Abstract
To investigate UV light response mechanisms in higher plants, we isolated a UV light–sensitive mutant, rev3-1, in Arabidopsis. The root growth of rev3-1 was inhibited after UV-B irradiation under both light and dark conditions. We found that chromosome 1 of rev3-1 was broken at a minimum of three points, causing chromosome inversion and translocation. A gene disrupted by this rearrangement encoded the catalytic subunit of DNA polymerase ζ (AtREV3), which is thought to be involved in translesion synthesis. The rev3-1 seedlings also were sensitive to γ-rays and mitomycin C, which are known to inhibit DNA replication. Incorporation of bromodeoxyuridine after UV-B irradiation was less in rev3-1 than in the wild type. These results indicate that UV light–damaged DNA interrupted DNA replication in the rev3-1 mutant, leading to the inhibition of cell division and root elongation.
INTRODUCTION
Plants spend all their lives at the position at which they sprouted; thus, they cannot easily escape from a stressful environment. Even sunlight, which is essential for photosynthesis, is stressful. Too strong light damages the photosystems in the chloroplasts by the overproduction of reactive oxygen species (Asada, 1999). UV light, which corresponds to a small fraction of the total solar radiation, exerts serious effects on plants. UV light damages various cellular compounds, including DNA, membranes, photosystems, and phytohormones, either directly or indirectly through the induction of reactive oxygen species (Rozema et al., 1997). In addition, DNA damage caused by the formation of cyclobutane pyrimidine dimers (CPDs) and pyrimidine (6-4) pyrimidone photoproducts [(6-4) photoproducts] is quite toxic to plant cells because this type of damage prevents DNA replication and transcription and also causes mutations.
Plant UV light tolerance systems have been studied for years. The best known system is the synthesis of secondary metabolites that absorb UV photons or free radicals produced by UV radiation (Harborne and Williams, 2000). These secondary metabolites serve as a shield and protect DNA from damage. Another mechanism is to enzymatically repair DNA damaged by UV light. Plants possess a pair of powerful repair enzymes called photoreactivating enzymes or photolyases. Using the photon energy from visible light, CPD photolyase repairs CPDs (Ahmad et al., 1997) and (6-4) photolyase repairs (6-4) photoproducts (Nakajima et al., 1998). It was suggested recently that other enzymes participate in nucleotide excision repair (NER) of UV light–damaged DNA (Xu et al., 1998; Gallego et al., 2000; Liu et al., 2000, 2001).
On the other hand, some organisms have a mechanism, called the damage-tolerance pathway, that increases their ability to tolerate unrepaired DNA lesions. In yeast, for example, the stalling of replication caused by unrepaired DNA lesions is overcome by the activities of a series of proteins encoded by genes belonging to the so-called RAD6 epistasis group (Haynes and Kunz, 1981). Translesion synthesis (TLS) is one of the activities performed by the damage-tolerance pathways. In TLS, DNA damage is bypassed and the nascent DNA strand is extended by specialized DNA polymerases. If the formation of a pyrimidine dimer or other damage is not repaired and remains on the replication fork, it distorts the DNA structure and inhibits the synthesis of the daughter strand by replication-type polymerases. The TLS-type polymerases, which are more tolerant of abnormal template structure, replace the normal polymerase and insert a nucleotide opposite the position of the DNA lesion. The TLS activities have been studied in many organisms, including yeast (Nelson et al., 1996a, 1996b; Johnson et al., 1999), Escherichia coli (Reuven et al., 1999; Tang et al., 1999, 2000; Wagner et al., 1999), and human (Gibbs et al., 1998; Lin et al., 1999; Matsuda et al., 2000). Although several bioinformatics analyses predicted the presence of a TLS pathway in higher plants (Lin et al., 1999; Lawrence et al., 2000; Kimura et al., 2002), such a pathway has not been found experimentally.
Here, we report the isolation of a UV light–sensitive mutant, rev3-1, from Arabidopsis by applying an ion beam mutagenesis technique (Tanaka et al., 1997). We found that rev3-1 was disrupted in a gene named AtREV3, a homolog of yeast REV3 (Morrison et al., 1989) and of REV3 in other organisms (Gibbs et al., 1998; Van Sloun et al., 1999; Eeken et al., 2001; Sakai et al., 2002) that are known to be involved in TLS. The UV light–sensitive phenotype of rev3-1 can be a malfunction of TLS. Our findings suggest the presence of a TLS mechanism in a higher plant.
