Abstract
In chloroplasts, stromal and thylakoid-bound ascorbate peroxidases (tAPX) play a major role in the removal of H2O2 produced during photosynthesis. Here, we report that hexaploid wheat (Triticum aestivum) expresses three homeologous tAPX genes (TaAPX-6A, TaAPX-6B, and TaAPX-6D) mapping on group-6 chromosomes. The tAPX activity of a mutant line lacking TaAPX-6B was 40% lower than that of the wild type. When grown at high-light intensity photosystem II electron transfer, photosynthetic activity and biomass accumulation were significantly reduced in this mutant, suggesting that tAPX activity is essential for photosynthesis. Despite the reduced tAPX activity, mutant plants did not exhibit oxidative damage probably due to the reduced photochemical activity. This might be the result of a compensating mechanism to prevent oxidative damage having as a consequence a decrease in growth of the tAPX mutant plants.
Plant cells are continuously exposed to reactive oxygen species (ROS) generated as by-products of fatty acid β-oxidation, photorespiration, and photosynthesis. Both biotic and abiotic stresses usually lead to an enhanced ROS production, therefore ROS-scavenging mechanisms acting in different organelles play a major role in plant survival and productivity, particularly in extreme environments. Under ROS-generating stress conditions, many antioxidative enzymes such as catalase (CAT), monodehydroascorbate reductase (MDHAR), dehydroascorbate reductase (DHAR), glutathione S-transferase (GST), glutathione reductase (GR), superoxide dismutase (SOD), and ascorbate peroxidase (APX) counteract the otherwise uncontrolled oxidation of cellular components (Noctor and Foyer, 1998). Among the enzymes involved in ROS-removal, APX (EC 1.11.1.11) plays a major role in H2O2-scavenging in plants (Asada, 1992). Three major groups of APX isoenzymes (i.e. chloroplastic, cytosolic, and glyoxysomal APXs) have been identified based on their subcellular location (Mittler and Zilinkas, 1992; Miyake and Asada, 1992; Bunkelmann and Trelease, 1996). Cytosolic APXs (cAPX) are likely to be involved in pathogen response, whereas glyoxysomal APXs remove the H2O2 generated by fatty acid β-oxidation and photorespiration (Bunkelmann and Trelease, 1996; Mittler et al., 1998). Chloroplastic APX (chAPX) composed of thylakoid-bound (tAPX) and stromal (sAPX) isoforms scavenge the H2O2 generated during photosynthesis, which is a major H2O2-producing metabolic process in green tissues (Nakano and Asada, 1981).
Detoxification of ROS is required to avoid damage to the photosynthetic machinery. In chloroplasts, large amounts of O-2 are produced by the transfer of electrons from the donor side of photosystem I (PSI) to O2 (Asada, 1994). Dismutation of O-2 to H2O2 occurs either spontaneously or by a SOD-catalyzed reaction. Subsequent reduction of H2O2 by chAPX produces water and the monodehydroascorbate radical, which can be regenerated to ascorbic acid (AA) by either reduced ferredoxin or NAD(P)H in a reaction catalyzed by MDHAR (Sano and Asada, 1994; Sano et al., 1995). This process, known as the water-water cycle, dissipates the excess of excitation energy incoming to photosystems through the transfer of electrons to molecular oxygen and, in addition, reinforces the trans-thylakoid pH gradient used for ATP synthesis (Asada, 1999). SOD and APX located at the stromal side of thylakoid membranes locally remove the ROS produced at the donor side of the PSI. However, tAPXs constitute themselves a primary target for inactivation by ROS under oxidative stress, such as that produced by methylviologen (MV) and excess of excitation energy (Mano et al., 2001).
Because tAPX mutant plants have not been generated and/or identified so far, the in vivo role of chAPX has been analyzed in transgenic plants overexpressing ROS-scavenging enzymes. Studies with transgenic tobacco (Nicotiana tabacum) plants that overexpress bacterial CAT in chloroplast demonstrated that chAPXs are strongly inhibited under oxidative stress, which suggests that these enzymes could be a limiting component of the plant antioxidative defense (Shikanai et al., 1998; Miyagawa et al., 2000). Therefore, the overexpression of chAPXs might protect the plants from oxidative damage under stress. Accordingly, cotton (Gossypium hirsutum) plants that overexpress recombinant cAPX in chloroplasts showed an enhanced resistance to chilling-associated oxidative stress (Payton at al., 2001). In addition, it has been recently shown that transgenic tobacco plants that overexpress tAPX are more resistant to MV and chilling than the wild-type plants (Yabuta et al., 2002). Overall, these results showed that tAPXs are key components of the chloroplastic antioxidant defenses in vivo.
In the present work, three tAPX genes from wheat (Triticum aestivum; TaAPX-6A, TaAPX-6B, and TaAPX-6D) were identified. A TaAPX-6B-deleted mutant line (S-SV8) exhibits reduced weight, size, and seed production relative to the parental near isogenic line (R-SV8). The comparison of APX activities and photosynthetic parameters between the R-SV8 and the S-SV8 isolines led us to conclude that a reduced tAPX activity leads to an impaired photosynthesis.
RESULTS
The Hexaploid Wheat Genome Contains Three Homeologous Genes Encoding tAPXs
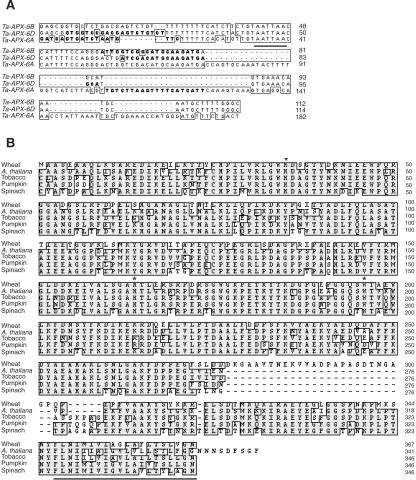
In an attempt to identify genes accountable for the phenotypic differences observed between R-SV8 and S-SV8, we isolated a cDNA for which the coding gene is absent in the mutant line. The translated sequence of this cDNA (clone TaRr16) did not show significant homology with any protein in nonredundant databases (Danna et al., 2002). However, after the 5′-RACE-mediated elongation of this sequence, we obtained a near full-length cDNA that encodes a protein highly identical to tAPX proteins from spinach (Spinacia oleracea) and pumpkin (Cucurbita pepo). Three cDNAs from wheat were identified after an extensive search based on 5′- and 3′-RACE procedures. Although their coding regions are highly similar (more than 95% identity), major differences were observed at their 3′-untranslated regions (UTRs; Fig. 1A). A comparison of their deduced amino acid sequences revealed high homology to various tAPXs from dicots (70%–80% identity). Proteins encoded by these tAPX genes from wheat show the hallmarks of prokaryotic class I peroxidases (Welinder, 1992). Their carboxy termini showed the predicted putative membrane-anchor domain characteristic of tAPXs (Ishikawa et al., 1997; Mano et al., 1997). Although predicted tAPX proteins from wheat are highly similar to other tAPXs from dicots, they have an insertion of 26 amino acid residues near the anchor domain (Fig. 1B).
Figure 1.
