Abstract
The molecular mechanism of mRNA degradation in the chloroplast consists of sequential events, including endonucleolytic cleavage, the addition of poly(A)-rich sequences to the endonucleolytic cleavage products, and exonucleolytic degradation. In spinach chloroplasts, the latter two steps of polyadenylation and exonucleolytic degradation are performed by the same phosphorolytic and processive enzyme, polynucleotide phosphorylase (PNPase). An analysis of its amino acid sequence shows that the protein is composed of two core domains related to RNase PH, two RNA binding domains (KH and S1), and an α-helical domain. The amino acid sequence and domain structure is largely conserved between bacteria and organelles. To define the molecular mechanism that controls the two opposite activities of this protein in the chloroplast, the ribonuclease, polymerase, and RNA binding properties of each domain were analyzed. The first core domain, which was predicted to be inactive in the bacterial enzymes, was active in RNA degradation but not in polymerization. Surprisingly, the second core domain was found to be active in degrading polyadenylated RNA only, suggesting that nonpolyadenylated molecules can be degraded only if tails are added, apparently by the same protein. The poly(A) high-binding-affinity site was localized to the S1 domain. The complete spinach chloroplast PNPase, as well as versions containing the core domains, complemented the cold sensitivity of an Escherichia coli PNPase-less mutant. Phylogenetic analyses of the two core domains showed that the two domains separated very early, resulting in the evolution of the bacterial and organelle PNPases and the exosome proteins found in eukaryotes and some archaea.
INTRODUCTION
Polynucleotide phosphorylase (PNPase) is an exoribonuclease that catalyzes the processive 3′ → 5′ phosphorolysis of RNA or a processive polymerization of this molecule (Littauer and Grunberg-Manago, 1999). In Escherichia coli, this enzyme is mostly active in 3′ → 5′ phosphorolysis during RNA degradation or processing (Grunberg-Manago, 1999; Jarrige et al., 2002). Limited polymerization activity can be detected in vivo under certain growth conditions or when the gene for the poly(A)-polymerase (PAP) is inactivated (Sarkar, 1997; Mohanty and Kushner, 2000). In E. coli, a small proportion of PNPase is a constituent of the degradosome, a multiprotein complex composed also of RNase E, RNA helicase, enolase, and possibly other molecules (Symmons et al., 2002). The association of PNPase with the RNA helicase RhlB alone also was observed recently (Liou et al., 2002). However, PNPase in the chloroplast was found to form a homotrimeric complex eluting from a size-exclusion column at 600 kD and lacks any known interactions with other proteins (Baginsky et al., 2001). PNPase also was reported to be a global regulator of virulence and persistence in Salmonella enterica (Clements et al., 2002). Recently, the human PNPase was identified in an overlapping-pathway screen to discover genes that displayed coordinated expression as a consequence of the terminal differentiation and cellular senescence of human melanoma cells (Leszczyniecka et al., 2002).
The amino acid sequences of different PNPases from bacteria, as well as from the nuclear genomes of plants, yeast, and mammals, display a high level of identity and feature similar structures composed of five motifs (Symmons et al., 2000, 2002; Zuo and Deutscher, 2001; Raijmakers et al., 2002). These consist of two core domains having different degrees of identity to the E. coli phosphorylase RNase PH, an α-helical domain between the two core domains, and two adjacent RNA binding domains (KH and S1) that also are found in other RNA binding proteins. X-ray crystallographic analysis was used to reveal the three-dimensional structure of the PNPase from the bacterium Streptomyces antibioticus. The enzyme is arranged in a homotrimeric complex forming a “doughnut” shape surrounding a central channel that could accommodate a single-stranded RNA molecule (Symmons et al., 2000, 2002). Similar structure and homology of the two core domains were assigned recently to the exosome, a multiprotein complex that functions in the 3′ → 5′ degradation of RNA in the cytoplasm and nucleus of eukaryotic cells (Aloy et al., 2002; Raijmakers et al., 2002).
The molecular mechanism of RNA degradation in the chloroplast has been elucidated and was found to be very similar to that of bacteria (Hayes et al., 1999; Schuster et al., 1999; Monde et al., 2000). In both bacteria and chloroplasts, the first event is endonucleolytic cleavage of the RNA molecule, followed by the addition of a poly(A) tail in bacteria and a poly(A) tail (Komine et al., 2000) or a poly(A)-rich tail (Lisitsky et al., 1996) in chloroplasts. The polyadenylated cleavage products then are directed to rapid exonucleolytic degradation by PNPase and RNase II in E. coli and by PNPase and possibly other exoribonucleases in the chloroplast (Lisitsky et al., 1997a, 1997b; Lisitsky and Schuster, 1999). Therefore, polyadenylation is part of the RNA degradation mechanism in bacteria, in chloroplasts, and possibly also in plant mitochondria (Carpousis et al., 1999; Coburn and Mackie, 1999; Gagliardi and Leaver, 1999; Hayes et al., 1999; Lupold et al., 1999; Schuster et al., 1999; Monde et al., 2000; Regnier and Arraiano, 2000; Kuhn et al., 2001).
As discussed above, polyadenylation in E. coli is performed mostly by PAP, with PNPase functioning as a polymerase only in the absence of PAP (Mohanty and Kushner, 2000). However, we recently found that no PAP can be detected in spinach chloroplasts, and thus both polyadenylation and degradation are performed by one enzyme, PNPase (Yehudai-Resheff et al., 2001). A similar situation also was found recently in cyanobacteria (Rott et al., 2003). To understand how the spinach chloroplast PNPase performs these opposing activities of polymerization and phosphorolysis, and because the activity of the full-length protein is mediated by the sum of activities of the different domains, we decided to analyze the different domains for polymerization, degradation, and RNA binding properties. We found that the core domains had distinct activities and that their cooperation was required to achieve their functionality.
RESULTS
The Spinach Chloroplast PNPase Structure Is Similar to That of the Bacterial Enzyme
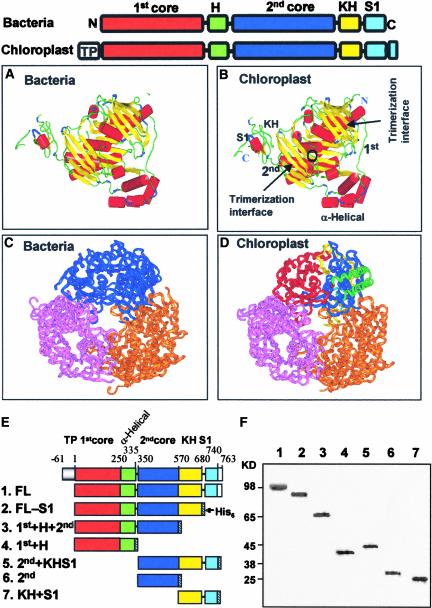
The spinach PNPase is homologous with the bacterial enzyme, with an additional N-terminal transit peptide of 61 amino acids and a 22–amino acid C-terminal extension (Figure 1, top). The transit peptide is cleaved off during translocation of the preprotein to the chloroplast; therefore, it is not present in the mature protein. Like the bacterial PNPases, the protein is composed of two core domains, the second of which is homologous with the RNase PH, another bacterial exoribonuclease that is a phosphorylase (first [1st] and second [2nd] cores in Figure 1). The two core domains share some degree of identity; therefore, low similarity between RNase PH and the 1st core domain also is observed (Symmons et al., 2002). The 1st core domain is followed by an α-helical domain, and the 2nd core domain is followed by the KH and S1 domains, which are predicted to function in RNA binding and are present in many RNA binding proteins (Symmons et al., 2000, 2002; Zuo and Deutscher, 2001) (Figure 1).
Figure 1.
Molecular Homology-Based Model of the Spinach Chloroplast PNPase, and Protein Constructs Used in This Work.
(A) to (D) The domain structures of the bacterial and chloroplast PNPases are presented schematically at top. The boxes represent the different domains, as indicated above. TP indicates the chloroplast transit peptide. The C-terminal 22–amino acid extension is unique to the chloroplast PNPase. The resolved structures ([A] and [C]) and predicted models ([B] and [D]) of the PNPase enzymes from S. antibioticus ([A] and [C]) and spinach chloroplast ([B] and [D]) are shown. The monomer structure is shown in (A) and (B), and the trimer structure is shown in (C) and (D). The different domains and the trimerization interfaces are indicated in the spinach chloroplast PNPase (B). The circle indicates the location of the phosphorylase activity site at the 2nd core domain as identified from tungsten binding of the S. antibioticus enzyme (B). The trimeric doughnut-like structure is presented in (C) and (D) in an orientation allowing easy observation of the middle channel. Each monomer is colored differently, and in one monomer of the spinach enzyme each domain is colored as outlined in the scheme presented at top. The homology-based modeling was performed using the 3D-PSSM Fold Recognition Server at http://www.sbg.bio.ic.ac.uk/~3dpssm/. The complex was built by applying the crystal symmetry of the structure using the Quanta program (Accelrys). The figure was constructed using Insight II (Accelrys).
(E) Scheme of the spinach PNPase protein. The full-length protein (FL) was produced in bacteria without the addition of a His6 tag (see text). The other versions were expressed in E. coli fused to the His6 tag at the C terminus, as shown by the hatched boxes.
(F) Silver-stained polyacrylamide gel profile of the recombinant proteins (35 to 50 ng) after expression in an E. coli strain lacking the endogenous PNPase. Proteins containing the His6 tag (lanes 2 to 7) were purified by affinity chromatography and anion-exchange (MonoQ) steps and loaded onto a 10% SDS-PAGE gel. The FL protein (lane 1) was purified biochemically on size-exclusion, heparin, and anion-exchange columns. Molecular mass markers are shown at left.