RESULTS
rev3-1 Is a UV-B Light–Hypersensitive Mutant
We screened ∼3000 M2 lines derived from ion beam–irradiated Arabidopsis seeds for UV-B light sensitivities under the dark condition. Thirteen M2 lines whose growth was inhibited by UV-B irradiation were selected as primary candidates for UV-B light–sensitive mutants. Based on the results of rough mapping and complementation tests, these candidate lines were categorized into six independent UV-B light–sensitive groups. One of these mutants, on which we focus here, was a mutation in a gene homologous with the yeast REV3 gene (see below). Thus, we named it rev3-1.
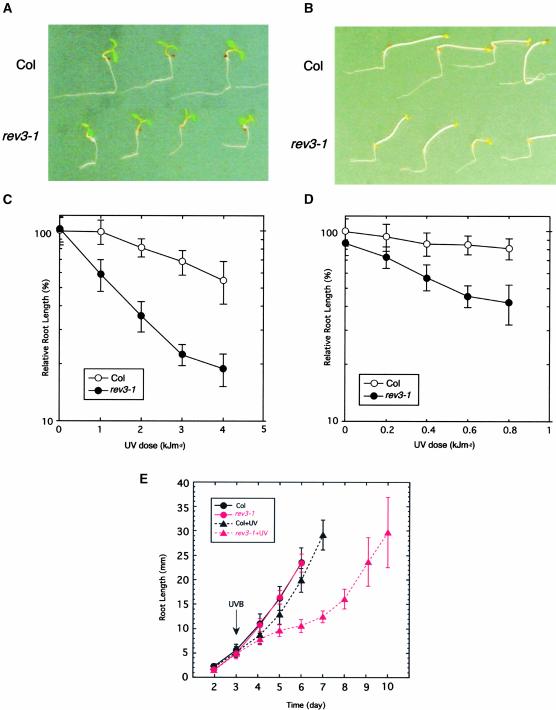
To characterize the rev3-1 mutant, the seedlings were irradiated with various doses of UV-B light, and their responses were analyzed by a root-bending assay (Britt et al., 1993). The root growth of the wild-type seedling was inhibited slightly by a dose of 1 kJ m−2, but it was inhibited at an increased UV-B light dose under the light condition (Figures 1A and 1C). Under the dark condition, the root growth of the wild-type plants was inhibited slightly (Figures 1B and 1D). On the contrary, the root growth of the rev3-1 mutant was reduced severely in a UV-B light dose-dependent manner not only under the dark condition (Figures 1B and 1D) but also under the light condition (Figures 1A and 1C), in which the photoreactivating enzymes should be active.
Figure 1.
UV-B Light Sensitivities of Wild-Type and rev3-1 Seedlings under Light and Dark Conditions.
(A) Root growth of wild-type (Columbia [Col]) and rev3-1 seedlings in the light condition. Three-day-old seedlings were exposed to 3 kJ m−2 UV-B light and then incubated under continuous white light for 3 days.
(B) Root growth of wild-type and rev3-1 seedlings in the dark condition. Seedlings were exposed to 0.6 kJ m−2 UV-B light and then incubated in the dark for 3 days.
(C) and (D) UV light dose-response curve for root growth in the wild type (open circles) and rev3-1 (closed circles) under light (C) and dark (D) conditions. Root growth after irradiation was measured using NIH Image. Each value represents an average of 18 to 26 measurements. Error bars indicate sd.
(E) Analysis of root growth rate after UV irradiation. Wild-type and rev3-1 seedlings were sown on a nutritive agar plate, and root growth was checked every day. Three days after the start of incubation, the seedlings were exposed to 2 kJ m−2 UV-B light (arrow). Each value represents an average of 15 to 21 measurements. Error bars indicate sd.