Sequence alignments of 3′-UTRs of tAPX cDNAs from wheat and proteins from different plant species. A, Alignment among 3′-UTRs of tAPX cDNAs from wheat. Nucleotide homology is boxed. Primers for each cDNA are indicated by bold type. The sense primer of TaAPX-6B is located over the coding region, hence, not shown (see “Materials and Methods”). Polyadenylation signals are underlined. B, Alignment among TaAPX-6B and other thylakoid-bound APX proteins from dicot species. Conserved amino acids are shown by a shadow box. Asterisks indicate essential amino acid residues for protein activity. Putative thylakoid membrane anchor domains are underlined.
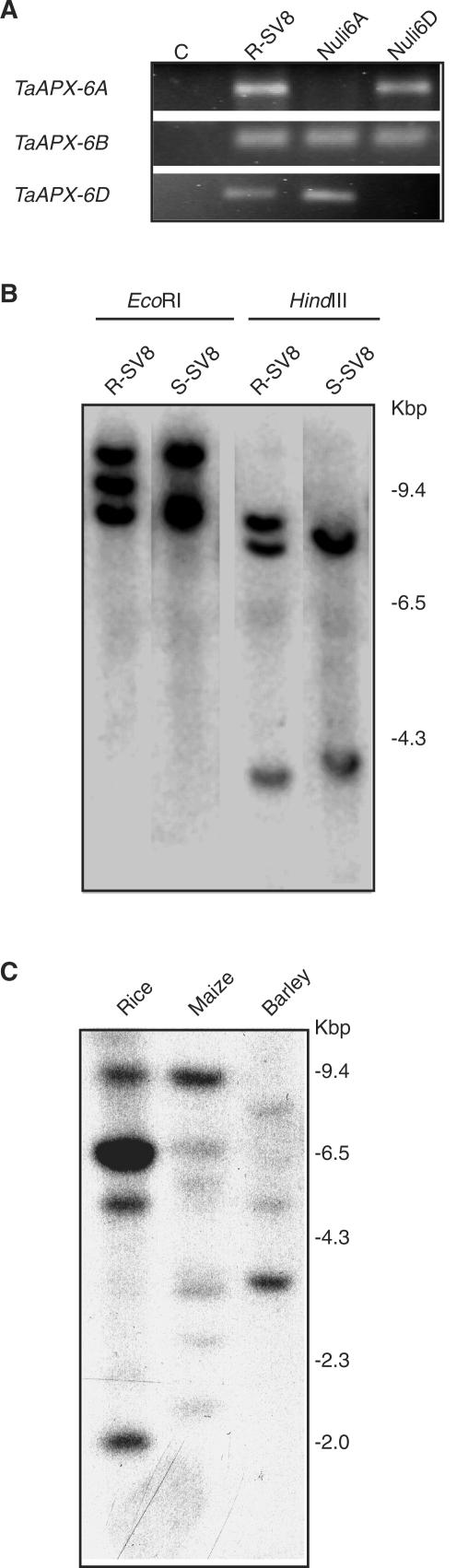
Because one of the three tAPX genes (clone TaRr16) mapped on the distal region of chromosome 6BL (Danna et al., 2002), this gene was named TaAPX-6B. To determine whether the tAPXs genes are located in group-6 chromosomes, genomic DNA from Nuli-Tetrasomic wheat lines (Chinese Spring Nuli6A-Tetra6D and Nuli6D in which 6A or 6D chromosomes are absent respectively) were analyzed by PCR. As shown in Figure 2A, these genes were located in chromosomes 6A and 6D, and consequently, they were named TaAPX-6A and TaAPX-6D. These results indicate that TaAPX-6A, TaAPX-6B, and TaAPX-6D are homeologous genes, located at chromosome 6 of the hexaploid wheat genome. High-stringency DNA hybridization analysis revealed the presence of three copies in the parental line and two in the S-SV8 mutant line (Fig. 2B). Low-stringency hybridization revealed the presence of six bands in the parental and five in the mutant (data not shown), indicating that TaAPX-6B is probably the only chAPX gene absent in the mutant line. Because chAPX genes have not been isolated from monocots so far, we used the coding region of TaAPX-6B to assess the copy number and homology in other monocot plants. A main single band was detected in maize, rice, and barley (Fig. 2C). Consistently, only one tAPX gene was identified in the complete rice genome database, which confirms the presence of a single tAPX gene in this species.
Figure 2.
Chromosomal location of tAPX genes. A, PCR amplification of tAPX genes from wheat genomic DNA obtained from R-SV8 (normal hexaploid genome), Nuli6A (a NuliA-TetraD genome), and Nuli6D (a NuliD-TetraA genome) plants. C, Negative PCR control. B, TaAPX-6B DNA gel-blot hybridization on genomic DNA from R-SV8 and S-SV8 plants digested with EcoRI or HindIII. C, TaAPX-6B DNA gel-blot hybridization on 10 μg of genomic DNA from rice (Oryza sativa), maize (Zea mays), and barley (Hordeum vulgare), digested with Hin-dIII. Results are representative of two independent experiments.
The Expression of tAPX Genes Overlaps in Most Tissues
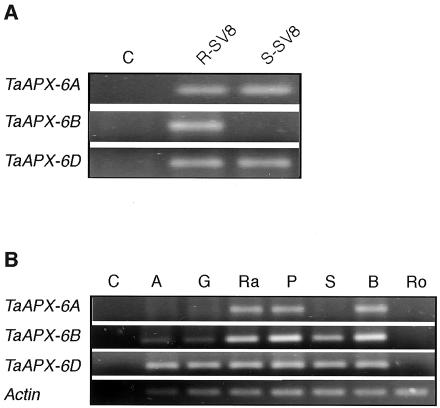
To analyze whether the chromosomal deletion affects the expression of the tAPX genes in the mutant line, TaAPX-6A and TaAPX-6D expression was further explored in both isolines. Whereas the three genes were expressed in the R-SV8 plants, TaAPX-6A and TaAPX-6D expression was detected in the mutant (Fig. 3A). To test whether or not the three genes have a similar function, their expression was studied in different organs of R-SV8 plants. The three genes are expressed in aerial organs but not in roots. Although their expression patterns essentially overlap in most tissues, there are some subtle differences in expression. Whereas TaAPX-6D and TaAPX-6B expression was detected in green tissues and reproductive organs, TaAPX-6A expression was only detected in green tissues excluding sheaths (Fig. 3B). These results indicate that tAPX genes are controlled by similar but not identical regulatory elements.
Figure 3.
Expression of tAPX genes. A, Reverse transcriptase (RT)-PCR amplification of tAPX genes (TaAPX-6A, TaAPX-6B, and TaAPX-6D) from leaves of R-SV8 and S-SV8 plants. C, Negative PCR control. B, RT-PCR amplification of tAPX genes from different organs of R-SV8 plants. C, Negative PCR control; A, anthers; G, gynoecium; Ra, floral rachis; P, glumes and paleas; S, foliar sheaths; B, leaf blades; and Ro, roots. Results are representative of two independent experiments.
TaAPX-6B Encodes a Protein with APX Activity
The cDNA region encoding the putative mature protein of TaAPX-6B was subcloned into plasmids for protein expression. Two versions of the recombinant protein were overexpressed in Escherichia coli: a long version containing the so-called membrane anchor domain (rAPX-1) and a short version in which this region was deleted (rAPX-2). Both rAPX-1 and rAPX-2 showed their predicted molecular weights in SDS-PAGE gels. The anchor-containing protein yielded an insoluble form, whereas the anchor-deleted form produced a soluble protein (data not shown). The insoluble form (rAPX-1) did not show any APX activity, but the soluble protein (rAPX-2) exhibited APX activity at 26.4 μmol AA min-1 mg-1 of recombinant protein in the supernatant (data not shown). The specific activity of the recombinant TaAPX-6B was similar to the activity reported for a recombinant cAPX from soybean (Glycine max) overexpressed in E. coli (Dalton et al., 1996). This result confirms that TaAPX-6B encodes an active APX protein.