Because the amino acid sequence of the spinach chloroplast PNPase shares a high degree of identity with that of the S. antibioticus enzyme, we used the coordinates of the bacterial enzyme to build a putative structure of the spinach PNPase. Figures 1A and 1B present the predicted monomeric structure of the chloroplast PNPase compared with the S. antibioticus enzyme. The structures are very similar with respect to the relative positions of the various subdomains. Both the 1st and 2nd core domains provide the β-sheet strands, which function as the trimerization interfaces, and the vicinity of the phosphorolytic catalytic site (binding of the tungsten) known for the 2nd domain (Symmons et al., 2000, 2002). The α-helical domain is situated separately from the main structure of the 1st and 2nd core domains, and although it is arranged similarly, it is not identical for the bacterial and chloroplast proteins. The KH and S1 domains, as well as the extra amino acids at the C terminus of the chloroplast enzyme, also are separated from the core domains.
The arrangement of three PNPase polypeptides in the doughnut-shaped trimer is shown in Figures 1C and 1D. The formation of the trimers creates a central channel that is of the correct dimensions for a single-stranded RNA molecule (Symmons et al., 2000, 2002). The KH and S1 domains of the three subunits all point in one direction, suggesting the possibility of binding the RNA molecule before it enters the channel and undergoes degradation there. Indeed, the spinach chloroplast PNPase was purified as a homomultimer that fractionated on a size-exclusion column at 600 kD (Baginsky et al., 2001). It is not known whether the PNPase is a trimer that is eluted at this size or whether two trimers join to form a hexamer.
Preparation of Recombinant PNPase and the Different Fragments
The molecular analysis of RNA polyadenylation and degradation in spinach chloroplasts revealed that both activities are performed by PNPase (Yehudai-Resheff et al., 2001). To better understand how the enzyme activity is shifted to favor polymerization or degradation, we analyzed the activity of the protein and its component parts in detail. To do so, we prepared the PNPase recombinant protein lacking the chloroplast transit peptide as described in Methods, as well as several deletion proteins containing different domains (Figure 1E). The recombinant proteins were purified and analyzed by SDS-PAGE to determine their apparent molecular mass and purity (Figure 1F). In addition, all proteins were verified by immunoblot analysis using antibodies against either the PNPase or the His6 tag (data not shown).
RNA Degradation and Polyadenylation Activities of the Spinach Chloroplast PNPase and Its Domains
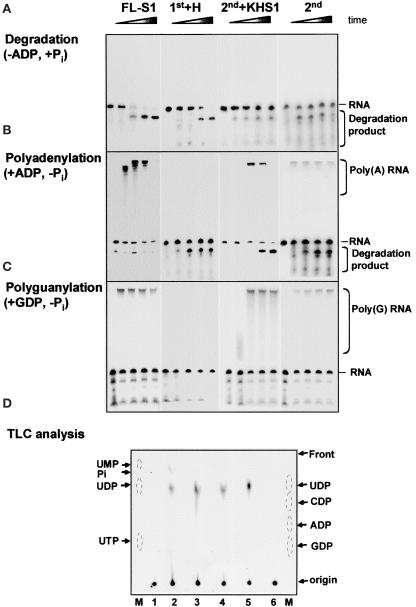
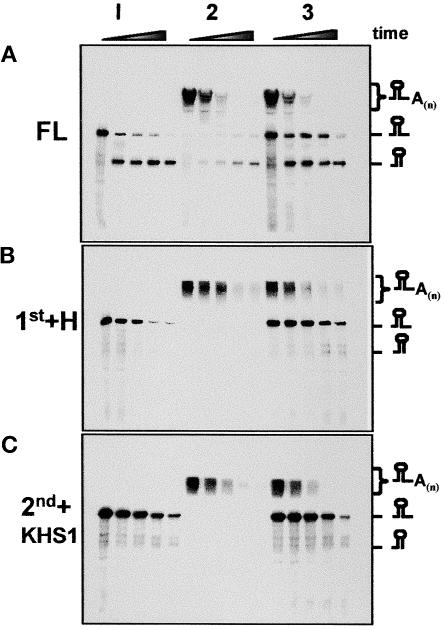
As discussed above, structural analysis of the S. antibioticus enzyme predicted that only the 2nd core would be active in phosphorolysis. This conclusion was strengthened recently by mutational analysis of the E. coli PNPase, in which most of the mutations eliminating the phosphorolysis activity were located in the 2nd core domain in the vicinity of the tungsten binding site in the S. antibioticus PNPase (Jarrige et al., 2002). Here, we characterized the chloroplast PNPase and its fragmented domains for RNA degradation and phosphorolysis activities. The degradation activity was assayed by incubating the corresponding protein with 32P-labeled RNA in the presence of 10 mM Pi (Figure 2A). Indeed, the full-length (FL; data not shown) and FL-S1 proteins rapidly degraded the RNA (Figure 2A). Interestingly, even though the RNA substrate used here was chosen because it does not contain a predicted stem-loop structure, the enzyme paused at a certain sequence and a degradation product accumulated. No differences were observed when the activity of the recombinant FL protein was compared with that of purified PNPase from spinach chloroplasts (Yehudai-Resheff et al., 2001) (data not shown). The 1st+H protein, composed of the 1st core and the α-helical domains, also was active in RNA degradation ( Figure 2A). This result was surprising, because previous data did not predict activity, given the tungsten binding data and the mutagenesis of the bacterial PNPases (Symmons et al., 2000; Jarrige et al., 2002). Therefore, the chloroplast enzyme may differ from the bacterial PNPases in the activity of the 1st core domain.
Figure 2.
Polymerization and Degradation Activities of the Different Proteins.
(A) to (C) Synthetically transcribed 32P-RNA corresponding to part of the chloroplast gene petD (petD-Dra) was incubated with the proteins and 10 mM Pi to determine the presence of degradation activity (A). The Pi was replaced with 1 mM ADP or GDP to measure the polyadenylation and polyguanylation activities ([B] and [C], respectively). Samples were withdrawn at 0, 15, 35, 60, and 90 min, and the RNA was analyzed by denaturing PAGE and autoradiography. The proteins used are indicated at top, and the input RNA, as well as the polymerized and degradation products, are indicated at right.
(D) Thin layer chromatography (TLC) analysis of the degradation products. Proteins were incubated with uniformly 32P-UTP–labeled RNA. After the incubation, the reaction products were spotted onto a polyethyleneimine TLC plate, which was developed with LiCl, dried, and autoradiographed. Lane 1, no protein; lane 2, FL-S1; lane 3, 2nd+KHS1; lane 4, 1st+H; lane 5, E. coli PNPase; lane 6, KH+S1. Monophosphate, diphosphate, and triphosphate nucleotides of A, G, C, and U were analyzed on the same TLC plate and visualized by fluorescence quenching (lanes M). 32P-Pi also was separated on this plate as a marker. The migration patterns of the markers are indicated with circles.
Even more surprising was the observation that the 2nd core domain, which was predicted to harbor the active site either with or without the KH and S1 domains, displayed very low RNA degradation activity even in the presence of 10 mM Pi (Figure 2A). Only a small amount of the substrate RNA was digested, compared with that in the 1st core and the FL proteins. However, this issue was resolved when RNA polyadenylation was assayed in the presence of ADP without the addition of Pi. Under these conditions, the 2nd core displayed both polymerization and RNA degradation activities (Figure 2B). The polyadenylation activity of the 2nd core was transient and preceded degradation, as observed previously for the FL protein (Yehudai-Resheff et al., 2001). No polyadenylation activity was obtained under these conditions with the 1st core (Figure 2B). These results suggested that the degradation activity of the 2nd core is dependent on previous polyadenylation, again similar to what we observed in the lysed chloroplast system (Yehudai-Resheff et al., 2001).
In addition, there also is the possibility that polyadenylation preceding degradation occurs with the 1st core but is too rapid or highly transient and thus not detected here. To analyze whether or not the 1st domain expresses polymerization activity, the polymerization assay was repeated with GDP replacing ADP. Under these conditions, the RNA is polyguanylated by PNPase, and because poly(G) forms a strong tertiary structure that efficiently inhibits the exonuclease activity, only the polymerization activity is observed (Sundquist, 1993; Yehudai-Resheff et al., 2001). In addition, such an assay enabled us to determine whether the polymerization activity of the 2nd core is required for the subsequent degradation activity, because if it is, only polymerization would be obtained with GDP. Indeed, when the polyguanylation assay was performed, polymerization activities were obtained with the FL-S1, 2nd+KHS1, and 2nd domains, and RNA degradation activity was inhibited (Figure 2C). No polymerization activity was observed with the 1st+H protein, suggesting that the 1st domain is active only in RNA degradation and not in polymerization. Because the RNA degradation and polymerization activities of the 2nd+KHS1 and the 2nd domains alone were very similar, we concluded that, as predicted, these activities were located at the 2nd core domain.
To analyze the degradation products generated by the exoribonuclease activities of the two domains, thin layer chromatography was performed (Figure 2D). As expected from a phosphorylase, the degradation activities of all proteins examined resulted in the formation of nucleoside diphosphates. Therefore, the product detected with 32P–UTP-RNA was 32P-UDP and that detected with 32P–ATP-RNA was 32P-ADP (Figure 2D).
Together, the results of the experiments shown in Figure 2 demonstrate that for the spinach chloroplast PNPase, the 1st core domain is active in RNA degradation but not in polymerization, whereas the 2nd core domain is active in polyadenylation-dependent RNA degradation.
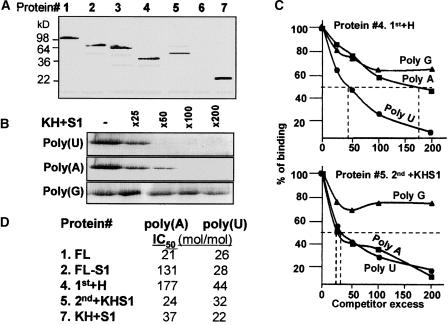
The High-Affinity Poly(A) Binding Site Is Located in the S1 Domain
The bacterial and spinach PNPase proteins were characterized previously using the UV light cross-linking assay as RNA binding proteins (Lisitsky et al., 1997b; Lisitsky and Schuster, 1999). In these experiments, high affinity for poly(A) and poly(U) was observed, suggesting an explanation for how this enzyme competes in bacteria or within the chloroplast for polyadenylated RNA over nonpolyadenylated RNA. We wished to determine whether this protein contains more than one RNA binding site and in which domain(s) high-affinity poly(A) binding is located. First, RNA binding was tested by UV light cross-linking, in which the protein is incubated with 32P-RNA followed by UV irradiation, digestion of the RNA with ribonucleases, and analysis by PAGE and autoradiography. Figure 3A presents the results of this experiment using an RNA corresponding to the chloroplast psbA gene. All of the proteins, except for the one composed of only the 2nd core domain, bound RNA. This result suggested that the 2nd core domain, although harboring a phosphorolysis active site, does not bind RNA in such a way that it could be detected by the UV light cross-linking assay used here. However, the 1st+H protein bound RNA. As expected, the protein composed of the KH and S1 domains, which were predicted to be RNA binding domains, bound RNA (Figure 3A).
