 5-min wash in 100% (v/v) ACN, and vacuum dried. For in-gel proteolysis, dry gel pieces were incubated for 16 h at 37°C in 60 μL of 40 μg mL-1 trypsin (Promega, Madison, WI) dissolved in 25 mm ammonium bicarbonate (pH 8.0). For the detection of phosphopeptides, the trypsin digestion was supplemented with 1 μg of barium hydroxide (Aldrich Chemical, Milwaukee, WI). Trypsin-digested proteins were eluted with 75 μL of 50% (v/v) ACN and 5% (v/v) trifluoroacetic acid (TFA) and vacuum dried. Samples digested with trypsin in the presence of barium hydroxide were resuspended in 10 μL of 0.1% (v/v) TFA and were purified using a reverse-phase ZipTip C18 column (Millipore). Each column was equilibrated with 10 μL of 100% (v/v) ACN followed by three washes with 10 μL of 0.1% (v/v) TFA. Peptides were bound to the column by four cycles of aspiration and dispension. After binding, columns were washed three times with 10 μL of 0.1% (v/v) TFA, and peptides were eluted with 50 μL of 50% (v/v) ACN and 5% (v/v) TFA and vacuum dried. The peptides were cocrystalized in 9 μL of α-cyano-4-hydroxycinnamic acid (Sigma-Aldrich) saturated with 50% (v/v) ACN, 0.1% (v/v) TFA, and 1 μL of 100% (v/v) ACN and were used for MALDI-TOF mass spectrometry (Voyager-DE STR mass spectrometer, PerSeptive Biosystems, Foster, CA). Mass to charge ratio (m/z) data were collected in the positive ionization mode. The m/z value of monoisotopic peaks with single charged ions was selected, and background peaks were removed by filtering with the Voyager Data Explorer Software. Internal calibration of peptide mass (D) was performed with two trypsin autodigestion product peaks with m/z values corresponding to 842.5100 and 2,211.1046 D. Proteins were identified by searching against the Tr-EMBL database using PeptIdent algorithm (http://ca.expasy.org/tools/peptident.html). The mass tolerance parameter was set at 80 or 200 ppm, where ppm equals ([observed mass (D) - theoretical mass]/theoretical mass), when two or one trypsin autodigestion products was detected, respectively. Phosphopeptides were identified by use of an algorithm prepared to identify carboxy-terminal peptides of RPS6 modified by barium-hydroxide modification of phospho-residues (I.-F. Chang and J. Bailey-Serres, unpublished data). Barium-hydroxide treatment of phosphorylated Ser and Thr results in removal of H3PO4 by β-elimination, converting phospho-Ser and phospho-Thr to dehydro-Ala and dehydroamino 2-butyric acid, respectively. This resulted in an 18-D reduction in mass relative to the unmodified residue. A database was made of theoretical masses of the 25 possible peptide variants for trypsin-cleaved peptides for the carboxy-terminal region Arg-235 to Ala-251. Peptides with mass values with a Δ mass [observed mass - theoretical mass] less than ±1.0 D were identified.
Acknowledgments
We are grateful to R. Miranda and M. Bryant for assistance with the statistical analysis; to K. Szick-Miranda, H. Gydee, S. Manjunath, J. Traugh, and L. Walling for numerous discussions; and to members of the J. Bailey-Serres laboratory for critical reading of the manuscript.
This work was supported by the U.S. Department of Agriculture/National Research Initiative Competitive Grants Program (grant no. 97–35100–4191 to J.B.S.). A.J.W. was supported by a National Science Foundation Graduate Research Traineeship (grant no. DGE–9355042), and I.-F.C. was supported by the Ministry of Education, Republic of China, Taiwan.