The Photon Flux Density (PFD) Applied during Growth of Plants Modulates TaAPX-6B Expression
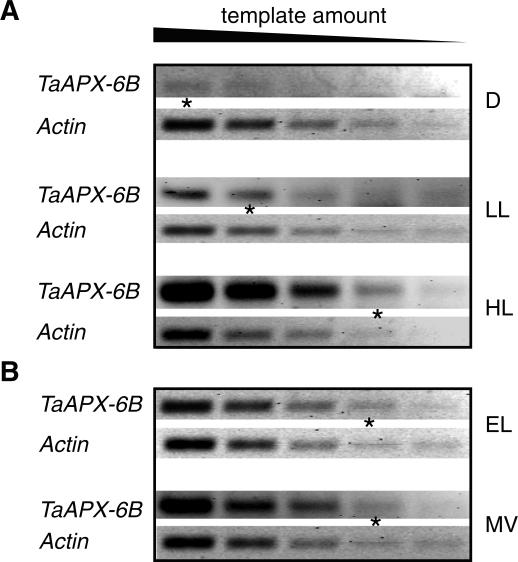
To determine whether the expression of tAPX genes is regulated by the PFD, gene expression in leaf blades of R-SV8 plants grown in the dark, in low-light condition (LL), or in high-light condition (HL) was analyzed. In darkness, TaAPX-6B expression was nearly undetectable (Fig. 4A). A 2-fold increase in the TaAPX-6B mRNA steady-state level was observed in plants grown at LL (50–100 μmol photons m-2 s-1). Plants grown at HL (700–1,000 μmol photons m-2 s-1) showed 4-fold higher TaAPX-6B expression than etiolated plants (Fig. 4A). The expression of TaAPX-6A and TaAPX-6D, roughly undetectable in the dark, increased 2-fold in LL-grown plants, but no further induction was detected in HL-grown plants (data not shown). This expression analysis indicates that tAPX genes are light-inducible but only TaAPX-6B is modulated by the PFD used for plant growth.
Figure 4.
Semiquantitative RT-PCR analysis of TaAPX-6B mRNA accumulation in R-SV8 plants. A, Analysis of steady-state mRNA accumulation was carried out with plants grown in the dark (D), at 50 to 100 μmol photons m-2 s-1 (LL), or at 700 to 1,000 μmol photons m-2 s-1 (HL). B, Analysis of the short-term mRNA accumulation in plants suddenly exposed to 1,800 μmol photons m-2 s-1 (EL) combined with the application of MV for 4 h. In this case, plants were previously grown at 200 to 400 μmol photons m-2 s-1. From left to right, a base-two serial dilution of 100 and 10 ng of total cDNA was used as template for the amplification of TaAPX-6B and Actin, respectively. Asterisks indicate the last dilution at which TaAPX-6B was detected. Actin was detected up to the fifth cDNA dilution in every sample. Results are representative of two independent experiments.
The short-term effect of either excess excitation energy (EL) or MV-mediated oxidative stresses on tAPX gene expression was also investigated. R-SV8 plants grown at 200 to 400 μmol photons m-2 s-1 were sprayed with 20 μm MV and were exposed to EL (1,800 μmol photons m-2 s-1). No differences in TaAPX-6B expression were detected either after EL or MV combined with EL treatments, indicating that TaAPX-6B is not regulated by a sudden oxidative stress in a 4-h temporal window (Fig. 4B). Likewise, neither MV nor EL modifies the mRNA steady-state levels of TaAPX-6A and TaAPX-6D (data not shown). As previously reported for tAPX genes from spinach (Yoshimura et al., 2000), the mRNA level of tAPX genes from wheat is not regulated by oxidative stress.
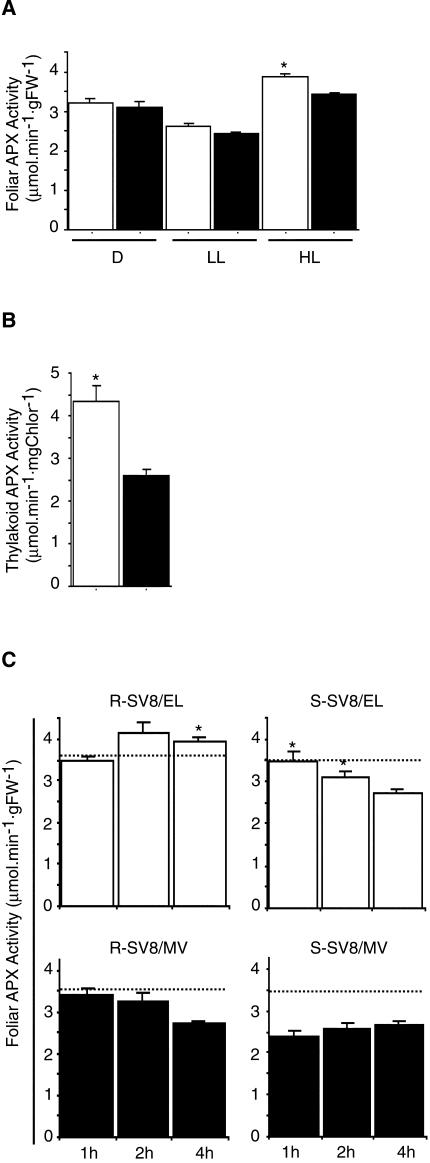
APX Activity of Mutant Plants Is Reduced at High and Excess PFD
Because TaAPX-6B is deleted in S-SV8 plants, studies were carried out to test whether these plants have a disturbed APX activity. No significant differences in foliar APX activity were detected between R-SV8 and S-SV8 plants grown in the dark or at LL conditions. However, S-SV8 plants grown at HL, showed a 15% lower APX activity than that of the R-SV8 plants (Fig. 5A). Although these measurements cannot distinguish among different isoenzymes contributing to the foliar APX activity, the reduced activity of the mutant is likely to be due to the lack of TaAPX-6B. Because TaAPX-6B is likely to encode a tAPX enzyme, APX activity in thylakoid membranes was measured. As shown in Figure 5B, HL-grown S-SV8 plants showed a 40% lower activity in the thylakoids than HL-grown R-SV8 plants. This result is consistent with the proposed subcellular location of TaAPX-6B in thylakoid membranes and indicates that TaAPX-6A and TaAPX-6D did not fully compensate the lost of tAPX activity in the mutant line.
Figure 5.
APX activity in R-SV8 and S-SV8 plants. A, Foliar APX activity in R-SV8 (white bars) and S-SV8 (black bars) 2-week-old plants grown in the dark (D), at 50 to 100 μmol photons m-2 s-1 (LL), and at 700 to 1,000 μmol photons m-2 s-1 (HL). Bars are mean values ± se of three independent experiments, each one consisting of 10 replicates. B, Thylakoid-bound APX activity in HL-grown R-SV8 (white bars) and S-SV8 plants (black bars). Bars are mean values ± se of two experiments, each one consisting of three replicates. C, Foliar APX activity in R-SV8 (left panels) and S-SV8 plants (right panel) suddenly exposed to 1,800 μmol photons m-2 s-1 (white bars) and MV (black bars). Plants were previously grown at 200 to 400 μmol photons m-2 s-1. Bars are mean values ± se of two independent experiments, each one consisting of 10 replicates. Horizontal lines indicate APX activity at time 0. Asterisks indicate significant differences (t test, P < 0.05) between EL and MV treatments for a given genotype.