Figure 3.
RNA Binding Properties of the Derivative Proteins.
(A) The proteins (10 ng each) FL (lane 1), FL-S1 (lane 2), 1st+H+2nd (lane 3), 1st+H (lane 4), 2nd+KHS1 (lane 5), 2nd (lane 6), and KH+S1 (lane 7) (as shown in Figure 1) were analyzed for RNA binding by the UV light cross-linking assay as described in Methods.
(B) Competition of ribohomopolymers for RNA binding of the KH+S1 protein (protein 7 in Figure 1) was performed using 32P-psbA RNA in a UV light cross-linking competition assay. The numbers at top indicate the molar excess of the ribohomopolymer.
(C) Competition of ribohomopolymers for RNA binding of the 1st+H and 2nd+KHS1 proteins (proteins 4 and 5 in Figure 1, respectively). The UV light cross-linking competition assays were performed as described for (B), and the intensity of the UV light cross-linking band without competitor was defined as 100%. The data shown are averages of at least three independent experiments. IC50 values (competitor excess that resulted in 50% inhibition of the UV light cross-linking signal) are indicated by dashed lines.
(D) UV light cross-linking competition assays were performed as described for (B) and (C). IC50 values for binding the ribohomopolymers poly(A) and poly(U) by the different proteins are presented.
Next, we wanted to locate the high-affinity site for poly(A) binding. To this end, the UV light cross-linking competition assay was used. In this assay, the signal obtained by the UV light cross-linking of 32P-RNA to protein is competed by adding increasing amounts of the tested nonradioactive RNA, in this case, ribohomopolymers. The efficiency with which an RNA competes for UV light cross-linking reflects its affinity for the protein. The IC50 was defined as the molar excess of the competitor RNA that resulted in a 50% reduction in the radioactive UV light cross-linking signal (Lisitsky et al., 1994). The lower the IC50 value for a specific RNA, the higher its affinity for the protein. An example of such a UV light competition assay is presented in Figure 3B, and quantification of RNA binding to the different proteins is shown in Figures 3C and 3D. A high binding affinity of the FL protein was observed for both poly(A) and poly(U), as reported previously for the purified protein (Lisitsky et al., 1997b). Interestingly, all of the proteins (except the 2nd core, which did not bind RNA in the UV light cross-linking assay and therefore was not analyzed here) bound poly(U) with high affinity. However, high affinity for poly(A) was observed only with proteins containing the S1 domain. The affinity for poly(A) was reduced sixfold compared with the FL protein when only the S1 domain was eliminated, and it was reduced ninefold compared with the 1st+H protein. Together, the results of the RNA binding analysis suggest that the poly(A) high-binding-affinity site is located in the S1 domain and that other RNA binding sites of the PNPase possess a high affinity for poly(U).
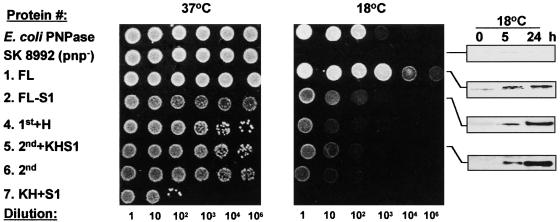
The Spinach Chloroplast PNPase and Its Active Fragments Complement the Growth of an E. coli PNPase- and RNase PH–less Strain at 18°C
The E. coli strain SK 8992 contains an insertion of the Tn5 transposable element into the pnp gene encoding the PNPase and also lacks the other Pi-dependent exoribonuclease RNase PH. Although this strain lacks the PNPase, it is viable, probably because RNase II can compensate for the PNPase RNA degradation activity. However, this strain is sensitive to cold and cannot grow at 18°C (Yancey and Kushner, 1990; Craven et al., 1992; Zhou and Deutscher, 1997; Beran and Simons, 2001). We used SK 8992 to reveal whether the spinach chloroplast PNPase could complement its E. coli counterpart and, if so, which domains of the protein also could do so. To this end, the FL spinach chloroplast PNPase and its derivatives were cloned into the PT7-7 vector (Citovsky et al., 1990), which enables the expression of recombinant proteins without the addition of other amino acids. The plasmids then were introduced into SK 8992 cells. SK 8992 is PNPase- and RNase PH–less but does not contain the T7 RNA polymerase encoded in the chromosome. We had to shift to this strain because the ENS134 strain containing the T7 RNA polymerase used for the expression of the recombinant protein grew at 18°C when the PT7-7 plasmid alone, which does not express any part of the PNPase, was introduced (data not shown). As described above, shifting to SK 8992, which does not contain the T7 RNA polymerase gene, solved this problem.
Taking this into account, and because it remained unclear how the expression of the recombinant proteins was established, their expression was verified by immunoblot analysis of proteins from bacteria grown at 18°C (Figure 4, right). As expected, the SK 8992 cells grew well at 37°C but not at 18°C, compared with the control cells, which were transformed with a plasmid expressing the E. coli PNPase (Figure 4, top two rows). Interestingly, full complementation was obtained with the FL protein, indicating that the chloroplast PNPase can efficiently restore the growth defect at 18°C of the E. coli strain lacking PNPase and RNase PH (Figure 4, third row). In addition, all of the deletion mutants of the chloroplast PNPase containing at least one core domain were able to partially complement the cold growth defect. The proteins containing only one core were less efficient than those containing both cores, but even the protein composed of only the 2nd core domain partially complemented the temperature-sensitive growth defect. These results support the biochemical assay showing that the 1st core domain of the chloroplast PNPase is active in RNA degradation, an activity that probably is required to restore growth at 18°C (Zhou and Deutscher, 1997).
Figure 4.
Complementation of Growth of the E. coli pnp− Strain at 18°C by the Chloroplast PNPase.
SK 8992 cells were transformed with a plasmid expressing the E. coli enzyme as a positive control (E. coli PNPase), the plasmid vector PT7-7 [SK 8992 (pnp−)], and the different derivatives of the spinach chloroplast PNPase as indicated. This strain also lacks the other Pi-dependent exoribonuclease, RNase PH. The cells were grown overnight at 37°C and then spotted onto l-agar/ampicillin plates at the dilutions shown at bottom. The plates were incubated for 16 and 48 h at 37 and 18°C, respectively. To verify expression of the corresponding proteins, the bacteria also were grown in cultures at 18°C and analyzed for recombinant protein expression by immunoblotting using the PNPase antibodies (gels at right).
Although the biochemical data suggested that this domain does not express polyadenylation activity, polyadenylation could be performed in E. coli by PAP I. RNase PH alone (containing only one core domain) was shown previously to complement the growth of the double mutant (PNPase and RNase PH) at 31°C (Zhou and Deutscher, 1997) but not at 15°C (Beran and Simons, 2001). Therefore, although it seems unlikely given that 18°C was used in our experiments, the possibility cannot be excluded that each of the spinach PNPase core domains alone actually complemented the RNase PH and not the PNPase function and that this complementation enabled slow growth at 18°C.
In contrast to the proteins containing the core domains, expression of the KH+S1 protein did not restore growth at 18°C (Figure 4, bottom row). Moreover, the expression of this protein inhibited bacterial growth even at 37°C, suggesting that the RNA binding properties of this protein, when not connected to the core domains, are deleterious to the bacteria.
Together, these results showed that the chloroplast PNPase could fully complement the cold defect of E. coli lacking PNPase and RNase PH. Each part containing one of the core domains can partially complement this defect, but the protein composed of only the KH+S1 domain inhibits growth even at 37°C.
Unlike the FL PNPase, the Proteins That Include Only One Core Domain Do Not Pause at a Stem-Loop Structure
One of the well-known characteristics of the PNPase enzyme is its pausing at a stem-loop structure when processively degrading RNA (Hayes et al., 1996; Blum et al., 1999; Liou et al., 2002). This phenomenon is very important for the 3′ end processing of bacterial and chloroplast transcripts, because the 3′ end of most transcripts is characterized by a stable stem-loop structure formed by the exonucleolytic trimming of a longer precursor (Carpousis et al., 1999; Monde et al., 2000). Because homotrimer formation is dependent on the interfaces formed by the two core domains (Figure 1), no trimers are formed in the absence of one of them. Indeed, evidence that each of the domains alone could not form a high-molecular-weight complex was obtained experimentally when the recombinant proteins were fractionated by size-exclusion chromatography or nondenaturing polyacrylamide gels (data not shown). Because both core domains were found to be active in RNA degradation, we asked whether each core, which probably does not form the trimeric doughnut/channel conformation, pauses at the stem-loop structure, like the FL enzyme.
To answer this question, an RNA molecule corresponding to the 3′ end of the spinach chloroplast psbA transcript and containing a stem-loop structure was incubated with the FL enzyme and the fragments corresponding to the 1st+H and 2nd+KHS1 domains. As presented in the left lanes of Figures 5A to 5C (lanes 1), the RNA was degraded promptly by the FL and 1st+H proteins and very slowly by the 2nd+KHS1 proteins, as shown in Figure 2. However, the product containing the stem-loop structure at the 3′ end accumulated only for the FL protein. Therefore, this result indicates that only the protein containing two RNase PH core domains paused at a stem-loop structure. However, because the activity of the 2nd core on nonpolyadenylated RNA is very low, we repeated the experiment using a polyadenylated version of the same substrate. Indeed, as observed in Figure 2, the polyadenylated molecule, unlike the nonpolyadenylated RNA, was degraded at a similar rate by the FL, 1st+H, and 2nd+KHS1 proteins. Also in this case, a product produced as a result of pausing at the stem-loop structure was detected only for the FL protein and not for either of the two fragmented proteins (Figure 5, lanes 2). As reported previously, the accumulation of this product was reduced significantly when the substrate was polyadenylated compared with the nonpolyadenylated version (Lisitsky et al., 1996, 1997b). Because the 3′ ends of most transcripts are characterized by stem-loop structures formed by exonucleolytic processing involving PNPase (Walter et al., 2002), this observation suggests a functional explanation for a PNPase enzyme harboring two RNase PH domains.
