Abstract
In a differential screening between Arabidopsis plants pretreated with the resistance-inducer β-aminobutyric acid and untreated control plants, we have identified a gene encoding a novel lipase-like protein, PRLIP1. The abundance of PRLIP1 mRNAs in Arabidopsis leaves was up-regulated by application of β-aminobutyric acid, salicylic acid (SA), and ethylene as well as by various pathogens. Induction of PRLIP1 depended on a functioning SA and ethylene signal transduction pathway but was independent of jasmonate signaling. This novel pathogenesis-related (PR) gene of Arabidopsis belongs to a gene family consisting of six (PRLIP1, PRLIP2, PRLIP4, PRLIP5, PRLIP6, and PRLIP7) closely related members in tandem position on chromosome 5. Among these genes, PRLIP2 also was induced in leaves by SA and infections by pathogens but on a much lower level than PRLIP1. The PRLIP1 family showed a tissue-specific expression pattern. Both PRLIP1 and PRLIP2 were specifically expressed in leaves and siliques, PRLIP1 additionally in stems and flowers. The expression of PRLIP6 and PRLIP4 was root specific, whereas mRNA of PRLIP5 and PRLIP7 were not detected in any of these tissues. The more distantly related genes PRLIP3, PRLIP9, and PRLIP8 were found on chromosomes 2, 4, and 5, respectively. The expression level of PRLIP3 was checked and found constitutive during the different stress conditions tested. The PRLIP1 gene was overexpressed in Escherichia coli, and the resulting PRLIP1 protein showed esterase activity on p-nitrophenyl-butyrate and allowed the growth of the bacteria on lipidic substrates such as Tween20 or Tween80.
During evolution, plants have developed various mechanisms to cope with numerous biotic and abiotic stresses. Beside constitutive barriers, plants can recognize a microbial invader early in the interaction and activate adequate defense responses. Rapid recognition of a pathogen is based on the presence of resistance (R) genes in the host, the products of which are presumed to interact with the products of avirulence (Avr) genes of the pathogen. This activates multiple signal transduction pathways finally leading to the induction of plant defenses (Dangl and Jones, 2001). At the cellular level, the result of pathogen recognition often becomes apparent as a necrosis localized at the site of attack, called the hypersensitive response, that is accompanied by various cellular changes such as an oxidative burst leading to the release of active oxygen species (ROI), a rise in salicylic acid (SA) levels, and the induction of defense genes such as those coding for pathogenesis-related (PR) proteins (Doke et al., 1996; Mittler and Lam, 1996; Dempsey et al., 1999; van Loon and van Strien, 1999). Such a localized induced defense response often leads to the activation of systemic resistance mechanisms effective against a large number of pathogens (Sticher et al., 1997). This type of resistance can be achieved by a pretreatment with either necrotizing pathogens or chemical inducers of resistance (systemic acquired resistance [SAR]; Ryals et al., 1996) as well as by nonpathogenic rhizobacteria (induced systemic resistance [ISR]; van Loon et al., 1998). SAR and ISR can be distinguished with respect to the signal transduction pathway required for the induction of defense. SAR is SA-dependent leading to the induction of PR genes, whereas ISR functions via a jasmonate (JA)/ethylene (ET)-dependent pathway (Pieterse et al., 1998). The latter pathway is also necessary for the induction of defense genes such as plant defensins (Penninckx et al., 1996).
Recently, genes with homology to lipases were found to be required for SA-dependent induction of defense responses. For instance, the enhanced disease susceptibility 1 (EDS1) gene encodes a protein that has homology to the catalytic site of eukaryotic lipases (Falk et al., 1999). EDS1 has been shown to function upstream of SA-dependent PR1 accumulation but it is not required for JA-induced PDF1.2 expression. Inactivation of EDS1 by mutation leads to enhanced susceptibility, loss of SA accumulation, and PR genes expression (Falk et al., 1999). The phytoalexin deficient 4 (PAD4) gene has originally been identified as playing a role in multiple defense responses including the synthesis of camalexin (the phytoalexin of Arabidopsis; Glazebrook et al., 1996). PAD4 also displays sequence similarity to lipases and accumulates after infection with avirulent bacteria or treatment with SA (Jirage et al., 1999). EDS1 and PAD4 operate upstream of pathogen-induced SA accumulation, yet their expression can be enhanced by exogenous applications of SA (Falk et al., 1999; Jirage et al., 1999). This finding provides evidence for a SA-dependent positive feedback loop that may potentiate plant defense (Shirasu et al., 1997; Falk et al., 1999; Jirage et al., 1999). Both pad4 and eds1 mutations affect the TIR-NB-LRR type of R gene functions, but the loss of resistance in pad4 is typically not as complete as in eds1 (Feys et al., 2001).
In addition to SA, JA and ET also play important roles as signal molecules mediating disease resistance, especially in response to necrotrophic fungal pathogens (Penninckx et al., 1996; Reymond and Farmer, 1998) or in ISR (Pieterse and van Loon, 1999). Most of the JA/ET-dependent defense responses analyzed to date are SA independent. For instance, the plant defensin gene PDF1.2 is induced concomitantly by the JA- and ET-signaling pathways but not by SA. However, its induction is enhanced in transgenic NahG plants (Penninckx et al., 1998), indicating that SA may down-regulate the JA/ET signaling. In eds1 and pad4 mutant plants with reduced SA levels, elevated JA-dependent gene expression in response to inducers was also observed, providing genetic evidence for the interference of SA with JA-dependent signaling (Gupta et al., 2000). Additional indication for an interaction of the SA and JA/ET pathways is provided by the cpr6 and acd2 mutations that cause constitutive expression of the JA/ET-responsive PDF1.2 gene as well as the SA-responsive PR genes (Penninckx et al., 1996; Clarke et al., 1998). Novel PR genes and other marker genes, therefore, may provide insight to the regulation of these pathways and their interactions.
Arabidopsis plants pretreated with the non-protein amino acid β-aminobutyric acid (BABA) exhibit enhanced resistance against pathogens (Zimmerli et al., 2000). This protective effect of BABA is based on a more rapid and efficient defense response upon exposure to pathogen attack. This phenomenon is known as potentiation or priming (Conrath et al., 2002) and BABA is able to potentiate SA-dependent or -independent defenses. To gain insight in the molecular mechanisms behind BABA-induced priming, we have performed a screening for differentially expressed genes after BABA treatment in Arabidopsis.