To determine whether the response to oxidative stress is compromised by the tAPX deficiency of the S-SV8 line, plants grown at 200 to 400 μmol photons m-2 s-1 were subjected to EL (1,800 μmol photons m-2 s-1) and MV. Foliar APX activity increased in R-SV8 while it decreased in S-SV8 plants 4 h after exposure to EL (Fig. 5C). Addition of MV under EL conditions resulted in a sharp reduction of APX activity in R-SV8 plants 4 h after treatment, whereas a drastic reduction in APX activity was observed in the mutant as soon as 1 h after this treatment. It is noteworthy that the magnitude of the APX inhibition in S-SV8 plants 4 h after EL was similar to that observed 1 h after MV. These results suggest that an elevated tAPX activity may be required to avoid foliar APX inhibition under a sudden oxidative stress.
The Ascorbate-Glutathione Cycle Is Not Altered in tAPX Mutant Plants
Transgenic plants overexpressing antioxidative enzymes usually show altered activities of ROS-scavenging endogenous enzymes (Sen Gupta et al., 1993a, 1993b). To determine whether the tAPX deficiency of S-SV8 plants causes any alteration in the activity of other antioxidative enzymes involved in the ascorbate-glutathione cycle, DHAR, MDHAR, and GR activities were assayed. No differences were detected between R-SV8 and S-SV8 plants growing at HL (nonstressing PFD), indicating that tAPX deficiency has no effect on the activity of these enzymes (Table I). Although HL-grown mutant plants had roughly 40% lower tAPX activity than R-SV8 plants, the level of AA was similar in both the isolines (Table I).
Table I.
Foliar AA content and ascorbate-glutathione cycle enzyme activities and AA in leaves of plants grown at 700 to 1,000 μmol photons m-2 s-1
Mean values of three measurements (plants) from four independent experiments. se is shown in parentheses.
| Plant | AA Content | DHAR | MDHAR | GR |
|---|---|---|---|---|
| μmol g dry wt-1 | nmol mg protein-1min-1 | |||
| R-SV8 | 16.9 (1.7) | 193.4 (9.7) | 91.5 (9.4) | 77.0 (6) |
| S-SV8 | 19.8 (0.8) | 191.3 (8.1) | 83.2 (2.7) | 77.4 (6) |
tAPX Mutant Plants Show Impaired Electron Transport and Photosynthetic Activity and Reduced Growth under Normal PFD
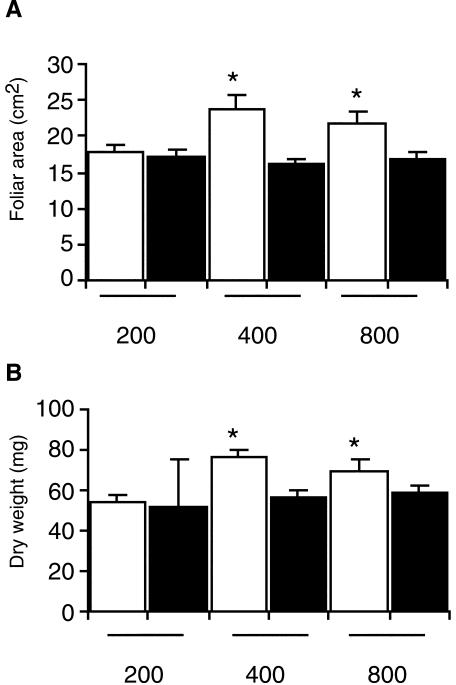
Because tAPXs remove H2O2 generated by dismutation of O-2 produced at the donor side of PSI, the possible role of TaAPX-6B in the protection of the photosynthetic apparatus was investigated by comparing CO2 assimilation and chlorophyll fluorescence parameters in both isolines. HL-grown S-SV8 plants displayed a significantly lower photosynthetic activity than that of R-SV8 plants. The quantum yield of PSII (φPSII) and photochemical quenching (qP) were also reduced in the mutant, but there were no significant differences in non-photochemical quenching between both isolines (Table II). The reduced φPSII in the mutant plants was only observed at high PFD (800 μmol photons m-2 s-1), suggesting that the mutant is more susceptible to photodamage than the parental line (Table III). To study the effect of the reduced PSII electron transfer and photosynthetic activity on growth and biomass accumulation in S-SV8 plants, leaf area and dry weight of R-SV8 and S-SV8 plants were measured. R-SV8 and S-SV8 plants grown at a PFD of 200 μmol photons m-2 s-1 showed non-significant differences in dry matter and foliar area. However, at moderate (400 μmol photons m-2 s-1) and high (800 μmol photons m-2 s-1) PFD, the mutant plants developed a smaller leaf area and accumulated less dry matter than the parental plants (Fig. 6, A and B). To determine whether the reduced accumulation of biomass in the mutant could be due to photodamage, measurements of protein carbonylation, which indicates oxidative damage, were carried out. No significant differences were detected between the parental and the mutant (data not shown), indicating that mutant plants do not experience a chronic oxidative damage at LL or HL conditions. To determine whether the tAPX deficiency of S-SV8 plants leads to an enhanced susceptibility to photodamage under stress conditions, the response to a sudden increase in PFD was investigated. HL-grown mutant and parental plants were suddenly exposed for 4 h to EL. A decrease in Fv/Fm indicating PSII photoinhibition was detected in both the isolines, but this decrease was significantly higher in the mutant than in the parental plants (Table IV). In summary, these results suggest that a reduced tAPX activity leads to an increased susceptibility to an abrupt oxidative stress.
Table II.
Photosynthetic parameters
Photosynthetic activity (CO2-assimilation), quantum yield of PSII (ϕ PSII), photochemical quenching (qP), and non-photochemical (qNP) quenching of chlorophyll a fluorescence in leaves of 2-week-old plants grown at 700 to 1,000 μmol photons m-2 s-1. Mean values of 10 measurements (plants) from two independent experiments. se is shown in parentheses.
| Plant | Photosynthesisa | ϕPSII | qP | qNP |
|---|---|---|---|---|
| μmol CO2m-2s-1 | ||||
| R-SV8 | 36.4 (0.7) | 0.41 (0.03) | 0.56 (0.03) | 0.30 (0.05) |
| S-SV8 | 32.0b (1.3) | 0.32b (0.02) | 0.44b (0.04) | 0.36 (0.04) |
Actinic light, 750 μmol photons m-2 s-1
Significant difference (t test, P < 0.05)
Table III.
Dependency of ϕPSII on irradiance
Quantum yield measurements were performed in leaves of 2-week-old plants grown at 200, 400, or 800 μmol photons m-2 s-1. Mean values of eight measurements (plants) from two independent experiments. SE is shown in parentheses.
| Plant
|
μmol Photons m-2 s-1
|
||
|---|---|---|---|
| 200 | 400 | 800 | |
| R-SV8 | 0.65 (0.01) | 0.66 (0.02) | 0.53 (0.04) |
| S-SV8 | 0.64 (0.03) | 0.62 (0.02) | 0.47a (0.04) |
Significant differences (t test, P < 0.05)
Figure 6.