Figure 5.
Unlike the FL Protein, the Degradation Activity of Each Core Domain Is Not Inhibited by a Stem-Loop Structure.
32P-RNA corresponding to the 3′ end of the spinach chloroplast psbA gene (296 nucleotides) (lanes 1), or the same RNA that was first elongated with ∼200 adenosines (lanes 2), was used as a substrate for the PNPase and the two parts 1st+H and 2nd+KHS1. In lanes 3, half of the amount of RNA molecules incubated in lanes 1 and 2 were mixed together and incubated with the proteins. Samples were withdrawn at 0, 35, 60, 90, and 120 min and analyzed by denaturing PAGE and autoradiography. Schemes of the corresponding RNA molecules are shown at right.
Competition of PNPase and Each of the RNase PH Core Domains for Polyadenylated RNA
We reported previously that both the chloroplast and the E. coli PNPases compete for polyadenylated RNAs and that the biochemical reason is their high affinity for poly(A) (Lisitsky et al., 1997b; Lisitsky and Schuster, 1999). This competition was observed in experiments in which two RNA molecules, one polyadenylated and the other nonpolyadenylated, were incubated with the enzyme. When incubated separately, the degradation rates were similar. However, when mixed and incubated with limited amounts of enzyme, the nonpolyadenylated RNA was stable but the polyadenylated RNA was degraded rapidly (Lisitsky et al., 1997b; Lisitsky and Schuster, 1999). This result is interpreted most easily as preferential binding of the enzyme to the polyadenylated RNA, and because PNPase works processively, it is sequestered from the nonpolyadenylated RNA.
In the experiments reported here, we investigated whether the PNPase fragments also competed for polyadenylated substrates. Equal amounts of polyadenylated and nonpolyadenylated RNAs were mixed and incubated with the FL, 1st+H, and 2nd+KHS1 proteins. As shown in Figure 5A (lanes 3) for the FL protein, the degradation rates for the two RNAs were similar when incubated separately. However, when mixed, the degradation rate of the nonpolyadenylated RNA was reduced markedly. Stabilization of the nonpolyadenylated RNA also was observed with the 1st+H protein, but it was impossible to determine for the 2nd+KHS1 protein, because the degradation rate of the nonpolyadenylated RNA was very slow even when incubated separately, as described above ( Figures 2 and 5C). However, the results presented in Figure 5C clearly show that polyadenylated RNA is degraded by the 2nd+KHS1 protein much faster than by the nonpolyadenylated molecule, suggesting that high poly(A) affinity of the S1 domain is essential for efficient degradation activity. These results also suggest that the competition for polyadenylated RNA is performed by both the 1st+H and 2nd+KHS1 subdomains. Although the competition for polyadenylated RNA of the 2nd+KHS1 protein can be explained by the presence of the S1 domain (Figure 3), that of the 1st+H protein cannot be explained by the same mechanism.
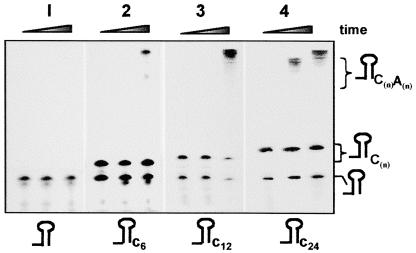
A Platform of 6 to 12 Nucleotides 3′ to the Stem Loop Is Required for RNA Polyadenylation by PNPase
Most chloroplast transcripts are characterized at the 3′ end by a stem-loop structure, and these RNA molecules are polyadenylated inefficiently by PNPase (Lisitsky et al., 1996; Schuster et al., 1999; Monde et al., 2000). We found previously that the E. coli PAP I also is inhibited by a stem-loop structure but that the addition of two nucleotides 3′ to the stem loop is sufficient to promote efficient polyadenylation (Yehudai-Resheff and Schuster, 2000). Here, we wanted to determine how many nucleotides 3′ to the stem-loop structure are required to promote polyadenylation by the chloroplast PNPase. RNA was prepared representing the E. coli malE-malF transcript, whose stable stem-loop structure was strengthened further by modifying position 5 from the base of the stem loop from A to C (Blum et al., 1999; Yehudai-Resheff and Schuster, 2000). The same RNA having 6, 12, or 24 nucleotides 3′ to the stem loop also was prepared, as shown schematically at the bottom of Figure 6. Each of the RNAs was incubated with the FL enzyme and ADP in a polyadenylation assay. The results showed no activity without the addition of nucleotides to the stem loop, very little polyadenylation activity with 6 nucleotides, and significant activity with 12 nucleotides. Therefore, the spinach chloroplast PNPase requires 6 to 12 nucleotides 3′ to the stem loop for efficient polyadenylation. Together with the results presented in Figure 5, it is evident that a stable RNA stem-loop structure inhibits the processive degradation and polymerization activity of the enzyme, clarifying its known function as a protective cis element.
Figure 6.
A Tail of 6 to 12 Nucleotides at the 3′ End of the Stem Loop Is Required for Polyadenylation by PNPase.
32P-RNAs corresponding to the 3′ end of the E. coli malE-malF mRNA with the addition of 6, 12, or 24 nucleotides 3′ to the stem loop were tested for polyadenylation using the FL PNPase and ADP. The reaction was stopped at 0, 35, and 60 min, and the RNA was purified and analyzed by denaturing PAGE and autoradiography. Some RNA molecules ending at the 3′ end at the stem-loop structure were produced as a result of the early termination of the T7 RNA polymerase in the in vitro transcription reaction.
Similarities and Differences between the Amino Acid Sequences of the Two Core Domains of PNPase and RNase PH
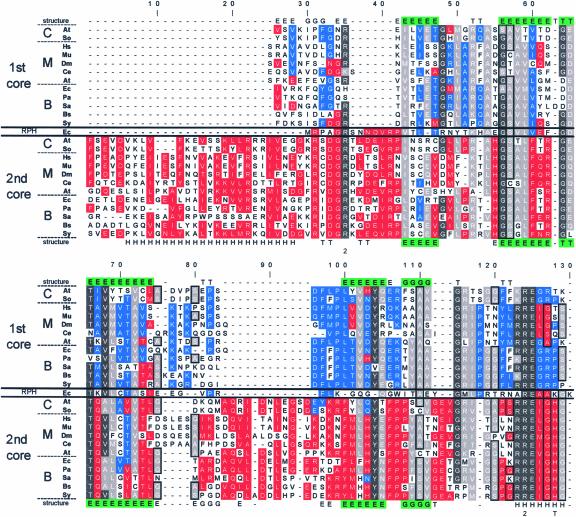
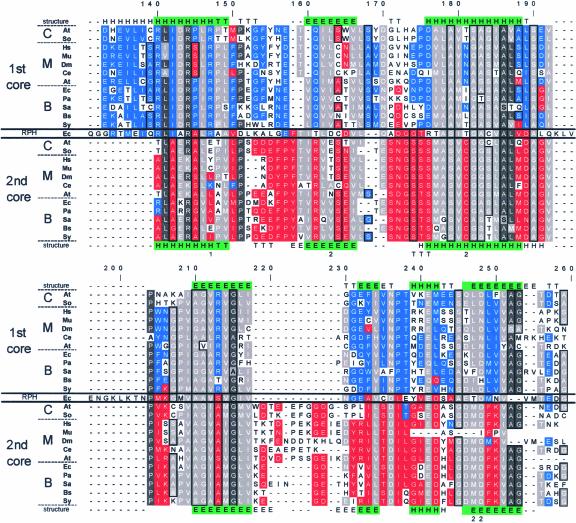
The observation that the chloroplast PNPase is composed of two active domains homologous with RNase PH raised the question of the similarity between the two domains and whether these domains are the result of a duplication event of a common RNase PH ancestor. Figure 7 presents a multiple sequence alignment of the two domains from several bacterial and eukaryotic nuclear genes that encode mitochondrial and chloroplast PNPases as well as E. coli RNase PH. First, each core domain was aligned to RNase PH of E. coli. In the second step, the two multiple alignments obtained were combined. Finally, the combined alignment was adjusted manually to give the best identity according to the crystallographic structure of the S. antibioticus PNPase (indicated in the first and last lines of Figure 7). Identical and similar amino acids between the two domains are indicated with dark and bright gray backgrounds, respectively, whereas homology restricted to the 1st or 2nd core domain is colored blue or red, respectively.
Figure 7.
Multiple Sequence Alignment of the 1st and 2nd Core Domains of PNPases.
The two core domain sequences of PNPases from chloroplast (C), mitochondria (M), and bacteria (B), as well as the E. coli RNase PH (RPH), were aligned to show modifications subsequent to the gene duplication fusion event. In addition, the known crystallographic structure of the S. antibioticus also was applied to fine-tune the alignment. In the lines marked “structure,” the secondary structure is indicated: H represents an α-helix, E represents a β-strand, T represents a turn with hydrogen bonding, and G represents a 310-helix. Green shading indicates identity in the structure of the two cores. The organisms are as follows: At, Arabidopsis thaliana; So, Spinacia oleracea; Hs, Homo sapiens; Mu, Mus musculus; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; Ec, Escherichia coli; Pa, Pseudomonas aeruginosa; Sa, Streptomyces antibioticus; Bs, Bacillus subtilis; and Sy, Synechocystis sp PCC6803. Amino acids were grouped by polarity as follows: (1) R and K; (2) D, E, Q, and N; (3) W, Y, and F; (4) I, V, L, M, and A; and (5) S and T. In addition, the amino acids A, S, and G were grouped by virtue of their small sizes and are framed in black. The gaps were introduced to allow maximum alignment of the two core and RNase PH domains. Locations at which >50% of the amino acids belong to the same group are boxed in gray. Similarities within the 1st and 2nd core domains are marked in red and blue, respectively. Locations of >75% identity are colored with dark gray. The numerals 1 and 2 below the alignment indicate the site-directed mutations in the 1st and 2nd cores of the E. coli PNPase, respectively, resulting in the inhibition of RNA degradation activity (Jarrige et al., 2002). The letter T below the alignment at positions 128, 174, 175, and 176 indicates the tungsten binding site of the S. antibioticus PNPase (Symmons et al., 2000).