References
- Bailey-Serres J (1998) Cytoplasmic ribosomes of higher plants. In J Bailey-Serres, DR Gallie, eds, A Look beyond Transcription: Mechanisms Determining mRNA Stability and Translation in Plants. American Society of Plant Physiologists, Rockville, MD, pp 125-144
- Bailey-Serres J, Freeling M (1990) Hypoxic stress-induced changes in ribosomes of maize seedling roots. Plant Physiol 94: 1237-1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballou LM, Jeno P, Thomas G (1988) Protein phosphatase 2A inactivates the mitogen-stimulated S6 kinase from Swiss mouse 3T3 cells. J Biol Chem 263: 1188-1194 [PubMed] [Google Scholar]
- Barth-Baus D, Stratton CA, Parrott L, Myerson H, Meyuhas O, Templeton DJ, Landreth GE, Hensold JO (2002) S6 phosphorylation-independent pathways regulate translation of 5′-terminal oligopyrimidine tract-containing mRNAs in differentiating hematopoietic cells. Nucleic Acids Res 30: 1919-1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrán-Peña E, Aguilar R, Ortiz-Lopez A, Dinkova TD, Sanchez de Jimenez ES (2002) Auxin stimulates S6 ribosomal protein phosphorylation in maize thereby affecting protein synthesis regulation. Physiol Plant 115: 291-297 [DOI] [PubMed] [Google Scholar]
- Bonham-Smith PC, Oancia TL, Moloney MM (1992) Cytoplasmic ribosomal protein S15a from Brassica napus: molecular cloning and developmental expression in mitotically active tissues. Plant Mol Biol 18: 909-919 [DOI] [PubMed] [Google Scholar]
- Cohen PTW (1997) Novel protein serine/threonine phosphatases: Variety is the spice of life. Trends Biochem Sci 22: 245-251 [DOI] [PubMed] [Google Scholar]
- Dennis PB, Fumagalli S, Thomas G (1999) Target of rapamycin (TOR): balancing the opposing forces of protein synthesis and degradation. Curr Opin Genet Dev 9: 49-54 [DOI] [PubMed] [Google Scholar]
- Dennis PB, Jaeschke A, Saitoh M, Fowler B, Kozma SC, Thomas G (2001) Mammalian TOR: a homeostatic ATP sensor. Science 294: 1102-1105 [DOI] [PubMed] [Google Scholar]
- Downing G, Kim S, Nakanishi S, Catt R (1996) Characterization of a soluble adrenal phosphatidylinositol 4-kinase reveals wortmannin sensitivity of type III phosphatidylinositol kinases. Biochemistry 35: 3587-3594 [DOI] [PubMed] [Google Scholar]
- Drøbak BK, Dewey RE, Boss WF (1999) Phosphoinositide kinases and the synthesis of polyphosphoinosidies in higher plant cells. Int Rev Cytol 189: 95-130 [DOI] [PubMed] [Google Scholar]
- Fennoy SL, Bailey-Serres J (1995) Post-transcriptional regulation of gene expression in oxygen-deprived roots of maize. Plant J 7: 287-295 [DOI] [PubMed] [Google Scholar]
- Ferrari S, Bannwarth W, Morley SJ, Totty NF, Thomas G (1992) Activation of p70s6k is associated with phosphorylation of four clustered sites displaying Ser/Thr-Pro motifs. Proc Natl Acad Sci USA 89: 7282-7285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S, Thomas G (2000) S6 phosphorylation and signal transduction. In JWB Hershey, MB Mathews, N Sonenberg, eds, Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 695-717
- Gao J, Kim S-R, Lee JM, An G (1994) Developmental and environmental regulation of two ribosomal protein genes in tobacco. Plant Mol Biol 25: 761-770 [DOI] [PubMed] [Google Scholar]
- Glisin V, Crkvenjakov V, Byus C (1974) Ribonucleic acid isolation by cesium chloride centrifugation. Biochemistry 13: 2633-2637 [DOI] [PubMed] [Google Scholar]
- Hartley D, Cooper GM (2002) Role of mTOR in the degradation of IRS-1: regulation of PP2A activity. J Cell Biochem 85: 304-314 [DOI] [PubMed] [Google Scholar]
- Jefferies HBJ, Fumagalli S, Dennis PB, Reinhard Ch, Pearson RB, Thomas G (1997) Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J 16: 3693-3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies HBJ, Thomas G (1996) Ribosomal protein S6 phosphorylation and signal transduction. In JWB Hershey, MB Mathews, N Sonenberg, eds, Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 389-409