PFD-dependent growth of parental and mutant plants. A, Leaf area of R-SV8 (white bars) and S-SV8 plants (black bars) grown at 200, 400, or 800 μmol photons m-2 s-1. B, Dry weight of second leaf stage R-SV8 and S-SV8 plants grown at 200, 400, or 800 μmol photons m-2 s-1. Bars are mean values ± se, corresponding to three independent experiments, each one consisting of four replicates (plants). Asterisks indicate significant differences between genotypes (t test, P < 0.05).
Table IV.
Photoinhibition of PSII measured as Fv/Fm
Measurements of Fv/Fm in 2-week-old plants grown at 700 to 1,000 μmol photons m-2 s-1 (HL) were suddenly transferred to 1,800 μmol photons m-2 s-1 (EL) for 4 h. Mean values of 10 measurements (plants) from two independent experiments. SE is shown in parentheses.
| Plant | HL | EL |
|---|---|---|
| R-SV8 | 0.819 (0.004) | 0.652 (0.018) |
| S-SV8 | 0.816 (0.004) | 0.540a (0.030) |
Significant differences (t test, P < 0.05)
DISCUSSION
Early studies postulated that thylakoid-bound APX enzymes play a key role in the scavenging of H2O2 in chloroplasts, helping plants to dissipate excess excitation energy through electron transfer from PSI to O2 (Asada and Takahashi, 1987). The rate of electron flow from PSI to O2 increases dramatically in plants exposed to various stresses, generating large quantities of ROS, which in turn lead to oxidative stress (Foyer et al., 1994). The relevance of tAPX activity in H2O2 removal under non-stressing and stressing conditions has been previously analyzed in transgenic plants (Shikanai et al., 1998; Miyagawa et al., 2000; Payton et al., 2001). Here, we report the identification of a tAPX mutant line of wheat that displays reduced tAPX activity and impaired photosynthesis.
tAPX Redundancy Allows Wheat Mutant Plants to Survive
Several tAPX cDNAs have been already isolated, and their encoded proteins have been characterized. Most of these studies have been carried out using pumpkin and spinach as models, which contain a single chAPX gene encoding both the tAPX and the sAPX by means of mRNA alternative splicing (Ishikawa et al., 1997; Mano et al., 1997). chAPX mutants have not been isolated in a nonredundant background genome so far, which suggests a conditional lethal nature for such a mutation. In fact, as reported recently, transgenic tobacco plants expressing tAPX transgene in the antisense orientation were not obtained most probably due to the lethal nature of the tAPX suppression (Yabuta et al., 2002). In Arabidopsis, tAPX and sAPX are encoded by two different loci (At1g77490 and At4g08390), and accordingly, T-DNA interrupted lines for each one of these genes were obtained (http://signal.salk.edu/cgi-bin/tdnaexpress). The hexaploid genome of wheat may facilitate the survival of a tAPX mutant and thereby allows determining the in vivo role of tAPX enzymes. DNA gel-blot hybridization analysis indicates that monocots like rice, maize, and barley contain a single tAPX gene (Fig. 2C). Unlike those species, three tAPX genes were detected in bread wheat mapping on the group-6 chromosomes. Because no additional copies of tAPX map on the deleted region of the mutant (Fig. 2B), its reduced tAPX activity could be attributed to the lack of TaAPX-6B (Fig. 5B). Moreover, in vitro activity assays of the recombinant protein confirmed that TaAPX-6B encodes a functional APX enzyme, providing additional evidence for the latter hypothesis. Because only TaAPX-6B expression is regulated by the PFD at non-stressing conditions (Fig. 4A), it seems possible that the lack of either TaAPX-6A or TaAPX-6D could have a minor effect on the phenotype of wheat plants. In this regard, it should be emphasized that no compensatory tAPX activity seems to be provided by TaAPX-6A and TaAPX-6D in the mutant (Fig. 5B). Although reproductive organs and sheaths do not express TaAPX-6A, the three tAPX genes are expressed in most aerial tissues (Fig. 3B). This overlapped pattern of gene expression could lead to functional redundancy, and such a redundancy could explain the remainder tAPX activity in S-SV8 plants (Fig. 5B) and probably the viability of the mutant.
Because S-SV8 plants bear a deletion in chromosome 6BL, we cannot rule out that the lack of other genes (not known so far) could be accountable for some of the physiological differences detected between these two near-isogenic lines. However, despite the deletion, previous analysis indicated that R-SV8 and S-SV8 lines are nearly identical at a genetic level. No RAPDs were detected among 2,400 genomic loci analyzed (F. Sacco, unpublished data) and only TaAPX-6B out of 6,000 genes analyzed was differentially expressed between these two isolines (Danna et al., 2002). Moreover, no disturbances in the ascorbate-glutathione cycle were detected in the mutant, suggesting that no general disorder of the anti-oxidant metabolism is contributing to the phenotype analyzed here. Further experiments on the molecular characterization of the distal region of chromosome 6BL will help to assess whether the phenotype of the mutant is attributable solely to the lack of TaAPX-6B.
tAPX Deficiency Leads to Oxidative Stress Susceptibility
Previous reports have demonstrated that chAPXs are highly sensitive to MV-induced oxidative stress (Shikanai et al., 1998; Mano et al., 2001). We observed that exposure to EL leads to reduced foliar APX activity in S-SV8 but not in R-SV8, indicating that the mutant is more susceptible than the parental to the oxidative stress produced by light. Although MV treatment had a similar effect on both the isolines, the inhibition of APX in the mutant was more dramatic and rapid. The inhibition of S-SV8 foliar APX activity at EL could be due to the reduced tAPX activity of the mutant at HL. Then, these plants would be susceptible to oxidative stress at the time they are subjected to EL. In this scenario, the rather limited tAPX activity in the mutant could be easily overwhelmed by the increased H2O2 production at high PFD, as previously suggested by Mano et al. (2001). Besides, upon transfer to EL, mutant plants showed a pronounced decrease of Fv/Fm, indicating photodamage to PSII, which might be the consequence of the increased H2O2 production at the donor side of PSI (Maxwell and Johnson, 2000; Tjus et al., 2001). A recent work reported that transgenic plants that overexpress tAPX in chloroplasts had improved resistance to oxidative stress (Yabuta et al., 2002). Accordingly, our observations indicate that an impaired tAPX activity leads to photodamage at the time that plants are exposed to a sudden oxidative stress. On the contrary, HL-grown mutant plants, which have an impaired tAPX activity at HL steady-state condition, do not display a chronic oxidative damage (see below).