As observed previously, several regions highlight the homology of the two core domains with each other and with RNase PH (Zuo and Deutscher, 2001; Aloy et al., 2002; Raijmakers et al., 2002; Symmons et al., 2002). Some highly conserved regions were observed in each of the domains. For example, the residues spanning positions 170 to 192 (which actually continue at positions 204 to 217) are very conserved in all of the PNPase 2nd core domains and also contain the tungsten binding site and two residues in which mutations of the E. coli enzyme inhibited phosphorolysis activity (Symmons et al., 2000; Jarrige et al., 2002). Interestingly, four of the amino acids that eliminated degradation activity when mutated in the E. coli PNPase were conserved in both domains (residues 35, 124, 163, and 249) (Jarrige et al., 2002). Of the others, the 1st core mutation at position 147 eliminated degradation activity of the enzyme and was conserved completely in this domain but was not conserved in the 2nd domain. However, the functionally important residue at position 163 was conserved only in bacterial PNPases, and the residue at position 181 was conserved only in the 2nd domain. In addition, the amino acids that participate in the formation of the S. antibioticus activity site, as identified by the binding of tungsten to the protein (positions 128, 174, 175, and 176), were conserved only in the 2nd domain, in agreement with the biochemical observation of this work that only the 2nd core is active in polymerization. Finally, it is clear from Figure 7 that the 1st core of the mitochondria enzymes is more different from its bacterial and chloroplast counterparts but is closer to the 2nd core consensus (e.g., positions 101, 126, 127, and 145).
The 1st domain was characterized as possessing different activities in diverse organisms, including the synthesis of the nucleotide ppGppp in S. antibioticus, the absence of this activity in E. coli (Symmons et al., 2000), and RNA degradation but not polymerization in spinach chloroplasts (Figure 2). As discussed above, the tungsten binding/phosphorolysis activity site of the S. antibioticus enzyme was highly conserved in the 2nd domain but shared only limited identity with the 1st core domain. Interestingly, alignment of the 1st and 2nd core sequences to the RNase PH disclosed that most of the amino acids conserved in the two cores also were conserved in RNase PH (Figure 7, middle line). However, certain sequences of the RNase PH were better conserved in the 1st domain, whereas others were much better conserved in the 2nd domain. As expected, the identity was best when comparing related enzymes in each group. For example, the human and mouse PNPases, which are nucleus encoded but probably targeted to mitochondria, share a very high degree of identity ( Figure 7). At several locations, both enzymes differ from the sequences conserved in most of the plant and bacterial enzymes (e.g., residues 48, 50, 114 to 118, 126, 127, and 134). Additional deletions and site-directed mutagenesis of each of the two domains is required to define exactly the degradation, phosphorolysis, polyadenylation, and ppGppp synthesis activity sites of PNPase in different organisms.
DISCUSSION
Activities of the Different Domains
PNPase plays a pivotal role in RNA degradation and polyadenylation in the chloroplast. After the initial cleavage by an endoribonuclease, which could be an RNase E/G protein, CSP41, or the 54 kD protein (Nickelsen and Link, 1993; Yang et al., 1996; V. Liveanu and G. Schuster, unpublished data), the cleavage product is modified by the addition of a poly(A)-rich tail, which can be several hundred nucleotides long. Only then is the polymerized RNA rapidly degraded exonucleolytically by PNPase and possibly by additional exoribonucleases such as RNase II/R. In spinach chloroplasts, both polymerization and degradation are performed by PNPase, because there is no evidence for PAP like that found in E. coli (Yehudai-Resheff et al., 2001). This also appears to be the situation in cyanobacteria, which is believed to be evolutionarily related to the chloroplast ancestor that invaded the primitive eukaryotic cell (Rott et al., 2003). However, only homogenous poly(A) tails were observed in Chlamydomonas reinhardtii chloroplasts, suggesting that perhaps a PAP exists there (Komine et al., 2000). In addition, the 3′ end processing of chloroplast RNA molecules was hampered in an Arabidopsis line in which the expression of PNPase was inhibited, indicating the importance of this enzyme for that process (Walter et al., 2002). Interestingly, under this condition, increased amounts of polyadenylated chloroplast RNAs were detected, suggesting that the remaining PNPase molecules are highly shifted to the polymerization mode of activity or that other polyadenylation activity could take place when PNPase is absent or reduced (Walter et al., 2002).
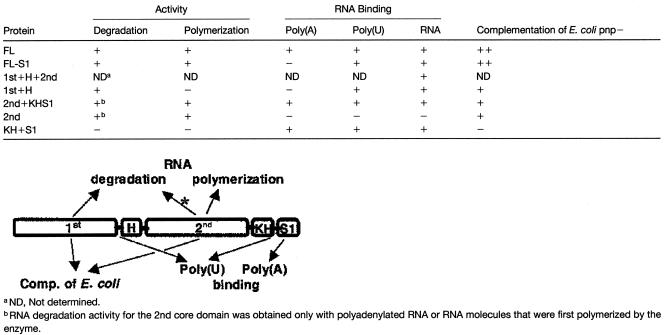
Because the PNPase conserved structure is composed of two RNase PH–like domains, two RNA binding domains (KH and S1), and one α-helical domain between the two RNase PH–related domains, we decided to study the RNA degradation and polymerization of the two RNase PH domains as well as the RNA binding properties of each domain. To this end, recombinant PNPase fragments were produced in an E. coli strain lacking the endogenous PNPase and their activities were characterized. The results are described in Table 1 and are discussed below.
Table 1.
Activities of the PNPase Domains
The 1st Core Domain
The 1st core domain did not bind the tungsten phosphate analog in the S. antibioticus PNPase, as seen in the crystal structure; therefore, it was not considered to be the location of the phosphorolysis catalytic domain (Symmons et al., 2000). However, one mutation of the E. coli enzyme in this domain eliminated catalytic activity, whereas several others reduced it markedly (Jarrige et al., 2002). In addition, compared with the 2nd core domain, the amino acid sequence is less conserved for the different species (Figure 7). Also, the PNPase activity of the synthesizing ppGppp component in S. antibioticus is believed to be localized in the 1st core domain, whereas no such activity is performed by the E. coli enzyme (Symmons et al., 2000). Therefore, the results presented here that this domain is active in RNA degradation but not in polymerization were somewhat surprising. If the lack of RNA degradation activity were found for this domain in other bacterial PNPases, it might suggest that the phosphorylase site was converted during evolution to perform other functions, such as ppGppp synthesis in S. antibioticus. In addition, this site could be converted in the spinach chloroplast PNPase to be active only in RNA degradation and not in polymerization. Indeed, we detected no polymerization activity of the 1st core domain even with GDP under conditions in which this activity was detected easily, presumably because of the lack of RNA degradation activity (Figure 2). Therefore, further studies are required to determine why the 1st core is active only in degradation and not in polymerization.
Activity of the 2nd Core Domain
The binding of the Pi analog tungsten only to the 2nd core domain in S. antibioticus PNPase, together with the greater similarity of the amino acid sequence of the 2nd core from different PNPases, made it the obvious candidate to harbor the phosphorolytic active site (Symmons et al., 2000). In addition, most of the mutations of the E. coli enzyme that inhibit this activity were located in this domain (Jarrige et al., 2002). Therefore, we were surprised initially to observe very little RNA degradation activity by the protein constructs harboring this and not the 1st domain (Figure 2). This discrepancy was resolved when these proteins were supplied with polyadenylated RNAs (Figure 5) or when nucleosides-diphosphate were added to the reaction mixture (Figure 2). Then, the degradation of polyadenylated RNA was at the same rate as that of the FL or the 1st core domain, and more interestingly, nonpolyadenylated RNAs were polymerized initially and only then degraded (Figure 2).
The observation that the phosphorolytic activity site of the spinach chloroplast PNPase is active only on polyadenylated RNAs, and is first polymerized transiently and only then degraded when nonpolyadenylated RNA is the substrate, is similar to the results obtained from RNA degradation assays performed with lysed chloroplasts. When lysed chloroplasts were supplied with 32P-RNA, it was initially polymerized transiently and only then degraded (Yehudai-Resheff et al., 2001). Moreover, similar to what was observed here for the 2nd core domain, the initial polymerization was required for the subsequent degradation step, because the addition of GDP resulted in polyguanylated RNA that was resistant to degradation by exoribonucleases (Lisitsky et al., 1997a; Yehudai-Resheff et al., 2001) ( Figure 2). Because the chloroplast tails are poly(A) rich and the S1 domain, characterized in this work to be the site of the high-affinity poly(A) binding, is located next to the 2nd core, it also probably is involved in the polymerization and degradation of the polyadenylated RNA. However, the mechanism may not be as simple as portrayed above, because the protein composed of only the 2nd core domain without the S1 also displayed this behavior (Figure 2). It will be interesting to investigate whether the 2nd domain of the bacterial and mitochondria PNPases, and possibly some of the corresponding proteins of the exosome, share this property.
PNPase Structure
The stem-loop structures located at the 3′ ends of most of the chloroplast transcripts play an important role as cis elements for 3′ end processing, protection from exoribonuclease attack, and the inhibition of polyadenylation that is followed by degradation (Monde et al., 2000). A well-known phenomenon of PNPase and RNase II is that they pause at sites containing double-stranded RNA, often being formed by the inverted repeats that form stem-loop structures. Here, we have shown that only the proteins that contain the two core domains pause at the stem-loop structures. Each independent domain completely degrades the RNA molecule without pausing at the stem-loop structure (Figure 5). This phenomenon is best explained by the mechanism already suggested whereby the RNA to be degraded enters the “hole” of the doughnut-shaped structure of the homotrimeric enzyme, which fits the size of a single-stranded RNA molecule (Symmons et al., 2000). A double-stranded RNA cannot enter this hole, and the enzyme is stuck when it reaches a stem-loop structure.