Here, we describe the isolation by subtraction suppression hybridization of a BABA-inducible gene, PRLIP1, and the biological and biochemical characterization of this gene and its product. The predicted PRLIP1 amino acid sequence has regions of similarity to Ser hydrolases (eukaryotic lipases and esterases) similar to PAD4 (Jirage et al., 1999) and EDS1 (Falk et al., 1999). PRLIP1 is a member of a unique Arabidopsis gene family with six members in tandem position on chromosome 5. The expression profile of this lipase-like gene family will also be described.
RESULTS
Description of the Gene Family
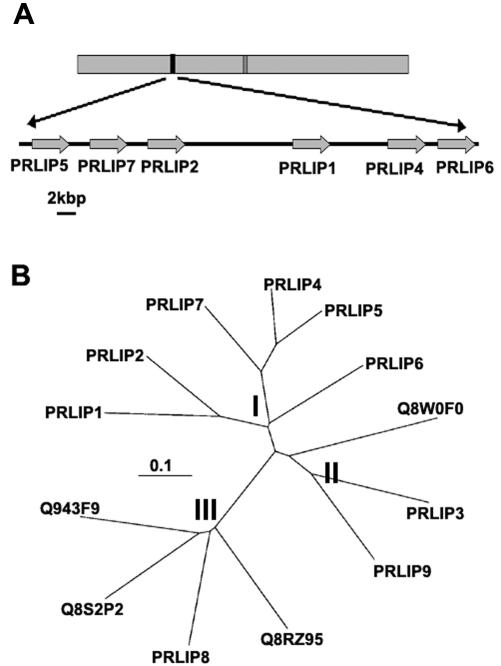
Subtractive suppression hybridization was used to detect changes in gene expression after treatment with BABA, an inducer of resistance to pathogens (Zimmerli et al., 2000). This approach yielded fragments representing differentially expressed mRNAs. They were used as probes to isolate the corresponding cDNAs from a cDNA library constructed from benzothiadiazole (BTH)-treated Arabidopsis Wassilewskija (WS) leaves (BTH is a functional analog of SA). One of the cDNAs showing the highest differential expression was designated PRLIP1, and its full-length cDNA corresponded to a gene localized on chromosome 5 (Fig. 1A). Using its amino acid sequence encoding a lipase-like protein in a BLAST search of the Arabidopsis genome yielded five other lipase-like genes in tandem arrangement with PRLIP1 on chromosome 5 (PRLIP2, PRLIP4, PRLIP5, PRLIP6, and PRLIP7), one localized on chromosome 2 (PRLIP3), one localized on chromosome 4 (PRLIP9), and one localized on another region of chromosome 5 (PRLIP8). A comparison of the amino acid sequences of the lipase-like proteins clustered the tandem PRLIP genes of chromosome 5 together, whereas PRLIP3, PRLIP8, and PRLIP9 had a distinctly different sequence (Fig. 1B). The similarity within the tandem family is high (63%–45% identity), whereas the homology to the other paralogs in the Arabidopsis genome is much lower. PRLIP1 shows only 36%, 35%, and 27% identity to PRLIP3, PRLIP9, and PRLIP8, respectively. According to their homology, we have divided the PRLIP proteins into subgroups. Subgroup I contains PRLIP1, PRLIP2, PRLIP4, PRLIP5, PRLIP6, and PRLIP7; subgroup II has PRLIP3 and PRLIP9; whereas PRLIP8 belongs to subgroup III. Predicted proteins from genomic sequences or expressed sequence tags showing homology to subgroup II or subgroup III were found in monocots like rice (Fig. 1B), wheat (Triticum aestivum), barley (Hordeum vulgare), rye (Secale cereale), and maize (Zea mays) and in dicots like tomato (Lycopersicon esculentum), potato (Solanum tuberosum), cotton (Gossypium hirsutum), lettuce (Lactuca sativa), soybean (Glycine max), Medicago truncatula, and Lotus japonicus, or even in loblolly pine (Pinus taeda; data not shown). However, no sequence showing homology to subgroup I proteins was detected in other plant species.
Figure 1.
Genomic positions of the PRLIP genes and their phylogenetic relationships. A, The arrangement of the PRLIP1 (At5g24210), PRLIP2 (At5g24200), PRLIP4 (At5g24220), PRLIP5 (At5g24180), PRLIP6 (At5g24230), and PRLIP7 (At5g24190) genes on chromosome 5 of Arabidopsis. A 37-kb region, where these genes are in tandem positions, is projected in a bigger scale. B, Unrooted phylogenetic tree of the Arabidopsis PRLIP proteins of the tandem repeat and three additional Arabidopsis paralogs (PRLIP3:At2g05260, PRLIP8: At5g50890, and PRLIP9:At4g10955) and their rice (Oryza sativa) homologs (Q943F9, Q8S2P2, Q8RZ95, and Q8W0F0). The protein sequences were aligned with ClustalW software, and an unrooted tree was constructed using the TreeView program. The scale value of 0.1 indicates 0.1 amino acid substitutions per site. I, subgroup I; II, subgroup II; and III, subgroup III.
The catalytic triad typical for the esterase and lipase activity, which includes the amino acids Ser, Asp, and His, is conserved among all the PRLIPs (Fig. 2), strongly suggesting that these proteins have esterase or lipase activity. Comparison of PRLIPs with other known or predicted Ser hydrolases and lipases placed them in a separated branch of the phylogenetic tree (data not shown), indicating that PRLIPs are representing a novel protein family.
Figure 2.
Alignment of PRLIP amino acid sequences. Identical amino acids are shown in black boxes, whereas conserved amino acid changes are shaded in gray. The arrowheads above the sequence show the conserved Ser (S), Asp (D), and His (H) residues that can form a putative lipase catalytic triad. The conserved secondary structure elements, a sharp turn of a β-sheet, and an α-helix around the S residue are indicated with a line above the sequence. The position of introns are indicated by lowercase letters. The first intron is present in position a in all genes except in PRLIP8 where it is shifted downstream to position b; the second intron of PRLIP9 is in position c; whereas all genes of subgroup I have the second intron in position d. PRLIP3 and PRLIP8 have no second intron.
The gene structure of the PRLIPs is also characteristic for each subgroup. The first intron of the coding region is always present and has a conserved position (Fig. 2) not only in Arabidopsis but also in the rice orthologs. The second intron, however, has a subgroup-specific characteristic. In all subgroup I genes, it has been observed in a conserved position of a variable region of the PRLIP proteins (Fig. 2). Subgroup II or III genes have no second intron in both Arabidopsis and rice with the only exception of PRLIP9 (Fig. 2).