For further characterization of the root growth of the mutant, we compared root growth rates of wild-type and rev3 plants under the light condition. We irradiated 3-day-old seedlings with 2 kJ m−2 of UV-B light (Figure 1E). Without UV-B light treatment, the profiles of the root growth of wild-type and rev3-1 plants were the same. One day after UV-B irradiation, the growth rates of both rev3-1 and wild-type roots were reduced by approximately the same amount compared with the growth rates of roots without irradiation. Two days after irradiation, the growth of wild-type roots recovered, becoming similar to the growth of the nonirradiated roots. The growth curves of the irradiated and nonirradiated wild-type plants indicated that the UV irradiation delayed root growth by ∼12 h. By contrast, the growth rate of UV-B–irradiated rev3-1 roots did not recover for several days after irradiation. Subsequently, some mutant roots started to regrow, whereas others did not regrow and sprouted secondary roots instead. Thus, we speculated that the root growth defect after UV treatment in the rev3-1 plants is attributable to a delay in the restart of cell division.
The response of the rev3-1 mutant to long-term UV-B irradiation also was examined (Table 1). The fresh weight of the rev3-1 mutant was slightly less than that of the wild-type plant without UV-B irradiation. After exposure to UV-B light for 2 weeks, the rev3-1 mutant showed a greater decrease in fresh weight than did the wild-type plant. The fresh weight of the rev3-1 mutant decreased by up to 50% in a UV-B light dose-dependent manner. Together, these results indicate that the rev3-1 mutant is significantly more sensitive to UV-B light than is the wild-type plant.
Table 1.
Effect of Long-Term UV-B Light Treatment on the Growth of Wild-Type Columbia and the rev3-1 Mutant
| UV Light Dose (kJ m−2) a | Fresh Weight b (mg)
|
rev3-1/Columbia (%) | |
|---|---|---|---|
| Columbia | rev3-1 | ||
| 0 | 26.66 ± 1.12 | 23.65 ± 2.19 | 88.73 |
| 7.6 | 8.64 ± 0.55 | 6.29 ± 0.42 | 72.78 |
| 14 | 1.66 ± 0.08 | 0.81 ± 0.05 | 48.88 |
For 2 weeks.
Average values of >20 plants ± se.
rev3-1 Possesses Normal DNA Damage-Repair Activity under Both Dark and Light Conditions
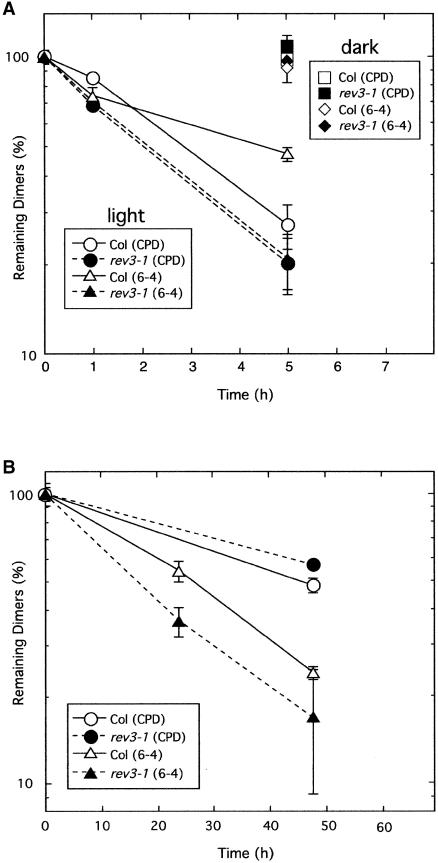
The results of the root-bending assay showed that rev3-1 plants were hypersensitive to UV-B irradiation under both dark and light conditions (Figure 1). This phenotype resembles those of other Arabidopsis mutants defective in the dark repair system (Jenkins et al., 1995). To determine whether the rev3-1 mutant is defective in the dark repair system, like other UV light–hypersensitive mutants, the reduction of CPDs and (6-4) photoproducts after UV-B light treatment was quantified in rev3-1 mutant plants by means of ELISA, as described previously (Tanaka et al., 2002). Immediately after UV-B irradiation (1 to 3 kJ m−2), the amounts of DNA damage in wild-type and rev3-1 plants were similar (data not shown). Then, UV-B light–treated plants were incubated under the dark or light condition, and the remaining DNA damage was quantified at several time points during incubation.