Continuing with this hypothesis, it is tempting to suggest that the formation of the doughnut-shaped structure of the homotrimer is related to the requirement of a system that should be inhibited at a stem-loop structure located at the 3′ ends of the chloroplast and perhaps prokaryotic mRNAs as well. A doughnut-shaped structure also is a characteristic of other processive enzymes. It will be interesting to analyze the biochemical properties of the exosome complex, because it has been suggested to share a similar structure (Aloy et al., 2002; Raijmakers et al., 2002; Symmons et al., 2002). In addition, a stem-loop structure is elongated inefficiently by PNPase and the E. coli PAP (Yehudai-Resheff and Schuster, 2000; Yehudai-Resheff et al., 2001; this work). A tail of 6 to 12 nucleotides 3′ to the stem is required for PNPase to polyadenylate the RNA ( Figure 6). Therefore, aside from protecting the transcript by inhibiting degradation activity, the stem-loop structure also protects the RNA by inhibiting polyadenylation.
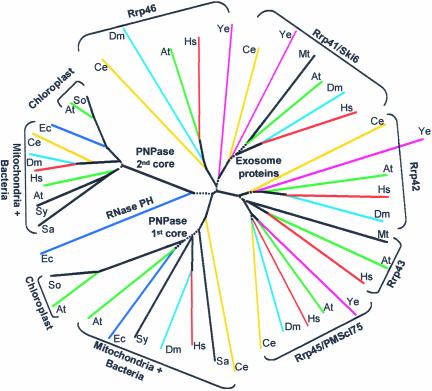
Evolution of the PNPase and the Exosome
The multiple sequence alignment of the PNPases from bacteria and organelles, as well as the exosome proteins, enabled the creation of a phylogenetic tree that clearly revealed four separate branches (excluding the single line of the E. coli RNase PH) (Figure 8). On the prokaryotic side, the duplicated ancient RNase PH domains remained linked in the same polypeptide, resulting in a single protein containing the two core domains followed by the two RNA binding domains. Three such proteins then could form the homotrimeric structure containing six core domains and three of each of the RNA binding KH and S1 domains. Interestingly, the first gene duplication that probably formed the PNPase ancestor occurred only once during evolution, and the PNPase proteins found today in the bacteria and organelles of all organisms originated from this event. The chloroplast enzymes are related more closely to each other than to the bacterial and mitochondria enzymes; therefore, they were resolved to different branches. The exosome structure was described recently as very similar to that of the PNPase trimer, as shown in Figure 1, suggesting functional and structural reasons for the “six-domain structure” composed of three PNPases or 10 to 11 exosome proteins (Aloy et al., 2002; Raijmakers et al., 2002; Symmons et al., 2002). The results of this analysis suggest close evolutionary relationships between the bacterial and organelle PNPases and the proteins of the exosome.
Figure 8.
Phylogenetic Tree of the RNase PH Domains of Bacterial and Organelle PNPases and Exosome Proteins.
The 1st and 2nd core domains of PNPases, the related exosome proteins, and the E. coli RNase PH were aligned using the CLUSTAL X multiple sequence alignment tool, and a phylogenetic tree was constructed as described in Methods. Proteins from the same organism are colored alike. The dotted lines indicate regions of the tree where the bootstrap value was <50%; therefore, the validity of these lines is low. The organisms are as follows: At, Arabidopsis thaliana; So, Spinacia oleracea; Hs, Homo sapiens; Ec, Escherichia coli; Sa, Streptomyces antibioticus; Sy, Synechocystis sp PCC6803; St, Staphylococcus aureus; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; Ye, Saccharomyces cerevisiae; and Mt, Methanobacterium thermoautotrophicum. In M. thermoautotrophicum, the proteins homologous with Rrp43 and Rrp41 are MTH682 and MTH683, respectively. Accession numbers are given in Table 3.
The related exosome proteins also were divided clearly into two completely separate branches, suggesting that each branch was either derived from one of the PNPase core domains or, alternatively, functionally related to it (Figure 8). Therefore, it is tempting to relate each branch to one of the PNPase core domains. However, the sequence alignment did not reveal a definite relationship of each branch to one of the PNPase core domains. Indeed, in previous alignments, Rrp42 was assigned to the 1st (Raijmakers et al., 2002) or 2nd (Aloy et al., 2002) core domain. A similar situation was described for PMScl75/Rrp45 (Aloy et al., 2002; Raijmakers et al., 2002). Here, the multiple alignment analysis and the derived phylogenetic tree, as presented in Figure 8, did not significantly favor the assignment of each branch of the exosome proteins to one of the PNPase core domains.
As with the PNPase branches, there is a complete separation between these two exosome proteins after the first duplication of the ancient RNase PH domain in all species, including yeast, plants, and mammals. Of evolutionary interest is the Methanococcus lineage of archaea, in which these proteins, although separated in the phylogenetic tree (as are the yeast, plant, and mammalian exosomes), are located on the same operon and possibly are derived from the same primary transcript (Koonin et al., 2001). It will be interesting to analyze the exosome of Methanococcus in light of the sequence data obtained to date from the Halobacterium lineage, in which no exosome proteins or PNPase were detected (Koonin et al., 2001). It also is interesting that although an exosome is present in yeast, no genomic or biochemical evidence for a mitochondrial PNPase was obtained (Figure 8). On the other hand, a complex composed of an RNase II–like ribonuclease and an RNA helicase was found (Dziembowski et al., 2002). By contrast, mitochondrial PNPases and RNA polyadenylation are present in mammals and plants (Figure 8) (Ojala et al., 1981; Gagliardi and Leaver, 1999; Lupold et al., 1999). Therefore, the Halobacterium lineage and Saccharomyces cerevisiae mitochondria might represent systems in which RNA degradation is dependent exclusively on a hydrolytic RNase II–like enzyme, perhaps without any phosphorylase or RNA polyadenylation involved (Dziembowski et al., 2002).
The recent analysis of the bacterial and organelle PNPases compared with the exosome proteins, together with the structural analysis of the two protein complexes, revealed a theme generally conserved in bacteria, chloroplasts, mitochondria, the cytoplasm, and the nucleus and possibly also in some archaea. In all of these systems, it probably is engaged in RNA degradation and processing. Analysis of the biochemical properties of each domain of the spinach chloroplast PNPase revealed unique features that may be related to the general function of this RNA degradation machine or specifically to the spinach chloroplast enzyme. For example, both complexes degrade RNA molecules containing poly(A) tails and display high affinity for A- and U-rich sequences (Mukherjee et al., 2002; this work). The molecular and biochemical analysis of additional PNPases and exosome-related proteins, as well as an additional analysis of the spinach PNPase, will reveal the molecular details of the mechanism of action of this evolutionarily conserved RNA degradation machine.
METHODS
Production of Recombinant PNPase and Its Fragmented Versions
The corresponding DNA sequences of the mature protein (without the transit peptide) and the different fragmented versions were amplified by PCR using the primers listed in Table 2, and spinach (Spinacia oleracea) oligo(dT)-primed cDNA was prepared as described previously (Baginsky et al., 2001). For expression in Escherichia coli, the PCR products were inserted into the Pet 20b vector (Novagen, Madison, WI) with the addition of a His6 tag to the C terminus (Figures 1E and 1F). Because the addition of the His6 tag to the N terminus of the E. coli PNPase had been reported to hamper its activity (Blum et al., 1999), we added this tag at the various C termini (Figure 1E). Nevertheless, we were unable to produce the FL protein (lacking only the transit peptide) with the His6 tag in a soluble form (Figure 1E). Therefore, this protein was expressed in the PT7-7 system (Citovsky et al., 1990) without the His6 tag and purified biochemically using a series of size-exclusion, heparin, and anion-exchange columns, resulting in a purified protein with a very low yield (Figure 1E). Fortunately, we were able to produce the other parts with the His6 tag, enabling rapid and easy purification, and at high yields (Figure 1F).
Table 2.
Oligonucleotides Primers Used for the Expression of PNPase Fragmented Proteins
| Protein | Name | Sequence |
|---|---|---|
| FL | F-172 | 5′-GGAATTCGCATATGGTTAGAGCTATGGCTCAA-3′ |
| R-2466 | 5′-CGGGATCCCCAAGCACGCCGACTAAGGC-3′ | |
| FL-S1 | F-172 | |
| R-2220 | 5′-CGGGATCCCCGGCCTTCGATTTCTCAAG-3′ | |
| 1st+H+2nd | F-172 | |
| R-1890 | 5′-CGGGATCCCCACAATCAGCATTTCCTGC-3′ | |
| 1st+H | F-172 | |
| R-1185 | 5′-CGGGATCCCCATCACCTTCATCAACTTCGCC-3′ | |
| 2nd+KHS1 | F-1227 | 5′-GGAATTCCATATGTTCTCTGAGGTAGATGTG-3′ |
| R-2400 | 5′-CGGGATCCCCTCCAACTTTAAATGCAT-3′ | |
| 2nd | F-1227 | |
| R-1890 | ||
| KH+S1 | F-1891 | 5′-GGAATTCCATATGGTTACAGCATTCCAAATG-3′ |
| R-2400 | ||
| E. coli PNPase | F | 5′-GGAATTCGCATATGATGCGCAGAAGATCGGGTAT-3′ |
| R | 5′-GCCCAAGCTTCTCGCCCTGTTCAGCAGCCG-3′ |
Restriction sites in the primer sequence are underlined (NdeI and BamHI). The name of the primer designates the direction (F, forward; R, reverse) and the number of the first nucleotide counting from the adenosine of the first ATG. If the primer was used several times, the nucleotide sequence is shown only once.