Response of the Different PRLIP Genes to Various Treatments
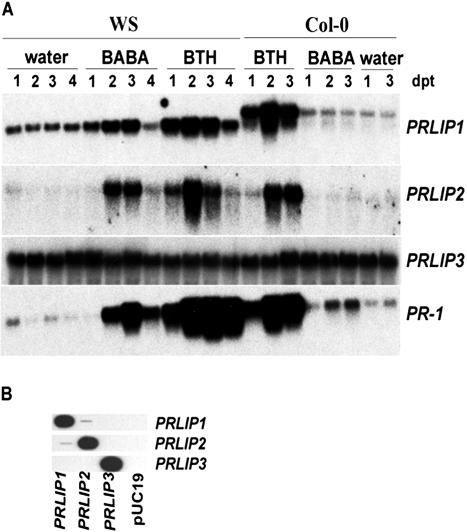
Increased expression of PRLIP1 and PRLIP2 genes after BABA and BTH treatments was found in northern blots (Fig. 3A) using sequence-specific probes (Fig. 3B). The mRNA levels of PRLIP4, PRLIP5, PRLIP6, and PRLIP7 in Arabidopsis leaves were too low to be detected. PRLIP1 had a basal overall mRNA level in non-treated leaves of both Arabidopsis accessions WS and Columbia (Col-0). Its expression was further induced by BTH in both accessions and additionally by BABA in WS. In Col-0 the mRNA of the PRLIP1 gene appeared to be longer. Besides a 19-nucleotide changes between the coding regions of the two accessions, sequence analysis revealed the presence of an additional intron in the 3′-untranslated region of Col-0 where the acceptor site was mutated (AG → GG), thus impairing its splicing (data not shown). The PRLIP2 gene had no detectable basal expression and was also inducible by BTH (both WS and Col-0) and BABA (WS; Fig. 3A), although to a much lower level than PRLIP1 (note the difference of the exposure times). Treatment with BTH led to an increase in expression of PRLIP1 and PRLIP2, and the kinetics of induction for both genes were comparable with that of PR-1. The effect of BABA on the induction of PRLIP1, PRLIP2, and PR-1 was much stronger in WS than in Col-0.
Figure 3.
Induction of the PRLIP1 and PRLIP2 genes in Arabidopsis accessions WS and Col-0 in response to BABA and BTH treatments. A, Four-week-old soil-grown plants were treated with BABA (300 μm) or BTH (300 μm) as soil drench. The plants were harvested 1 to 4 d post treatment, as indicated at the top of the figure. RNA blots were hybridized with the probes indicated on the right of the figure. The membranes were exposed to Biomax films (Eastman Kodak, Rochester, NY) for 12 h (PRLIP1, PRLIP3, and PR1) or for 48 h (PRLIP2). B, The specificity of the PRLIP probes was tested by slot blot of the cloned cDNA fragments. Plasmid DNA (1 ng of each) containing a fragment of the different PRLIP cDNAs, as indicated under the figure, was loaded in replicates. The blots were hybridized with the probes indicated on the right of the figure. DNA of the plasmid pUC19 (1 ng) was used as negative control.
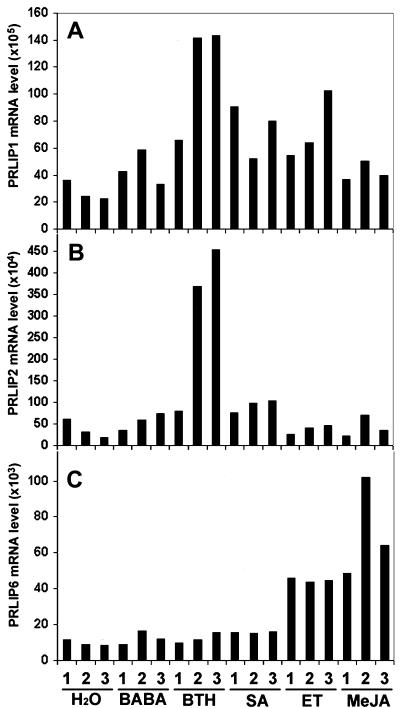
The expression pattern of the related gene from subgroup II, PRLIP3, localized at another genetic locus was tested and showed a constitutive expression level (Fig. 3A). Therefore further expression studies have been focused on the closely related genes of subgroup I. The induction of these PRLIP genes in Arabidopsis (Col-0) leaves after treatments with BABA, BTH, SA, ET, and methyl jasmonate (MeJA) was further investigated using real-time PCR to avoid the difficulties due to high sequence homologies and low mRNA levels (Fig. 4). The expression of PRLIP1 was induced by BTH, SA, and ET but not MeJA whereas PRLIP2 gene showed induction by BTH and SA only. A strong induction of both genes was observed after UV treatment but wounding had no effect (data not shown). PRLIP6 was induced by both ET and MeJA but not by SA and BTH (Fig. 4). Neither PRLIP4 nor PRLIP5 nor PRLIP7 mRNA was detected after these treatments.
Figure 4.
Expression pattern of PRLIP1, PRLIP2, and PRLIP6 genes in Arabidopsis (Col-0) leaves after treatment with different chemicals. Analyses were performed by real-time PCR with RNA extracted from plants after water, BABA, BTH, SA, ET, and MeJA treatment. Plant material was harvested 1, 2, or 3 d post treatment. The PCR reactions were performed with primers specific to the cDNA of PRLIP1 (A), PRLIP2 (B), or PRLIP6 (C). Results presented are typical data from three independent experiments and are expressed in mRNA level relative to the lowest detected PRLIP4 mRNA level in control leaves after normalization against Arabidopsis β-tubuline gene (see “Materials and Methods”).
Developmental Regulation of the PRLIP Genes
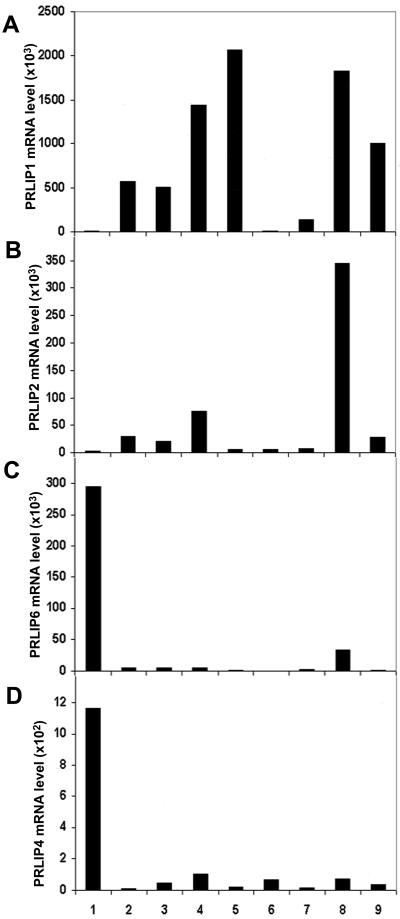
Real-time PCR was also used to determine the constitutive expression of the PRLIP genes in various organs of Arabidopsis plants (Fig. 5). Both PRLIP1 and PRLIP2 were expressed in 2-week-old Arabidopsis seedlings as well as in leaves and in siliques, and PRLIP1 was also highly expressed in stems. The expression level of PRLIP2 was always lower than PRLIP1; it was very weak in stems and flowers, and similarly to PRLIP1, it remained undetected in roots. In contrast, PRLIP6 was only weakly expressed in seedlings, leaves, stems, flowers, and siliques, but was highly expressed in cultured roots. The expression of the PRLIP4 gene was above detection level in root tissues only, whereas PRLIP5 and PRLIP7 were below detection level in all organs (data not shown). Thus, the expression of both PRLIP1 and PRLIP2 presented predominance in green tissues with a stronger overall expression of PRLIP1, whereas PRLIP4 and PRLIP6 showed root preference in their expression. Therefore, PRLIP1 and PRLIP2 were retained for further analyses, because both were induced in leaves.