Impairing the PSII Electron Transfer Would Impede Massive Oxidative Damage
The decreased photosynthetic activity of mutant plants is associated with both a low φPSII and a low qP. Because qP estimates the redox state of the primary acceptor of PSII, QA, we hypothesize that the impaired electron transfer of S-SV8 plants may be due to inefficient re-oxidation of QA. It has been reported that the amount of psaA protein (a PSI subunit) decreases faster than the amount of Rubisco, phosphoenolpyruvate carboxylase, and the PSII polypeptide D1 in maize plants after MV treatment (Kingston-Smith and Foyer, 2000), indicating that PSI may be quite sensitive to ROS produced at its donor side. Therefore, inactivation of PSI by ROS may reduce the rate of QA oxidation in tAPX mutant plants, thereby lowering qP and the overall photosynthetic activity of the leaves. The impaired PSII electron transfer of S-SV8 plants depends on the PFD, suggesting that the photosynthetic apparatus is upset only if the rate of H2O2 formation at the donor side of PSI exceeds the reduced tAPX activity of the mutant. In addition, a reduced biomass accumulation is observed at either moderate or high PFD, suggesting that the electron transfer and photosynthetic activity are affected by the reduced capacity of the mutant to cope with ROS. The impaired photosynthesis and the reduced accumulation of dry weight might explain the reduced size of mutant plants. It has been demonstrated that the overexpression of SOD in the chloroplasts of maize confers to transgenic plants a bigger size than wild-type plants, suggesting that an improved removal of ROS ameliorates plant growth and, conversely, that an impaired ROS detoxification limits biomass accumulation (Van Breusegem et al., 1999). Therefore, it seems reasonable to postulate that the reduced size of the mutant could be due to oxidative stress. However, the reduced PSII activity of the mutant may have a protective effect, because this would reduce the rate of electron transfer through PSI and thereby the rate of electron donation to molecular oxygen. A recent work reported that double antisense transgenic tobacco plants lacking cAPX and CAT are less susceptible to oxidative stress than the wild-type plants and that this improved resistance correlates with a reduced photosynthetic activity (Rizhsky et al., 2002). Likewise, although massive oxidative damage might be expected in tAPX mutant plants, no differences were detected in the level of protein carbonylation between R-SV8 and S-SV8 plants grown at LL and HL conditions, suggesting that the mutant is not suffering a massive oxidative damage. However, the reduced size of the tAPX mutant plants may be the cost for escaping oxidative damage by means of reducing photosynthetic electron transport.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The wheat (Triticum aestivum L. cv Sinvalocho MA) isolines R-SV8 and S-SV8 used in this study have been previously characterized at a genetic level. The mutant S-SV8 line originated spontaneously from the inbred R-SV8 line was propagated by self-fertilization (Sacco et al., 1995). Cytogenetic analysis showed a small deletion on the distal region of chromosome 6BL in the S-SV8 line (Sacco et al., 1998). Seeds were germinated on moistened filter paper in the dark. After germination, plants were grown in a greenhouse under a 14-h photoperiod at 18°C to 24°C. Etiolated plants were obtained by continuous exposure to dark. PFD was manipulated to produce LL- and HL-acclimatized plants. LL plants were cultured at 50 to 100 μmol photons m-2 s-1, whereas HL plants were grown at 700 to 1,000 μmol photons m-2 s-1. EL plants were obtained by 1, 2, or 4 h of direct exposure to sunlight (1,800 μmol photons m-2 s-1). Two-week-old plants were harvested, and fresh and dry weights were recorded.
Treatment with MV
Two-week-old plants (second leaf stage) grown at 200 to 400 μmol photons m-2 s-1 were transferred to EL and were immediately sprayed with a solution containing 20 μm MV and 0.01% (v/v) Tween 20. Control plants were sprayed with 0.01% (v/v) Tween 20. Leaf blade samples were obtained to determine foliar APX activity and TaAPX-6B mRNA accumulation.
RNA and DNA Isolation and Cloning of tAPX Genes
Genomic DNA was isolated as previously described (Danna et al., 2002). Total RNA was extracted from plant samples by using the Trizol-Reagent (Invitrogen, Carlsbad, CA). Poly(A) RNA was purified from total RNA by using the PolyA-Track Purification System (Promega, Madison, WI) following the manufacturer's indications. The TaRr16 cDNA previously identified (Danna et al., 2002) was elongated by 5′-RACE System (Invitrogen). First-strand cDNA synthesis was carried out at 65°C using ThermoScript reverse transcriptase (Invitrogen) on 50 ng of R-SV8 poly(A) RNA isolated from leaves. Sequence analysis revealed that the TaRr16 cDNA corresponds to the 3′-UTR of TaAPX-6B (AF532972). Because 3′-UTRs are usually less conserved than the coding regions, we used a specific primer matching the 5′ end of TaAPX-6B cDNA for 3′-RACE procedure to obtain a population of highly homologous cDNAs, thus yielding TaAPX-6A (AF532973) and TaAPX-6D (AF532974).
Gene Expression Studies in R-SV8 Plants
cDNAs were synthesized from total RNA extracted from different organs of R-SV8 plants. Genomic DNA contamination was tested by PCR amplification with specific primers for Actin as described below. Reverse transcription was carried out on 2 μg of RNA with oligo(dT18) oligonucleotide as a 3′ primer. PCR reactions were stopped at a fixed number of cycles during the exponential phase of amplification. Wheat Actin amplification was carried out using 5′-atgtggatatcaggaagga-3′ and 5′-ctcatacggtcagcaatac-3′ as sense and antisense primers, respectively.
To determine the organ-specific expression of tAPX genes, we performed RT-PCR studies. For detection of TaAPX-6B, we used the primers 5′-gcattcttgacgtctctggtc-3′ and 5′-catcttgcatgccgaccaat-3′ as sense and antisense specific primers, respectively. Detection of TaAPX-6A was carried out with 5′-gatcagtgatctaatgttct-3′ and 5′-aatgatgaaaacttaagaca-3′, whereas that of TaAPX-6D was performed with 5′-gtggtccgacgagtctgtct-3′ and 5′-tgctcatcttgcatgtcgat-3′ as sense and antisense primers, respectively. The annealing temperatures for the specific detection of TaAPX-6A, TaAPX-6B, and TaAPX-6D were 50°C, 65°C, and 69°C, respectively. PCR products were subjected to electrophoresis in a 3% (w/v) agarose ethidium bromide-stained gel and were photographed under UV light. Amplification specificity was confirmed by cloning and sequencing the PCR products.
DNA Gel-Blot Analysis and Chromosomal Location of tAPX Genes
Chromosome location was carried out using the aforementioned specific primers for the detection of each tAPX gene. Genomic DNA was isolated from Chinese Spring Nuli6A-Tetra6D and Nuli6D-Tetra6A wheat lines as described above. PCR specificity was confirmed by cloning and sequencing the PCR products. For DNA gel-blot hybridizations, genomic DNA (30 μg) was digested with different restriction enzymes, subjected to electrophoresis in 1× Tris-acetate EDTA 0.8% (w/v) agarose gels, and then transferred onto nylon membranes (Nylon-1, Invitrogen). Probes were labeled with [α-32P]dCTP. DNA blots were prehybridized, hybridized, and washed following the procedures formerly described (Santa-María et al., 1997). Filters were exposed to X-Omat AR films (Eastman Kodak, Rochester, NY) for 1 to 2 d at -70°C.