An additional obstacle in the purification of the overexpressed proteins in E. coli was that the recombinant proteins copurified with the E. coli PNPase because of the formation of heterotrimers between the chloroplast and bacterial subunits (data not shown). Use of the PNPase-less strain ENS134-3 containing the T7 RNA polymerase [BL21(DE3) (lacZ::Tn10 malPpΔ534::PT7lacZ-Arg5)(pnp::Tn5)] (Lopez et al., 1999), in which the E. coli PNPase is not expressed because of the insertion of the Tn5 transposable element (kindly obtained from Marc Dreyfus, Ecole Normale Supérieure, Paris, France), resolved this problem. Expression and purification were performed according to the manufacturer's protocol using a nitrilotriacetic acid agarose affinity column with an additional purification step using a MonoQ column (Pharmacia). The FL protein was expressed in the same cells, and the recombinant protein was purified biochemically using heparin and size-exclusion Superdex 200 and MonoQ columns (Pharmacia). All of the proteins were purified to one SDS-PAGE silver-stained band (Figure 1F) without any contamination activity of other ribonucleases detected.
Synthetic RNAs
The plasmids used for the in vitro transcription of parts of the spinach chloroplast psbA and petD-Dra were described previously (Yehudai-Resheff et al., 2001). The E. coli malE-malF intergenic region, which contained a stable stem loop in which the nucleotide at position 5 from the base of the stem loop was modified from A to C to ensure the stabilization of the stem-loop structure, also was described previously (Yehudai-Resheff and Schuster, 2000). RNAs were transcribed using T7 RNA polymerase and radioactively labeled with α-32P–UTP, and the FL transcription products were purified from 5% denaturing polyacrylamide gels (Lisitsky et al., 1996). For the preparation of transcripts terminated with the poly(A) sequence, the psbA transcription product was incubated with the yeast poly(A)-polymerase (Pharmacia) and 1 mM ATP for 30 min, and the elongated RNA was purified by denaturing PAGE (Lisitsky et al., 1996).
RNA Polyadenylation and Degradation Assays
Polyadenylation and degradation activities of the recombinant proteins were assayed as described previously (Yehudai-Resheff et al., 2001). Briefly, 32P-RNA (1 fmol) was incubated with the corresponding protein (1 fmol) in buffer E (20 mM Hepes, pH 7.9, 60 mM KCl, 12.5 mM MgCl2, 0.1 mM EDTA, 2 mM DTT, and 17% glycerol) at 25°C for the times indicated in the figures. When polyadenylation or polyguanylation were assayed, the corresponding nucleotide was added, also as indicated in the figures. For the degradation assay, Pi was added as indicated in the figures. After incubation, the RNA was isolated and analyzed by denaturing PAGE and autoradiography. For thin layer chromatography analysis of the degradation products, an aliquot of each sample was spotted on a polyethyleneimine thin layer chromatography plate, which then was developed with 1 M LiCl, dried, and exposed to autoradiography. Nucleosides mono-, di-, and three phosphates (5 μg of each) were separated on the same plate and visualized by fluorescence quenching.
UV Light Cross-Linking
UV light cross-linking of proteins to radiolabeled RNA was performed as described previously (Lisitsky et al., 1997b). The proteins (10 fmol) were mixed with 32P-RNA (10 fmol) in the buffer containing 10 mM Hepes-NaOH, pH 7.9, 30 mM KCl, 6 mM MgCl2, 0.05 mM EDTA, 2 mM DTT, and 8% glycerol and cross-linked immediately with 1.8 J of UV irradiation in a UV light cross-linker (Hoefer, San Francisco, CA). This was followed by digestion of the RNA with 10 μg of RNase A and 30 units of RNase T1 at 37°C for 1 h. The proteins then were fractionated by SDS-PAGE and analyzed by autoradiography. For the competition assay, the protein was mixed with ribohomopolymers for 5 min, and the radiolabeled RNA was added. An average length of 400 nucleotides was used to calculate the molar amount.
Complementation of the Growth of E. coli PNPase-less Mutants
The SK 8992 strain (thyA715, l-, rph-1, rna-19, pnp::Tn5, KanR), in which the gene encoding PNPase was inactivated by the insertion of the transposable element Tn5 and cannot grow at 18°C, was obtained from Sidney Kushner (Athens, GA). This strain differs from the ENS134 strain (pnp::Tn5) used for the expression of recombinant proteins because it is not a derivative of BL21(DE3) and therefore lacks the gene for the T7 RNA polymerase. We had to use this strain in the complementation experiments because we found that the insertion of any plasmid containing the T7 promoter sequence to the ENS134 strain resulted in the complementation of growth at 18°C (data not shown). In addition, this strain also lacks the other Pi-dependent exoribonuclease, RNase PH. Changing the E. coli strain to SK 8992 lacking the T7 RNA polymerase solved this problem. The different deleted PNPase constructs were cloned into the PT7-7 plasmid (Citovsky et al., 1990), transformed into this strain, and spotted on Luria-Bertani agar plates containing 100 μg/mL ampicillin and 25 μg/mL kanamycin. Because of the lack of the T7 RNA polymerase in this strain, it is unclear how abundantly the recombinant proteins were expressed. However, immunoblot analysis of protein extracts of the transformed bacteria clearly defined the accumulation of the recombinant proteins (Figure 4). As a positive control, the E. coli PNPase was amplified by PCR from genomic DNA using the primers listed in Table 1 and introduced into the same plasmid. Incubation time was 16 h at 37°C and 48 h at 18°C. Immunoblot analysis of the bacteria grown at either 37 or 18°C was used to follow the production of the corresponding proteins.
Structure Prediction, Complex Model Building, and Sequence Analysis
Homology-based modeling of the three-dimensional structure of the spinach PNPase was performed using the 3D-PSSM Fold Recognition Server at http://www.sbg.bio.ic.ac.uk/~3dpssm/. The visualization of the spinach PNPase trimer complex was performed by inserting the monomeric model described above into a pseudo rhombohedral (H32) space group with dimensions similar to those of the Streptomyces antibioticus crystal structure (PDB code 1E3H) using Quanta (Accelrys, San Diego, CA).
Sequence alignment of the different PNPases and the exosome components was performed by multiple alignment using CLUSTAL X and motif search for all of the proteins together and finally by manually improving the alignment. The phylogenetic tree was built using the neighbor-joining method with bootstrap (CLUSTAL X). Then, to improve the validity of the tree, the parsimony method with bootstrap (PAUP) was used.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact G. Schuster, gadis@tx.technion.ac.il.
Accession Numbers
The accession numbers of the different PNPase proteins shown in Figure 7 are as follows: At, Arabidopsis thaliana (NP_187021 chloroplast, NP_196962 mitochondria); So, Spinacia oleracea (AAC49669); Hs, Homo sapiens (XP_048088); Mu, Mus musculus (BAB23374); Ec, Escherichia coli (P05055); Pa, Pseudomonas aeruginosa (AAG08126); Sa, Streptomyces antibioticus (AAB17498); Bs, Bacillus subtilis (NP_657775); and Sy, Synechocystis sp PCC6803 (BAA16661).
Table 3.
Exosome Proteins That Display Homology with the Core Domains of PNPase
| Organism | Symbol | Protein | Accession No. |
|---|---|---|---|
| Arabidopsis thaliana | At | Rrp41 | NP_191721 |
| Rrp42 | AAF13093 | ||
| Rrp43 | NP_176216 | ||
| PM/Scl-75 | NP_566441 | ||
| Rrp46 | NP_190207 | ||
| Homo sapiens | Hs | Rrp41 | Q9NPD3 |
| Rrp42 | Q15024 | ||
| Rrp43 | Q96B26 | ||
| PM/Scl-75 | Q06265 | ||
| Rrp46 | Q9NQT4 | ||
| Caenorhabditis elegans | Ce | Rrp41 | Q17533 |
| Rrp42 | NP_508024 | ||
| PM/Scl-75 | T28842 | ||
| Rrp46 | NP_496284 | ||
| Drosophila melanogaster | Dm | Rrp41 | AAF53263 |
| Rrp42 | AAF58076 | ||
| PM/Scl-75 | AAF48665 | ||
| Rrp46 | AAF54530 | ||
| Saccharomyces cerevisiae | Ye | Ski6 | NP_011711 |
| Rrp42 | NP_010172 | ||
| Rrp45 | NP_010566 | ||
| Rrp46 | NP_011609 | ||
| Methanobacterium thermoautotrophicum | Mt | MTH682 | H69190 |
| MTH683 | NP_275826 |
Sequences of proteins homologous with one of the core domains of the spinach chloroplast PNPase were obtained from the National Center for Biotechnology Information database by Basic Local Alignment Search Tool (BLAST) search.
Acknowledgments
We thank Agamemnon J. Carpousis, Stanley Cohen, Marc Dreyfus, Sydney Kushner, and Philippe Regnier for the E. coli strains as well as for numerous discussions, encouragement, and valuable advice. We thank Varda Liveanu for her many suggestions and critical reading of the manuscript, Oded Beja for helping with the phylogenetic analysis, and David Stern for help in editing the manuscript. This work was supported by grants from the Israel Science Foundation and the Israel-U.S. Binational Agriculture Research and Development Foundation.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.013326.