Figure 5.
Tissue- and organ-specific expression pattern of PRLIP1, PRLIP2, PRLIP4, and PRLIP6 genes. Analyses were performed by real-time PCR with RNA extracted from cultured roots (1), 2-week-old seedlings (2), young leaves (3), fully expanded leaves (4), stems (5), young flowers (6), fully opened flowers (7), young siliques (8), and mature siliques (9). The PCR reactions were performed with primers specific to the cDNA of PRLIP1 (A), PRLIP2 (B), PRLIP6 (C), or PRLIP4 (D). Results presented were calculated from triplicate data and are expressed as mRNA level relative to PRLIP4 mRNA level in control leaves after normalization against Arabidopsis β-tubuline gene (see “Materials and Methods”).
Expression of PRLIP1 and PRLIP2 after Infection with Pathogens
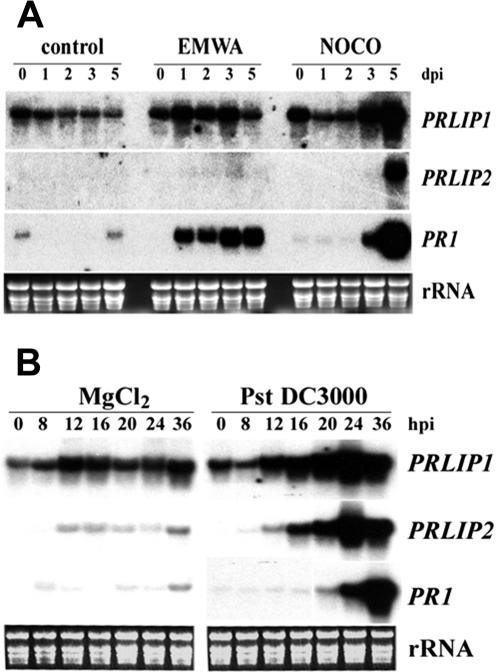
Inoculation of Arabidopsis (Col-0) with the avirulent Peronospora parasitica isolate EMWA led to a strong increase in the expression of PRLIP1 and PR1 that was detectable as early as 1 d after inoculation, whereas no change was observed in PRLIP2 expression (Fig. 6A). During the compatible interaction with P. parasitica isolate NOCO, an increase in expression of the three genes was only detected 3 to 5 d after inoculation (Fig. 6A). Infection with the virulent bacteria Pseudomonas syringae pv tomato (Pst) DC3000 induced PRLIP1 and PRLIP2 between 12 and 16 h postinoculation (hpi), whereas the expression of PR-1 was enhanced only 24 hpi (Fig. 6B).
Figure 6.
Induction of the PRLIP1 and PRLIP2 genes in Arabidopsis (Col-0) leaves in response to infection by pathogens. A, Three-week-old plants were inoculated with the incompatible isolate (EMWA) or the compatible isolate (NOCO) of P. parasitica, or sprayed with water (control). Plants were harvested for RNA isolation in a time course as indicated above the figure (dpi, days post inoculation). B, Leaves of 4-week-old plants were injected with the virulent bacterial pathogen Pst DC3000 or MgCl2 solution. Plant material was harvested for RNA isolation in a time course as indicated above the figure (hpi, hours postinoculation). RNA blots were hybridized with the probes indicated on the right of the figure. The membranes were exposed to Biomax films (Eastman Kodak) for 12 h (PRLIP1 and PR1) or for 48 h (PRLIP2). Equal loading is shown by rRNA stained with ethidium bromide.
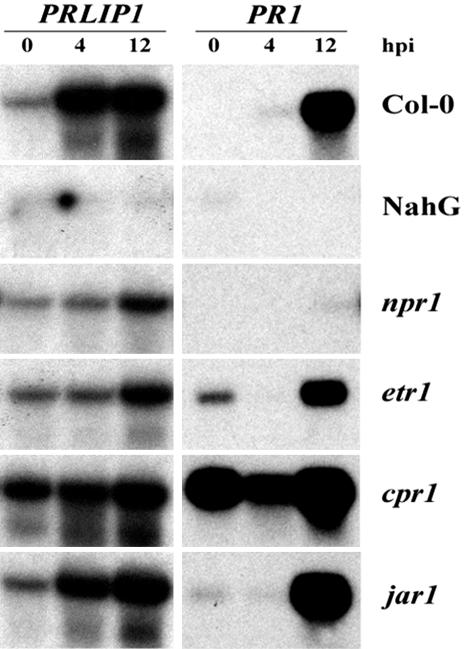
Inoculation with the avirulent bacteria Pst avrRpt2 led to an induction of PRLIP1 as early as 4 hpi, whereas the effect on PR-1 expression was apparent only after 12 hpi (Fig. 7). The response to avirulent bacteria was abolished in Arabidopsis overexpressing the NahG gene (Delaney et al., 1994) and was reduced in the SA-signaling mutant npr1 (Cao et al., 1994). The expression of the PRLIP1 gene was enhanced in cpr1, a mutant that constitutively overexpresses PR-1 and accumulates elevated levels of SA (Bowling et al., 1994). The expression of PRLIP1 was decreased in the ET-insensitive mutant etr1 (Chang et al., 1993) but not in the JA-insensitive mutant jar1 (Staswick et al., 1992; Fig. 7).
Figure 7.
Induction of the PRLIP1 gene in signaling mutants of Arabidopsis (Col-0) in response to infection with the avirulent bacterial pathogen, Pst DC3000 avrRpt2. Leaves of 4-week-old plants were injected with the bacterial cell suspension, and plant material was harvested 0, 4, or 12 h later as indicated above the figure (hpi, hours postinoculation). RNA was extracted from the wild-type, transgenic, and mutant lines (indicated on the right of the figure), and the blots were hybridized with PRLIP1- or PR1-specific probes (indicated on the top line).