- Jung J-Y, Kim Y-W, Kwak JM, Hwang J-U, Young J, Schroeder JI, Hwang I, Lee Y (2002) Phosphatidylinositol 3- and 4-phosphate are required for normal stomatal movements. Plant Cell 14: 2399-2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira R, Flotow H, Thomas G, Nosaka AY (1996) Solution structure of the phosphorylated sites of ribosomal protein S6 by 1H NMR spectroscopy. Int J Pept Protein Res 47: 282-288 [DOI] [PubMed] [Google Scholar]
- Kozma SC, Thomas G (2002) Regulation of cell size in growth, development and human diseases: PI3K, PKB and S6K. BioEssays 24: 65-71 [DOI] [PubMed] [Google Scholar]
- Krieg J, Olivier AR, Thomas G (1988) Analysis of 40S ribosomal protein S6 phosphorylation during the mitogenic response. Methods Enzymol 164: 575-581 [DOI] [PubMed] [Google Scholar]
- Manjunath S, Lee KH, VanWinkle P, Bailey-Serres J (1998) Molecular and biochemical characterization of cytosolic phosphoglucomutase in maize. Plant Physiol 117: 997-1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunath S, Sachs M (1997) Molecular characterization and promoter analysis of the maize cytosolic glyceraldehyde 3-phosphate dehydrogenase gene family and its expression during anoxia. Plant Mol Biol 33: 97-112 [DOI] [PubMed] [Google Scholar]
- Manjunath S, Williams AJ, Bailey-Serres J (1999) Oxygen deprivation stimulates Ca2+-mediated phosphorylation of mRNA cap-binding protein eIF4E in maize roots. Plant J 19: 21-30 [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Bassham D, Raikhel N, Nakamura K (1995) Different sensitivity to wortmannin of two vacuolar sorting signals indicates the presence of distinct sorting machineries in tobacco cells. J Cell Biol 130: 1307-1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C (2002) Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA 99: 6422-6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O, Hornstein E (2000) Translational control of TOP mRNAs. In JWB Hershey, MB Mathews, N Sonenberg, eds, Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 671-693
- Millward TA, Zolnierowicz S, Hemmings BA (1999) Regulation of protein kinase cascades by protein phosphatase 2A. Trends Biochem Sci 24: 186-191 [DOI] [PubMed] [Google Scholar]
- Monroy AF, Sangwan V, Dhindsa RS (1998) Low temperature signal transduction during cold acclimation: protein phosphatase 2A as an early target for cold-inactivation. Plant J 13: 653-660 [Google Scholar]
- Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G (1999) Drosophila S6 kinase: a regulator of cell size. Science 285: 2126-2129 [DOI] [PubMed] [Google Scholar]
- Myers EW, Miller W (1988) Optimal alignments in linear space. Comput Appl Biosci 4: 11-17 [DOI] [PubMed] [Google Scholar]
- Novak-Hofer I, Thomas G (1985) Epidermal growth factor-mediated activation of an S6 kinase in Swiss mouse 3T3 cells. J Biol Chem 260: 10314-10319 [PubMed] [Google Scholar]
- Nygärd O, Nilsson L (1990) Translational dynamics: interactions between the translational factors, tRNA and ribosomes during eukaryotic protein synthesis. Eur J Biochem 191: 1-17 [DOI] [PubMed] [Google Scholar]
- Olivier AR, Ballou LM, Thomas G (1988) Differential regulation of S6 phosphorylation by insulin and epidermal growth factor in Swiss mouse 3T3 cells: insulin activation of type 1 phosphatase. Proc Natl Acad Sci USA 85: 4720-4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende M, Kozma SC, Jaquet M, Oorschot V, Burcelin R, Marchand-Brustel YL, Klumperman J, Thorens B, Thomas G (2000) Hypoinsulinaemia, glucose intolerance and diminished β-cell size in S6K1-deficient mice. Nature 408: 994-997 [DOI] [PubMed] [Google Scholar]