Expression of Recombinant TaAPX-6B
Two recombinant TaAPX-6B proteins were produced. The large recombinant TaAPX-6B contained the mature coding region of TaAPX-6B and was obtained by digesting TaAPX-6B with EcoRI and by subcloning the digestion product into pTrc-His-C (BD Biosciences Clontech, Palo Alto, CA) protein expression vector. The short version, lacking the putative membrane-anchor region, was obtained by PCR using the larger version as template and the 5′-ggaattccatatgaggatccgatgcatggcggcgt-3′ (BamHI and atg-containing primer) and 5′-tcattagaattctcattattcttgtaggagtatttggc-3′ (EcoRI-containing) as upper and lower primers, respectively. This PCR product was digested with BamHI and EcoRI and was subcloned into pTrc-His-C vector digested with the same enzymes. Escherichia coli XL-1-Blue cultures were induced for the expression of recombinant proteins. Induction of protein expression was carried out for 4 h in 25 mL of Luria-Bertani medium supplemented with 200 μg mL-1 ampiciline and 1 mm isopropylthio-β-galactoside at 37°C and 300 rpm. Culture replicates were grown and isopropylthio-β-galactoside-induced in the presence of 50 mm hemine (Sigma-Aldrich, St. Louis). Bacterial pellets were resuspended in 50 mm PO4K buffer (pH 7) and lysed by sonication at 4°C. E. coli lysates were centrifuged at 20,000g at 4°C for 30 min. Total protein (30 μg) from the lysate was separated by electrophoresis in 12% (w/v) SDS-PAGE gels and was stained with Coomassie Blue.
APX, MDHAR, DHAR, and GR Activities
Fresh leaves (100–200 mg fresh weight) were frozen in liquid N2, ground, and mixed with 2 mL of a 50 mm MES/KOH buffer (pH 7.0) containing 40 mm KCl, 2 mm CaCl2, 1% (w/v) poly-vinyl-pyrrolidone, and 1 mm AA. After thawing, 0.1% (v/v) Triton X-100 was added to the mixture, and tubes were gently mixed for 15 min at 4°C. Homogenates were then centrifuged at 4,500g for 2 min, and the supernatants were used to measure foliar APX activity. For thylakoid isolation, leaf homogenates were centrifuged at 3,000g for 10 min, the pellet was washed and centrifuged again at 3,000g for 10 min, and APX activity was measured in the pellet without addition of Triton X-100 (Guiamét et al., 2002). Recombinant APX activity was measured in 1 mL of buffer containing 20, 40, and 80 μg of total proteins from bacterial lysates (quantified spectrophotometrically by Bradford assay) as described below. Mass of rAPX-2 for specific in vitro activity was estimated by comparison with quantified bovine serum albumin in Coomassie-stained gels. APX was measured spectrophotometrically by a modification of the method of Nakano and Asada (1987). The reaction mixture (950 μL) contained 50 mm KH2PO4/K2HPO4 buffer (pH 7.0), 500 μm AA, and 0.1 mm H2O2. Foliar homogenates (50 μL) were added to the reaction mixture and were gently mixed. Oxidation of AA was followed by a decrease in A290 in a spectrophotometer (DU-650, Beckman Coulter, Fullerton, CA) at 30°C. The reaction rates measured were linear for at least 3 min and were corrected for AA auto-oxidation in the presence of 0.1 mm H2O2. APX activity was calculated using an extinction coefficient of 2.8 mm-1cm-1 for AA. For measurements of MDHAR (EC 1.6.5.4), DHAR (EC 1.8.5.1), and GR (EC 1.6.4.2) activities, leaves were ground in a medium containing 0.1 m Bicine (pH 7.5), 1 mm EDTA, 10% (w/v) glycerol, 4 mm Cys, and protease inhibitors (25 mm phenylmethylsulfonyl fluoride and 2 mm leupeptin). Homogenates were filtered through a 20-μm mesh and were centrifuged at 10,000g for 10 min. The supernatants were used for the determinations of enzyme activities. MDHAR and DHAR were measured essentially as by De Gara et al. (2000), and GR was measured as described previously (Bartoli et al., 1999). AA content was determined by HPLC as described by Iwase (1992).
Photosynthetic Parameters
Photosynthesis was measured with an infrared gas analyzer (LI-6250, LI-COR, Lincoln, NE) fitted in a 1-L assimilation chamber, at 750 μmol photons m-2 s-1, 25°C, and 340 to 360 ppm of CO2. Chlorophyll a fluorescence was measured with an FMS2 Fluorescence Monitoring System (Hansatech, King's Lynn, UK), and fluorescence parameters were calculated as by Maxwell and Johnson (2000). Fluorescence parameters in the light-adapted state (i.e. Fm′, Ft, and F0′) were measured in leaves exposed to the PFDs applied during growth, and then leaves were dark-adapted for 30 min before measuring Fm and F0.
Acknowledgments
We thank Prof. Juan José Cazzulo, Dr. Eyleen J. O'Rourke, and Dr. Federico Katzen for revising the English of the manuscript. We express our gratitude to Dr. Pedro M. Civello and Ariel Vicente for assistance in APX measurements. Chinese Spring Nuli-Tetrasomic plants were kindly provided by Dr. Enrique Suárez and Dr. Silvina Lewis.
This work was supported by the National Research Council of Argentina (CONICET; to R.A.U.) and by the Agencia Nacional de Promoción Científica y Tecnológica of Argentina (ANPCIT; grant no. PICT 01–06565 to R.A.U.). C.H.D. is recipient of a fellowship from CONICET. C.G.B., G.E.S.-M., and R.A.U. are career researchers of CONICET. J.J.G. is a researcher of the Comisión de Investigaciones Científicas-Provincia de Buenos Aires. L.R.I. is a recipient of a fellowship from ANPCIT. F.S. is a researcher of Centro Nacional de Investigaciones Agropecuarias-Instituto Nacional de Tecnología Agropecuana, Castelar.