Biochemical Function of PRLIP1
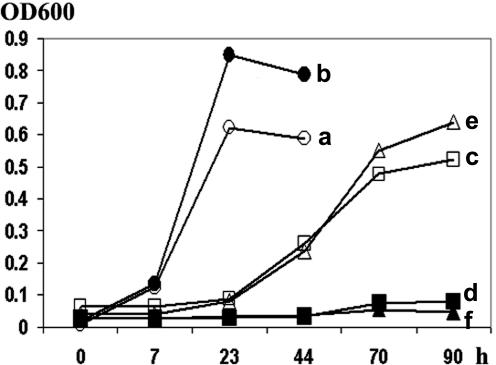
To study the enzymatic activity of PRLIP proteins, PRLIP1 was selected from this lipase-like gene family based on its expression pattern in response to various inducers. Its cDNA isolated from Arabidopsis (Col-0) was inserted into the multiple cloning site of the pGEX vector and expressed as a fusion protein with glutathione S-transferase (GST). Escherichia coli cells harboring this plasmid expressed a GST-PRLIP1 fusion protein of about 65 kD as shown by the presence of a band of the predicted Mr in homogenates of transformed bacteria (data not shown). To test for esterase activity, the potential of transformed bacteria to grow on Tween20 or Tween80 was compared between GST-PRLIP1-expressing cells and control cells expressing only GST (Hong et al., 2000). Figure 8 shows that only GST-PRLIP1-expressing bacteria were able to grow on a lipidic substrate, indicating that GST-PRLIP1 retained its esterase activity. GST-PRLIP1 could not be further analyzed, because the heterologous protein was associated with inclusion bodies and thus purification of the protein was not possible. Therefore, the PRLIP1 cDNA from both Col-0 and WS was also cloned into the bacterial expression vector pQE30 containing a 6His tag. The 6His-PRLIP1 was solubilized using an extraction buffer containing 8 m urea and purified and refolded on a cobalt column. By using the artificial substrate p-nitrophenyl-butyrate, an esterase activity of the purified and refolded protein was detected (Table I). Similar results were obtained with 6His-PRLIP1 protein of both Arabidopsis accessions. However, when the refolded proteins were tested with triolein, no triacylglycerol-lipase activity was detected (data not shown).
Figure 8.
Heterologous expression and in situ assay of lipolytic acyl hydrolase activity of PRLIP1 protein in E. coli cells. Cells harboring the fusion protein construct (GST-PRLIP1; a, c, and e) or the empty vector (pGEX-5X-1; b, d, and f) were cultured in basal salt medium (M9) supplemented with Glc (a and b), Tween20 (c and d), or Tween80 (e and f) and 1 mm isopropylthio-β-galactoside. The bacterial growth was determined by measurement of the optical density at 600 nm. The assay was performed three times with comparable results.
Table I.
Esterase activity assayed using p-nitrophenyl butyrate as substrate
Assays were performed in duplicates. Substrate conversion was linear during the time course of the experiment (90 min), and the result is expressed as nanomoles of p-nitrophenol formed per milligrams of protein per minute. Protein extracted from E. coli cells harboring either the empty pQE30 expression vector (pQE30) or the cDNA insert of PRLIP1 from Col-0 (PRLIP1-Col-0) or WS (PRLIP1-WS) was solubilized in 8 m urea and refolded before the activity assay.
| Construct | p-Nitrophenol |
|---|---|
| nmol mg-1min-1 | |
| pQE30 | 0.0014±0.0001 |
| PRLIP1-Col-0 | 3.735±0.065 |
| PRLIP1-WS | 10.475±0.365 |
DISCUSSION
Plants respond to the different biotic stresses by a local and systemic induction of various novel proteins referred to as PR proteins (van Loon and van Strien, 1999). Some PR proteins may be effective in inhibiting pathogen growth, multiplication and/or spread, whereas others may be responsible for maintenance of SAR (Ryals et al., 1996). Most PR proteins often are members of gene families and exist in both basic and acidic isoforms (Samac et al., 1990; Brederode et al., 1991). Acidic PR proteins are predominantly extracellular, and their expression is characteristically induced after the accumulation of SA (Malamy et al., 1990; Métraux et al., 1990).
In contrast to their acidic counterparts, many basic PR proteins do not accumulate in response to SA or during the establishment of SAR. The expression of basic PR proteins can be triggered by ET (Brederode et al., 1991; Eyal et al., 1993) and/or by JA (Penninckx et al., 1996; Reymond and Farmer, 1998) and is under the control of an organ-specific expression program (Eyal et al., 1993; Thomma and Broekaert, 1998). The different PRLIP genes presented here show expression patterns characteristic for PR genes.
First of all, the expression of PRLIP1 and PRLIP2 was induced in Arabidopsis leaves infected with P. parasitica, P. syringae pv tomato DC3000 (Fig. 6), and Phytophthora brassicae (data not shown), whereas no induction was observed after Botrytis cinerea infection (data not shown). The expression of PRLIP1 but not PRLIP2 was induced during the incompatible interaction with by P. syringae pv tomato avrRpt2, and the accumulation of mRNA preceded that of PR-1 (Fig. 7), a widely used marker for the SA pathway (Sticher et al., 1997), indicating an earlier role of the PRLIP1 protein in SA-mediated signaling and gene expression.
Like other PRs, the PRLIP genes were also induced in Arabidopsis upon ectopic treatments with various plant defense signals such as SA, ET, or JA. For instance, PRLIP1 and PRLIP2 were induced by SA in leaves, whereas PRLIP6 was induced in the same tissue by ET and MeJA (Fig. 4). The expression pattern of PRLIP1 placed this gene among the few ones known to be influenced by both SA and ET (Gu et al. 2000; Onate-Sanchez and Singh, 2002). This property places PRLIP1 at the crossroads of two signaling pathways adding additional specificity to its regulation. The accumulation of PRLIP1 was decreased in plants unable to accumulate SA (NahG plants) or mutants impaired in their response to SA (npr1) or ET (etr1), confirming the SA- and ET-dependent expression of this gene. On the basis of these results, we concluded that the PRLIP genes of subgroup I could form a bona fide PR gene family with members whose expression is regulated by SA (PRLIP2), SA and ET (PRLIP1), or ET and JA (PRLIP6). PRLIP genes encode proteins structurally belonging to the α/β-Ser hydrolase enzymes, and we have demonstrated that at least one of them, PRLIP1, has an esterase activity on lipidic substrates.