- Perez L, Aguilar R, Mendez A, Sanchez deJimenez E (1990) Phosphorylation of ribosomal proteins induced by auxins in maize embryonic tissues. Plant Physiol 94: 1270-1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RT, Desai BN, Hardwick JS, Schreiber SL (1999) Protein phosphatase 2A interacts with the 70-kDa S6 kinase and is activated by inhibition of FKBP12-rapamycin-associated protein. Proc Natl Acad Sci USA 96: 4438-4442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RT, Schreiber SL (1998) Translation control: connecting mitogens and the ribosome. Curr Biol 8: R248-R250 [DOI] [PubMed] [Google Scholar]
- Petritsch C, Beug H, Balmain A, Oft M (2000) TGF-β inhibits p70 S6 kinase via protein phosphatase 2A to induce G1 arrest. Genes Dev 14: 3093-3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino M, Mudd J, Bailey-Serres J (1995) Ozone-induced alterations in the accumulation of newly synthesized proteins in leaves of maize. Plant Physiol 108: 777-785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radimerski T, Mini T, Schneider U, Wettenhall R, Thomas G, Jeno P (2000) Identification of insulin-induced sites of ribosomal protein S6 phosphorylation in Drosophila melanogaster. Biochemistry 39: 5766-5774 [DOI] [PubMed] [Google Scholar]
- Reinhard C, Fernandez A, Lamb NJC, Thomas G (1994) Nuclear localization of p85–s6k: functional requirement for entry into S phase. EMBO J 13: 1557-1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Sander C (1994) Combining evolutionary information and neural networks to predict protein secondary structure. Proteins 19: 55-72 [DOI] [PubMed] [Google Scholar]
- Scharf KD, Nover L (1982) Heat-shock-induced alterations of ribosomal protein phosphorylation in plant cell cultures. Cell 30: 427-437 [DOI] [PubMed] [Google Scholar]
- Schlatter S, Fussenegger M (2002) Novel CNBP- and La-based translation control systems for mammalian cells. Biotechnol Bioeng 81: 1-12 [DOI] [PubMed] [Google Scholar]
- Siegmann M, Thomas G (1987) Separation of multiple phosphorylated forms of 40 S ribosomal protein S6 by two-dimensional polyacrylamide gel electrophoresis. Methods Enzymol 146: 362-369 [DOI] [PubMed] [Google Scholar]
- Smith RD, Walker JC (1996) Plant protein phosphatases. Annu Rev Plant Physiol Plant Mol Biol 47: 101-125 [DOI] [PubMed] [Google Scholar]
- Stevenson J, Pepera I, Heilmann I, Persson S (2000) Inositol signaling and plant growth. Trends Plant Sci 5: 252-258 [DOI] [PubMed] [Google Scholar]
- Stolovich M, Tang H, Hornstein E, Levy G, Cohen R, Bae SS, Birnbaum MJ, Meyuhas O (2002) Transduction of growth or mitogenic signals into translational activation of TOP mRNAs is fully reliant on the phosphatidylinositol 3-kinase-mediated pathway but requires neither S6K1 nor rpS6 phosphorylation. Mol Cell Biol 22: 8101-8113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Kozma SC, Thomas G, Nagy F (1998) A heat-sensitive Arabidopsis thaliana kinase substitutes for human p70–S6k function in vivo. Mol Curr Biol 18: 2038-2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahost CJ, Matter W, Hui KY, Brown RF (1994) A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY 2944002). J Biol Chem 269: 5241-5248 [PubMed] [Google Scholar]
- Westphal RS, Coffee RL Jr, Marotta A, Pelech SL, Wadzinski BE (1999) Identification of kinase-phosphatase signaling modules composed of p70 S6 kinase-protein phosphatase 2A (PP2A) and p21-activated kinase-PP2A. J Biol Chem 274: 687-692 [DOI] [PubMed] [Google Scholar]
- Williams ME, Sussex IM (1995) Developmental regulation of ribosomal protein L16 genes in Arabidopsis thaliana. Plant J 8: 65-76 [DOI] [PubMed] [Google Scholar]
- Yakovleva LA, Kulaeva ON (1987) The effect of phytohormones on phosphorylation of ribosomal proteins in detached pumpkin cotyledons. Biochem Physiol Pflanzen 182: 359-365 [Google Scholar]