References
- Aloy, P., Ciccarelli, F.D., Leutwein, C., Gavin, A.C., Superti-Furga, G., Bork, P., Bottcher, B., and Russell, R.B. (2002). A complex prediction: Three-dimensional model of the yeast exosome. EMBO Rep. 3, 628–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginsky, S., Shteiman-Kotler, A., Liveanu, V., Yehudai-Resheff, S., Bellaoui, M., Settlage, R.E., Shabanowitz, J., Hunt, D.F., Schuster, G., and Gruissem, W. (2001). Chloroplast PNPase exists as a homo-multimer enzyme complex that is distinct from the Escherichia coli degradosome. RNA 7, 1464–1475. [PMC free article] [PubMed] [Google Scholar]
- Beran, R.K., and Simons, R.W. (2001). Cold-temperature induction of Escherichia coli polynucleotide phosphorylase occurs by reversal of its autoregulation. Mol. Microbiol. 39, 112–125. [DOI] [PubMed] [Google Scholar]
- Blum, E., Carpousis, A.J., and Higgins, C.F. (1999). Polyadenylation promotes degradation of 3′-structured RNA by the Escherichia coli mRNA degradosome in vitro. J. Biol. Chem. 274, 4009–4016. [DOI] [PubMed] [Google Scholar]
- Carpousis, A.J., Vanzo, N.F., and Raynal, L.C. (1999). mRNA degradation, a tale of poly(A) and multiprotein machines. Trends Genet. 15, 24–28. [DOI] [PubMed] [Google Scholar]
- Citovsky, V., Knorr, D., Schuster, G., and Zambryski, P. (1990). The P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell 60, 637–647. [DOI] [PubMed] [Google Scholar]
- Clements, M.O., Eriksson, S., Thompson, A., Lucchini, S., Hinton, J.C., Normark, S., and Rhen, M. (2002). Polynucleotide phosphorylase is a global regulator of virulence and persistency in Salmonella enterica. Proc. Natl. Acad. Sci. USA 99, 8784–8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn, G.A., and Mackie, G.A. (1999). Degradation of mRNA in Escherichia coli: An old problem with some new twists. Prog. Nucleic Acids Res. 62, 55–108. [DOI] [PubMed] [Google Scholar]
- Craven, M.G., Henner, D.J., Alessi, D., Schauer, A.T., Ost, K.A., Deutscher, M.P., and Friedman, D.I. (1992). Identification of the rph (RNase PH) gene of Bacillus subtilis: Evidence for suppression of cold-sensitive mutations in Escherichia coli. J. Bacteriol. 174, 4727–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski, A., Piwowarski, J., Hoser, R., Minczuk, M., Dmochowska, A., Siep, M., Van Der Spek, H., Grivell, L., and Stepien, P.P. (2002). The yeast mitochondrial degradosome: Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J. Biol. Chem. 278, 1603–1611. [DOI] [PubMed] [Google Scholar]
- Gagliardi, D., and Leaver, C.J. (1999). Polyadenylation accelerates the degradation of the mitochondrial mRNA associated with cytoplasmic male sterility in sunflower. EMBO J. 18, 3757–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunberg-Manago, M. (1999). Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 33, 193–227. [DOI] [PubMed] [Google Scholar]
- Hayes, R., Kudla, J., and Gruissem, W. (1999). Degrading chloroplast mRNA: The role of polyadenylation. Trends Biochem. Sci. 24, 199–202. [DOI] [PubMed] [Google Scholar]
- Hayes, R., Kudla, J., Schuster, G., Gabay, L., Maliga, P., and Gruissem, W. (1996). Chloroplast mRNA 3′-end processing by a high molecular weight protein complex is regulated by nuclear encoded RNA binding proteins. EMBO J. 15, 1132–1141. [PMC free article] [PubMed] [Google Scholar]
- Jarrige, A., Brechemier-Baey, D., Mathy, N., Duche, O., and Portier, C. (2002). Mutational analysis of polynucleotide phosphorylase from Escherichia coli. J. Mol. Biol. 321, 397–409. [DOI] [PubMed] [Google Scholar]
- Komine, Y., Kwong, L., Anguera, M., Schuster, S., and Stern, D.B. (2000). Polyadenylation of three classes of chloroplast RNA in Chlamydomonas reinhardtii. RNA 6, 598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin, E.V., Wolf, Y.I., and Aravind, L. (2001). Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 11, 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, J., Tengler, U., and Binder, S. (2001). Transcript lifetime is balanced between stabilizing stem-loop structures and degradation-promoting polyadenylation in plant mitochondria. Mol. Cell. Biol. 21, 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leszczyniecka, M., Kang, D.C., Sarkar, D., Su, Z.Z., Holmes, M., Valerie, K., and Fisher, P.B. (2002). Identification and cloning of human polynucleotide phosphorylase, hPNPase old-35, in the context of terminal differentiation and cellular senescence. Proc. Natl. Acad. Sci. USA 99, 16636–16641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou, G.G., Chang, H.Y., Lin, C.S., and Lin-Chao, S. (2002). DEAD box RhlB RNA helicase physically associates with exoribonuclease PNPase to degrade double-stranded RNA independent of the degradosome-assembling region of RNase E. J. Biol. Chem. 277, 41157–41162. [DOI] [PubMed] [Google Scholar]
- Lisitsky, I., Klaff, P., and Schuster, G. (1996). Addition of poly(A)-rich sequences to endonucleolytic cleavage sites in the degradation of spinach chloroplast mRNA. Proc. Natl. Acad. Sci. USA 93, 13398–13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisitsky, I., Klaff, P., and Schuster, G. (1997. a). Blocking polyadenylation of mRNA in the chloroplast inhibits its degradation. Plant J. 12, 1173–1178. [Google Scholar]
- Lisitsky, I., Kotler, A., and Schuster, G. (1997. b). The mechanism of preferential degradation of polyadenylated RNA in the chloroplast: The exoribonuclease 100RNP/PNPase displays high binding affinity for poly(A) sequence. J. Biol. Chem. 272, 17648–17653. [DOI] [PubMed] [Google Scholar]
- Lisitsky, I., Liveanu, V., and Schuster, G. (1994). RNA-binding activities of the different domains of a spinach chloroplast ribonucleoprotein. Nucleic Acids Res. 22, 4719–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisitsky, I., and Schuster, G. (1999). Preferential degradation of polyadenylated and polyuridinylated RNAs by bacterial exoribonuclease polynucleotide phosphorylase (PNPase). Eur. J. Biochem. 261, 468–474. [DOI] [PubMed] [Google Scholar]
- Littauer, U.Z., and Grunberg-Manago, M. (1999). Polynucleotide phosphorylase. In The Encyclopedia of Molecular Biology, T.E. Creighton, ed (New York: John Wiley & Sons), pp. 1911–1918.
- Lopez, P.J., Marchand, I., Joyce, S.A., and Dreyfus, M. (1999). The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol. Microbiol. 33, 188–199. [DOI] [PubMed] [Google Scholar]
- Lupold, D.S., Caoile, A.G.F.S., and Stern, D.B. (1999). Polyadenylation occurs at multiple sites in maize mitochondrial cox2 mRNA and is independent of editing status. Plant Cell 11, 1565–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty, B.K., and Kushner, S.R. (2000). Polynucleotide phosphorylase functions both as a 3′ to 5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl. Acad. Sci. USA 97, 11966–11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monde, R.A., Schuster, G., and Stern, D.B. (2000). Processing and degradation of chloroplast mRNA. Biochimie 82, 573–582. [DOI] [PubMed] [Google Scholar]
- Mukherjee, D., Gao, M., O'Connor, J.P., Raijmakers, R., Pruijn, G., Lutz, C.S., and Wilusz, J. (2002). The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21, 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickelsen, J., and Link, G. (1993). The 54 kDa RNA-binding protein from mustard chloroplasts mediates endonucleolytic transcript 3′ end formation in vitro. Plant J. 3, 537–544. [DOI] [PubMed] [Google Scholar]
- Ojala, D., Montoya, J., and Attardi, G. (1981). tRNA punctuation model of RNA processing in human mitochondria. Nature 290, 470–474. [DOI] [PubMed] [Google Scholar]
- Raijmakers, R., Egberts, W.V., van Venrooij, W.J., and Pruijn, G.J. (2002). Protein-protein interactions between human exosome components support the assembly of RNase PH-type subunits into a six-membered PNPase-like ring. J. Mol. Biol. 323, 653–663. [DOI] [PubMed] [Google Scholar]
- Regnier, P., and Arraiano, C.M. (2000). Degradation of mRNA in bacteria: Emergence of ubiquitous features. Bioessays 22, 235–244. [DOI] [PubMed] [Google Scholar]
- Rott, R., Zipor, G., Portnoy, V., Liveanu, V., and Schuster, G. (2003). RNA polyadenylation and degradation in cyanobacteria are similar to the chloroplast but different from Escherichia coli. J. Biol. Chem. 278, 15771–15777. [DOI] [PubMed] [Google Scholar]
- Sarkar, N. (1997). Polyadenylation of mRNA in prokaryotes. Annu. Rev. Biochem. 66, 173–197. [DOI] [PubMed] [Google Scholar]
- Schuster, G., Lisitsky, I., and Klaff, P. (1999). Update on chloroplast molecular biology: Polyadenylation and degradation of mRNA in the chloroplast. Plant Physiol. 120, 937–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist, W.I. (1993). Conducting the G-quartet. Curr. Biol 3, 893–895. [DOI] [PubMed] [Google Scholar]
- Symmons, M.F., Jones, G.H., and Luisi, B.F. (2000). A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure 8, 1215–1226. [DOI] [PubMed] [Google Scholar]
- Symmons, M.F., Williams, M.G., Luisi, B.F., Jones, G.H., and Carpousis, A.J. (2002). Running rings around RNA: A superfamily of phosphate-dependent RNases. Trends Biochem. Sci. 27, 11–18. [DOI] [PubMed] [Google Scholar]
- Walter, M., Kilian, J., and Kudla, J. (2002). PNPase activity determines the efficiency of mRNA 3′-end processing, the degradation of tRNA and the extent of polyadenylation in chloroplasts. EMBO J. 21, 6905–6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancey, S.D., and Kushner, S.R. (1990). Isolation and characterization of a new temperature-sensitive polynucleotide phosphorylase mutation in Escherichia coli K-12. Biochimie 72, 835–843. [DOI] [PubMed] [Google Scholar]
- Yang, J., Schuster, G., and Stern, D.B. (1996). CSP41, a sequence-specific chloroplast mRNA binding protein, is an endoribonuclease. Plant Cell 8, 1409–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff, S., Hirsh, M., and Schuster, G. (2001). Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Mol. Cell. Biol. 21, 5408–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff, S., and Schuster, G. (2000). Characterization of the E. coli poly(A)-polymerase: Specificity to nucleotides, RNA-binding affinities and RNA-structure dependence activity. Nucleic Acids Res. 28, 1139–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., and Deutscher, M.P. (1997). An essential function for the phosphate-dependent exoribonucleases RNase PH and polynucleotide phosphorylase. J. Bacteriol. 179, 4391–4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, Y., and Deutscher, M.P. (2001). Exoribonuclease superfamilies: Structural analysis and phylogenetic distribution. Nucleic Acids Res. 29, 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]