The lipase-like proteins EDS1 and PAD4 have been shown to be important in pathogen-induced accumulation of SA (Falk et al., 1999; Jirage et al., 1999). This suggests that lipid-derived molecules can be involved in processes that control the production of SA, although the exact biochemical function of neither EDS1 nor PAD4 has been determined.
More is known about the role of lipases in JA-dependent defense signaling. JA is produced through the octadecanoid pathway and is initiated by the addition of molecular oxygen to linolenic acid (Schaller, 2001). Lipases, namely phospholipase D (PLD), have been postulated to produce linolenic acid from membrane phospholipids (Schaller, 2001). Antisense suppression of a PLD gene in Arabidopsis revealed the involvement of PLD in wound-induced accumulation of JA (Wang et al., 2000). Induction of genes encoding PLD and lysophospholipase (lysoPL1) in Arabidopsis leaves after P. syringae infection has also been reported (de Torres Zabela et al., 2002). PLDγ 1 and lysoPL1 are specifically upregulated during gene-for-gene interactions and upon SA treatment.
Recently, a phospholipase A1, defective in anther dehiscence1 (DAD1) was identified and shown to be involved in JA biosynthesis (Ishiguro et al., 2001). The expression of DAD1, however, is restricted to the stamen filaments making it unlikely to participate in wound- or pathogen-induced JA production in other tissues.
An ET-inducible gene has been reported recently in carnation encoding a protein capable of deesterifying fatty acids from p-nitrophenyl palmitate, tri-linolein, soybean phospholipids, and Tween (Hong et al., 2000). Inhibition of its Arabidopsis homolog by an antisense construct led to delayed senescence (Thompson et al., 2000), suggesting that lipases may also have important physiological role in that process.
Recent studies suggest that cross-talk between SA-, JA- and ET-dependent signaling pathways fine-tunes plant defense responses (Glazebrook, 2001). Emerging evidence indicates that membrane lipids not only serve as a passive, hydrophobic barrier for compartmentalization, but also play important and active roles in cellular processes such as signal transduction cascades and enzyme activation (Munnik, 2001; Laxalt and Munnik, 2002). Determination of the in vivo substrate(s) of the newly recognized pathogen-inducible PRLIP Ser hydrolase proteins may provide further insight into these signaling processes during plant-pathogen interactions.
MATERIALS AND METHODS
Biological Material
Arabidopsis mutants npr1, cpr1, jar1, and etr1 (all in Col-0 background) were obtained from X. Dong (Duke University, Durham, NC), P.E. Staswick (University of Nebraska, Lincoln), and the Nottingham Arabidopsis Stock Center (UK), respectively. A transgenic line of Arabidopsis (Col-0) harboring the NahG gene (Delaney et al., 1994) was provided by J. Ryals (Novartis, Research Triangle Park, NC). Arabidopsis wild-type accessions WS and Col-0 were obtained from Lehle Seeds (Round Rock, TX). Plants were grown in a steam-sterilized soil mix of commercial potting soil:perlite (3:1, v/v) at 22°C day and 18°C night temperature with 12 h of light per 24 h. Root cultures were maintained in Arabidopsis root culture medium according to Czakó et al. (1993).
Pathogen Maintenance and Plant Inoculation
Strain DC 3000 of Pseudomonas syringae pv tomato and the isogenic strain carrying the Avr gene avrRpt2 were cultivated at 28°C, 235 rpm in King's medium B containing rifampicin for selection as described earlier (Zimmerli et al., 2000).
For bacterial inoculation, cells were collected by centrifugation, resuspended in 10 mm MgCl2 to an approximate concentration of 105 colony forming units mL-1. Three leaves per plant were infiltrated using a 1-mL syringe without needle. Tissue samples were harvested from inoculated leaves at the indicated time points, flash frozen in liquid N2, and kept at -80°C until further use.
Plants were inoculated with Peronospora parasitica by spraying with a suspension of 105 conidia mL-1 water until shortly before run-off occurred. Inoculated plants were kept at 20°C in a 12-h/12-h light/dark cycle and during the 1st d of the growth cycle at 100% relative humidity to ensure infection. To produce new inoculum, 6 d after inoculation, plants were placed back to 100% relative humidity for 1 d to induce sporulation.
Chemical Treatment
BABA (Fluka, Buchs, Switzerland) and BTH (Novartis) were dissolved in water (300 μm) and applied as soil drench as described earlier (Zimmerli et al. 2000). SA (Fluka) was dissolved in water (1 mm) and sprayed onto the leaves. MeJA was applied as a droplet on a filter paper and allowed to evaporate in airtight containers to give a final concentration of 0.25 μL L-1. ET gas was injected into an airtight container to a concentration of 100 μL L-1.
RNA Extraction and Analysis
RNA was isolated from frozen tissue samples as described previously (Zimmerli et al., 2000). Total RNA samples (10 μg) were separated in formaldehyde-agarose (1.2%) gels and were blotted to a nylon membrane (Hybond-N, Amersham Biosciences, Little Chalfont, UK). [32P]cDNA probes were synthesized by random priming of the isolated DNA fragments using the random primers DNA labeling kit (RadPrime DNA Labeling System, Life Technologies, Merelbeke, Belgium). To obtain the gene-specific DNA fragments, PCR amplification was used with the following primers: PRLIP1, PRLIP1fw: 5′-GCAGTGCGGTTACAATCAC and PRLIP1rev: 5′-AGCATATGATTCCACCATTG; PRLIP2, PRLIP2fw: 5′-GCAGCTAGTGGAGAGTGAG and PRLIP2rev: CACCATTGATGAATCCCATG; and PRLIP3, PRLIP3fw: 5′-GGGTTAGGCACGGGATTAG and PRLIP3rev: 5′-GGTCATCTCTCCACCATTG.
Subtraction Suppression Hybridization
The PCR-Select cDNA Subtraction kit (BD Biosciences Clontech, Palo Alto, CA) was used according to manufacturer's instructions. The obtained PCR products were separated on sequencing PAGE, and the bands were stained by silver. The selected bands showing differential expression were excised and incubated in 100 μL of TE buffer overnight. The redissolved DNA was than reamplified using the same primer pair it was obtained with.