References
- Asada K (1992) Ascorbate peroxidase: a hydrogen peroxide-scavenging enzyme in plants. Physiol Plant 85: 235-241 [Google Scholar]
- Asada K (1994) Production and action of active oxygen species in photosynthetic tissues. In CH Foyer, PM Mullineaux, eds, Causes of Photooxidative Stress and Amelioration of Defenses Systems in Plants. CRC Press, Boca Raton, FL, pp 77-104
- Asada K (1999) The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol 50: 601-639 [DOI] [PubMed] [Google Scholar]
- Asada K, Takahashi M (1987) Production and scavenging of active oxygen in photosynthesis. In DJ Kyle, CB Osmond, CJ Arntzen, eds, Photoinhibition. Elsevier, Amsterdam, pp 227-287
- Bartoli CG, Simontacchi M, Tambussi E, Beltrano J, Montaldi E, Puntarulo S (1999) Drought and watering-dependent oxidative stress: effect on antioxidant content in Triticum aestivum L. leaves. J Exp Bot 50: 375-383 [Google Scholar]
- Bunkelmann JR, Trelease RN (1996) Ascorbate peroxidase: a prominent membrane protein in oilseed glyoxysomes. Plant Physiol 110: 589-598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton AD, Díaz del Castillo L, Kahn ML, Joyner SL, Chatfield JM (1996) Heterologous expression and characterization of soybean cytosolic ascorbate peroxidase. Arch Biochem Biophys 328: 1-8 [DOI] [PubMed] [Google Scholar]
- Danna CH, Sacco F, Ingala LR, Saione HA, Ugalde RA (2002) Cloning and mapping of genes involved in wheat-leaf rust interaction through gene-expression analysis using chromosome-deleted near-isogenic wheat lines. Theor Appl Genet 105: 972-979 [DOI] [PubMed] [Google Scholar]
- De Gara L, Paciolla C, De Tullio MC, Motto M, Arrigoni O (2000) Ascorbate-dependent hydrogen peroxide detoxification and ascorbate regeneration during germination of a highly productive maize hybrid: evidence of an improved detoxification mechanism against reactive oxygen species. Physiol Plant 109: 7-13 [Google Scholar]
- Foyer CH, Lelandais M, Kunert KJ (1994) Photooxidative stress in plants. Physiol Plant 92: 696-717 [Google Scholar]
- Guiamét JJ, Tyystjärvi E, Tyystjärvi T, John I, Kairavuo M, Pichersky E, Noodén LD (2002) Photoinhibition and loss of photosystem II reaction center proteins during senescence of soybean leaves: enhancement of photoinhibition by the “stay-green” mutation cytG. Physiol Plant 115: 468-478 [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Yoshimura K, Tamoi M, Takeda T, Shigeoka S (1997) Alternative mRNA splicing of 3′-terminal exons generates ascorbate peroxidase isoenzymes in spinach (Spinacia oleracea) chloroplasts. Biochem J 328: 795-800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase H (1992) Determination of ascorbic acid in elemental diet by high-performance liquid chromatography with electrochemical detection. J Chromatogr 606: 277-280 [DOI] [PubMed] [Google Scholar]
- Kingston-Smith AH, Foyer CH (2000) Bundle sheath proteins are more sensitive to oxidative damage than those of the mesophyll in maize leaves exposed to paraquat or low temperatures. J Exp Bot 51: 123-130 [PubMed] [Google Scholar]
- Mano J, Ohno C, Domae Y, Asada K (2001) Chloroplastic ascorbate peroxidase is the primary target of methylviologen-induced photooxidative stress in spinach leaves: its relevance to monodehydroascorbate radical detected with in vivo ESR. Biochim Biophys Acta 1504: 273-287 [DOI] [PubMed] [Google Scholar]
- Mano S, Yamaguchi K, Hayashi M, Nishimura M (1997) Stromal and thylakoid-bound ascorbate peroxidases are produced by alternative splicing in pumpkin. FEBS Lett 413: 21-26 [DOI] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN (2000) Chlorophyll fluorescence: a practical guide. J Exp Bot 51: 659-668 [DOI] [PubMed] [Google Scholar]
- Mittler R, Feng X, Cohen M (1998) Post-transcriptional suppression of cytosolic ascorbate peroxidase expression during pathogen-induced programmed cell death in tobacco. Plant Cell 10: 461-473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Zilinkas BA (1992) Molecular cloning and characterization of a gene encoding pea cytosolic ascorbate peroxidase. J Biol Chem 267: 21802-21807 [PubMed] [Google Scholar]
- Miyagawa Y, Tamoi M, Shigeoka S (2000) Evaluation of the defense system in chloroplast to oxidative stress caused by paraquat using transgenic tobacco plants expressing catalase from Escherichia coli. Plant Cell Physiol 33: 311-320 [DOI] [PubMed] [Google Scholar]
- Miyake C, Asada K (1992) Thylakoid-bound ascorbate peroxidase in spinach chloroplasts and photoreduction of its primary oxidation product monodehydroascorbate radical in thylakoids. Plant Cell Physiol 33: 541-553 [Google Scholar]
- Nakano Y, Asada K (1981) Hydrogen peroxide is scavenged by ascorbate peroxidase in spinach chloroplasts. Plant Cell Physiol 22: 867-880 [Google Scholar]
- Nakano Y, Asada K (1987) Purification of peroxidase in spinach chloroplasts: its inactivation in ascorbate-depleted medium and re-activation by monoedhydroascorbate radical. Plant Cell Physiol 28: 131-140 [Google Scholar]
- Noctor G, Foyer CH (1998) Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49: 246-279 [DOI] [PubMed] [Google Scholar]
- Payton P, Webb R, Kornyeyev D, Allen R, Holaday AS (2001) Protecting cotton photosynthesis during moderate chilling at high light intensity by increasing chloroplastic antioxidant enzyme activity. J Exp Bot 52: 2345-2354 [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Hallak-Herr E, Van Breusegem F, Rachmilevitch S, Barr JE, Rodermel S, Inzé D, Mittler R (2002) Double antisense plants lacking ascorbate peroxidase and catalase are less sensitive to oxidative stress than single antisense plants lacking ascorbate peroxidase or catalase. Plant J 32: 329-332 [DOI] [PubMed] [Google Scholar]
- Sacco F, Favret EA, Suárez EY, Solari RM, Saione HA (1995) Spontaneous genetic variation for leaf rust reaction of Sinvalocho MA. J Phytopathol 143: 251-255 [Google Scholar]
- Sacco F, Suárez EY, Naranjo T (1998) Mapping of the leaf rust resistance gene Lr3 on the chromosome 6B of Sinvalocho MA wheat. Genome 41: 686-690 [Google Scholar]
- Sano S, Asada K (1994) cDNA cloning of monodehydroascorbate radical reductase from cucumber: a high degree of homology in terms of amino acid sequence between this enzyme and bacterial flavoenzymes. Plant Cell Physiol 35: 425-437 [PubMed] [Google Scholar]
- Sano S, Miyake C, Mikami B, Asada K (1995) Molecular characterization of monodehydroascorbate radical reductase from cucumber highly expressed in Escherichia coli. J Biol Chem 270: 21354-21361 [DOI] [PubMed] [Google Scholar]
- Santa-María GE, Rubio F, Dubcovsky J, Rodríguez-Navarro A (1997) The HAK1 gene of barley is a member of a large gene family and encodes a high-affinity potassium transport. Plant Cell 9: 2281-2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen Gupta A, Heinen JL, Holaday AS, Allen RD (1993a) Increased resistance to oxidative stress in transgenic plants that overexpress chloroplastic Cu/Zn superoxide dismutase. Proc Natl Acad Sci USA 90: 1629-1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen Gupta A, Webb RP, Holaday AS, Allen RD (1993b) Overexpression of superoxide dismutase protects plants from oxidative stress. Plant Physiol 103: 1067-1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T, Takeda T, Yamauchi Y, Sano S, Tomizawa K, Yokota A, Shigeoka S (1998) Inhibition of ascorbate peroxidase under oxidative stress in tobacco having bacterial catalase in chloroplasts. FEBS Lett 428: 47-51 [DOI] [PubMed] [Google Scholar]
- Tjus SE, Scheller HV, Andersson B, Møller BL (2001) Active oxygen produced during selective excitation of photosystem I is damaging not only to photosystem I, but also to photosystem II. Plant Physiol 125: 2007-2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Breusegem F, Slooten L, Stassart JM, Moens T, Botterman J, Van Montagu M, Inzé D (1999) Overproduction of Arabidopsis thaliana FeSOD confers oxidative stress tolerance to transgenic maize. Plant Cell Physiol 40: 515-523 [DOI] [PubMed] [Google Scholar]
- Welinder KG (1992) Superfamily of plant, fungal and bacterial peroxidases. Curr Opin Struct Biol 2: 388-393 [Google Scholar]
- Yabuta Y, Motoki T, Yoshimura K, Takeda T, Ishikawa T, Shigeoka S (2002) Thylakoid membrane-bound ascorbate peroxidase is a limiting factor of antioxidative systems under photo-oxidative stress. Plant J 32: 915-925 [DOI] [PubMed] [Google Scholar]
- Yoshimura K, Yabuta Y, Ishikawa T, Shigeoka S (2000) Expression of ascorbate peroxidase isoenzymes in response to oxidative stress. Plant Physiol 123: 223-233 [DOI] [PMC free article] [PubMed] [Google Scholar]