Real-Time Quantitative PCR
The ABI Prism 7700 Sequence Detection System (Applied Biosystems, Foster City, CA) was used according to the manufacturer's instructions. The reverse transcription reactions were performed using 2 μg of total RNA, oligo(dT)18 primer, and the Omniscript kit (Qiagen, Hilden, Germany). The resulting cDNA solution was then diluted with water up to 1 mL. In the PCR reactions, 10 μL of this cDNA solution, 300 nm of each primers, and SYBR Green PCR Master Mix (Applied Biosystems) were used in a 50-μL final volume. The β-tubuline cDNA (At5g44340) was used as an endogenous control for equal loading, and the lowest detected PRLIP4 level was used as baseline for relative quantification. The gene-specific primer pairs used for the PCR are listed: β-tubuline, RTbtubfw: 5′-CCTCAAGCTCGCTAATCCTACCTTTGG and RTbtubrev: 5′-AGAAGTGAAGCCTTGGGAATGGGA; PRLIP1, LIP1fw: 5′-CCAAAGTCGTAGACCTTGATGAGGGTC and LIP1rev: 5′-CTCGAAATCTCACCGAGTCCCGC; PRLIP2, LIP2fw: 5′-CATAGCAAGAGTCCTAGACCTTAATGAGGGTC and LIP2rev: 5′-AACCTCTCGATCCTACCCAGTTGCAG; PRLIP4, LIP4fw: 5′-CGTAGCTATGGTCTTAACCGATCTACAGGTTC and LIP4rev: 5′-TTTCCCATCAATAGACTCCTAACTGACTTTCC; PRLIP5, LIP5fw: 5′-CCATGGCCTTAACCGATCTACAGGTTC and LIP5rev: 5′-CTTCCCACCAATAGACTCCTAACTGATTTTCT; PRLIP6, LIP6fw: 5′-GTTAAGGGTCGCCACCATAATAAGGGTC and LIP6rev: 5′-CAATTCTTTCGATTTTACCGGCTCCG; and PRLIP7, LIP7fw: 5′-GTTACTATGGCCTTAACTGATCAACGGGTTC and LIP7rev: 5′-CTCGATGTGGCTTGCTCCGATCTT. To avoid the amplification of contaminating genomic DNA, forward primers were designed to span over exon-exon junctions with their last four nucleotides.
Expression of PRLIP Protein in Escherichia coli
For heterologous expression of the GST-PRLIP1 fusion protein in E. coli, the pGEX-5X-1 vector (Amersham Biosciences, Uppsala) was used. The full-length coding region of PRLIP1 cDNA was obtained by PCR amplification using a 1:1 mixture of the Pfu and TaqDNA polymerases (Promega, Madison, WI) and the primers PRLIP1/5′Bam: 5′-CGGATCCACGAAAATTGGAAGGAGG and PRLIP1/3′Bam: 5′-CGGATCCTTAACAAATTATTCAGTTGACAAG. The fragment was inserted at the BamHI site of the vector and the construct was verified by sequencing (Microsynth, Balgach, Switzerland). To obtain 6x-His-tagged PRLIP1 protein, the cDNA was reinserted into the BamHI site of the pQE-30 vector (Qiagen).
E. coli BL-21 cells were transformed with these constructs, and protein was extracted from the sonicated bacteria by B-PER Bacterial Protein Extraction Reagent (Pierce, Rockford, IL). To purify the 6x-His-PRLIP1, protein inclusion bodies were solubilized using B-PER supplemented with 8 m urea. The 6x-His-tagged protein was then bound on a TALON Resin column (BD Biosciences Clontech) in the same buffer. The refolding of the denatured protein was done on the column by stepwise reduction of the concentration of urea to 0 m. The folded protein was eluted with 100 mm imidazole in 0.1 m phosphate buffer, pH 7. Protein concentrations were measured according to Bradford (1976), with bovine serum albumin as a standard. Esterase activity was measured spectrophotometrically using p-nitrophenyl-butyrate (Sigma-Aldrich, St. Louis) as substrate and monitoring the change in the OD400, corresponding to the absorption maximum of p-nitrophenolate anion with a molar extinction coefficient of 14,000 (Shirai and Jackson, 1982).
Protein Gel Electrophoresis
SDS-PAGE was performed according to Laemmli (1970), using a 5% (w/v) stacking gel and a 13% (w/v) running gel in a Mini-Protean II electrophoresis cell (Bio-Rad Laboratories, Richmond, CA). The run was carried out at 150 V of constant voltage. Proteins were detected in gels by Coomassie Brilliant Blue R-250 staining, and their molecular masses were determined by referring to the mobility of known molecular mass standards (Broad Range, Bio-Rad Laboratories).
Growth Assays on Different Substrates
Bacteria were grown in M9 minimal medium (Sambrook et al., 1989) supplemented with 1 mm isopropylthio-β-galactoside, 100 μg mL-1 ampicillin, and either 0.2% (w/v) of Tween20, Tween80, or Glc. The growth rates of the bacterial suspensions were determined at OD600 using BioPhotometer (Eppendorf, Boulder, CO).
Triacylglycerol Lipase Activity Assay
Triacylglycerol lipase activity was tested according to MacKenzie et al. (1967). In brief, 20 μL of refolded protein extract (100 μg protein) was placed on 2% (w/v) agarose plates containing 0.1 m Tris-HCl, pH 8, 25 mg mL-1 triolein (Fluka), and 0.02% (w/v) rhodamine B and incubated for 2 to 48 h at room temperature in the dark. Plates were then observed under UV light (365 nm) for the typical orange fluorescence. Hog pancreas lipase (Fluka) served as a positive control.
Acknowledgments
We thank J. Ryals (Novartis) for the cDNA of PR-1, and we are grateful to C. Nawrath (University of Fribourg, Switzerland) for the organ-specific RNA samples of Arabidopsis. We thank L. Sticher and F. Mauch (University of Fribourg, Switzerland) for the critical reading of this manuscript.
This work was supported by the Swiss National Foundation (grant nos. 3100–049279.96 to B.M.M. and 3100–055662.98 to J.-P.M.).
References
- Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X (1994) A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6: 1845-1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254 [DOI] [PubMed] [Google Scholar]
- Brederode FT, Linthorst HJ, Bol JF (1991) Differential induction of acquired resistance and PR gene expression in tobacco by virus infection, ethephon treatment, UV light and wounding. Plant Mol Biol 17: 1117-1125 [DOI] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong XN (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583-1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262: 539-544 [DOI] [PubMed] [Google Scholar]
- Clarke JD, Liu Y, Klessig DF, Dong X (1998) Uncoupling PR gene expression from NPR1 and bacterial resistance: characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell 10: 557-569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath U, Pieterse CM, Mauch-Mani B (2002) Priming in plant-pathogen interactions. Trends Plant Sci 7: 210-216 [DOI] [PubMed] [Google Scholar]
- Czakó M, Wilson J, Yu X, Marton L (1993) Sustained root culture for generation and vegetative propagation of transgenic Arabidopsis thaliana. Plant Cell Rep 12: 603-609 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826-833 [DOI] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gutrella M, Kessmann H, Ward E et al. (1994) A central role of salicylic acid in plant disease resistance. Science 266: 1247-1250 [DOI] [PubMed] [Google Scholar]
- Dempsey DA, Shah J, Klessig DF (1999) Salicylic acid and disease resistance in plants. Crit Rev Plant Sci 18: 547-575 [Google Scholar]
- de Torres Zabela M, Fernandez-Delmond I, Niittyla T, Sanchez P, Grant M (2002) Differential expression of genes encoding Arabidopsis phospholipases after challenge with virulent or avirulent Pseudomonas isolates. Mol Plant-Microbe Interact 15: 808-816 [DOI] [PubMed] [Google Scholar]
- Doke N, Miura Y, Sanchez LM, Park HJ, Noritake T, Yoshioka H, Kawakita K (1996) The oxidative burst protects plants against pathogen attack: mechanism and role as an emergency signal for plant bio-defence. A review. Gene 179: 45-51 [DOI] [PubMed] [Google Scholar]
- Eyal Y, Meller Y, Lev-Yadun S, Fluhr R (1993) A basic-type PR-1 promoter directs ethylene responsiveness, vascular and abscission zone-specific expression. Plant J 4: 225-234 [DOI] [PubMed] [Google Scholar]
- Falk A, Feys BJ, Frost LN, Jones JD, Daniels MJ, Parker JE (1999) EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc Natl Acad Sci USA 96: 3292-3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J 20: 5400-5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J (2001) Genes controlling expression of defense responses in Arabidopsis: 2001 status. Curr Opin Plant Biol 4: 301-308 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM (1996) Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143: 973-982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YQ, Yang C, Thara VK, Zhou J, Martin GB (2000) Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 12: 771-786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Willits MG, Glazebrook J (2000) Arabidopsis thaliana EDS4 contributes to salicylic acid (SA)-dependent expression of defense responses: evidence for inhibition of jasmonic acid signaling by SA. Mol Plant-Microbe Interact 13: 503-511 [DOI] [PubMed] [Google Scholar]
- Hong Y, Wang TW, Hudak KA, Schade F, Froese CD, Thompson JE (2000) An ethylene-induced cDNA encoding a lipase expressed at the onset of senescence. Proc Natl Acad Sci USA 97: 8717-8722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K (2001) The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13: 2191-2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J (1999) Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci USA 96: 13583-13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680-685 [DOI] [PubMed] [Google Scholar]
- Laxalt AM, Munnik T (2002) Phospholipid signalling in plant defence. Curr Opin Plant Biol 5: 332-338 [DOI] [PubMed] [Google Scholar]
- MacKenzie RD, Blohm TR, Auxier EM, Luther AC (1967) Rapid colorimetric micromethod for free fatty acids. J Lipid Res 8: 589-597 [PubMed] [Google Scholar]
- Malamy J, Carr JP, Klessig DF, Raskin I (1990) Salicylic-acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science 250: 1002-1004 [DOI] [PubMed] [Google Scholar]
- Métraux JP, Signer H, Ryals J, Ward E, Wyss-Benz M (1990) Increase in salicylic-acid at the onset of systemic acquired resistance in cucumber. Science 250: 1004-1006 [DOI] [PubMed] [Google Scholar]
- Mittler R, Lam E (1996) Sacrifice in the face of foes: pathogen-induced programmed cell death in plants. Trends Microbiol 4: 10-15 [DOI] [PubMed] [Google Scholar]
- Munnik T (2001) Phosphatidic acid: an emerging plant lipid second messenger. Trends Plant Sci 6: 227-233 [DOI] [PubMed] [Google Scholar]
- Onate-Sanchez L, Singh KB (2002) Identification of Arabidopsis ethylene-responsive element binding factors with distinct induction kinetics after pathogen infection. Plant Physiol 128: 1313-1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IA, Eggermont K, Terras FR, Thomma BP, De Samblanx GW, Buchala A, Métraux JP, Manners JM, Broekaert WF (1996) Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8: 2309-2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IA, Thomma BP, Buchala A, Métraux JP, Broekaert WF (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10: 2103-2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, van Loon LC (1999) Salicylic acid-independent plant defence pathways. Trends Plant Sci 4: 52-58 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, van Wees SCM, van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, van Loon LC (1998) A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10: 1571-1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Farmer EE (1998) Jasmonate and salicylate as global signals for defense gene expression. Curr Opin Plant Biol 1: 404-411 [DOI] [PubMed] [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD (1996) Systemic acquired resistance. Plant Cell 8: 1809-1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samac DA, Hironaka CM, Yallaly PE, Shah DM (1990) Isolation and characterization of the genes encoding basic and acidic chitinase in Arabidopsis thaliana. Plant Physiol 93: 907-914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schaller F (2001) Enzymes of the biosynthesis of octadecanoid-derived signalling molecules. J Exp Bot 52: 11-23 [PubMed] [Google Scholar]
- Shirai K, Jackson RL (1982) Lipoprotein lipase-catalyzed hydrolysis of p-nitrophenyl butyrate: interfacial activation by phospholipid vesicles. J Biol Chem 257: 1253-1258 [PubMed] [Google Scholar]
- Shirasu K, Nakajima H, Rajasekhar VK, Dixon RA, Lamb C (1997) Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell 9: 261-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH (1992) Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA 89: 6837-6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticher L, Mauch-Mani B, Métraux JP (1997) Systemic acquired resistance. Annu Rev Phytopathol 35: 235-270 [DOI] [PubMed] [Google Scholar]
- Thomma BP, Broekaert WF (1998) Tissue-specific expression of plant defensin genes PDF2.1 and PDF2.2 in Arabidopsis thaliana. Plant Physiol Biochem 36: 533-537 [Google Scholar]
- Thompson J, Taylor C, Wang TW (2000) Altered membrane lipase expression delays leaf senescence. Biochem Soc Trans 28: 775-777 [PubMed] [Google Scholar]
- van Loon LC, Bakker PAHM, Pieterse CMJ (1998) Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36: 453-483 [DOI] [PubMed] [Google Scholar]
- van Loon LC, van Strien EA (1999) The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol 55: 85-97 [Google Scholar]
- Wang C, Zien CA, Afitlhile M, Welti R, Hildebrand DF, Wang X (2000) Involvement of phospholipase D in wound-induced accumulation of jasmonic acid in Arabidopsis. Plant Cell 12: 2237-2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli L, Jakab G, Métraux JP, Mauch-Mani B (2000) Potentiation of pathogen-specific defense mechanisms in Arabidopsis by beta-aminobutyric acid. Proc Natl Acad Sci USA 97: 12920-12925 [DOI] [PMC free article] [PubMed] [Google Scholar]