Under the light condition, CPDs and (6-4) photoproducts were removed rapidly in both wild-type and mutant plants, probably because of the activities of the photoreactivating enzymes (Figure 2A). On the other hand, the dark repair activity in Arabidopsis was very low and hardly detectable within several hours (Figure 2A; cf. the dark repair and the light repair). Similar results were reported previously (Britt et al., 1993; Landry et al., 1997; Tanaka et al., 2002). When the plants were kept in the dark for up to 48 h, the amount of CPDs and (6-4) photoproducts per unit of DNA decreased slowly (Figure 2B). Although some cells in the seedling seemed to proliferate during the incubation, the increase in total DNA per seedling was <10% under these assay conditions (data not shown). Therefore, it is reasonable to suppose that the decreasing DNA damage is largely the result of the dark repair activity rather than of the dilution of CPDs and (6-4) photoproducts caused by the increase of DNA. These results suggest that rev3-1 has normal repair activities under both dark and light conditions.
Figure 2.
Elimination of Two Major Types of UV Damage in the Wild Type and the rev3-1 Mutant.
Five-day-old seedlings were irradiated with 3 kJ m−2 (A) or 1 kJ m−2 (B) UV-B light and then kept under white light (A) or in the dark ([A] and [B]). Seedlings were harvested after the indicated periods, and DNA was extracted from them. The amounts of CPDs or (6-4) photoproducts were determined by ELISA using specific antibodies. Each value represents an average of at least two independent measurements. Error bars indicate se. Col, Columbia wild type.
Genetic Analysis and Mapping of the rev3-1 Mutant Suggests an Unusual Organization of the rev3-1 Locus
To analyze the genetic traits of the rev3-1 mutant, the rev3-1 plant was backcrossed to the wild-type Columbia plant. Of 98 lines of F2 derived from this cross, 24 showed a UV light–sensitive phenotype by the root-bending assay. On the other hand, when the rev3-1 plant was crossed with the Landsberg erecta (Ler) plant, 145 of 611 lines of F2 derived from this cross showed a UV light–sensitive phenotype. In both segregation analyses, the segregation ratios were almost 3:1 (P < 0.05), indicating that rev3-1 is a single recessive mutant.
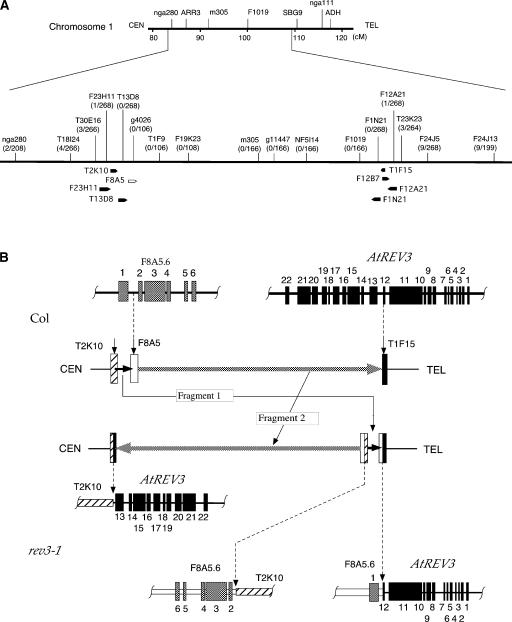
To identify the rev3-1 gene, homozygous UV-B light–sensitive F2 lines derived from a cross between rev3-1 and Ler were investigated. Our result showed that the rev3-1 mutation was linked closely to the bottom of chromosome 1. Of 58 chromosomes examined, six had recombinations at the nga111 (115.55 centimorgan) marker and none had recombinations at the nga280 (83.83 centimorgan) marker. To find the location of the fewest recombinations on chromosome 1, we selected five additional PCR-based markers (F19K23, g4026, m305a, g11447, and nF5I14) positioned between nga280 and nga111 and examined them in 166 chromosomes. However, none of these markers showed any evidence of recombination. Then, 12 new markers were designed based on 12 BAC sequences (Figure 3A; sequences are shown in the supplemental data online). Of 268 chromosomes analyzed, none showed any recombinations from T13D8 to F1N21 (Figure 3A). These results suggest that there is something unusual about the organization of chromosome 1 of the rev3-1 mutant.
Figure 3.
Unusual Recombination Rates and Chromosome Rearrangements in rev3-1.
(A) Recombination rates in F2 plants derived from a cross of rev3-1 with Ler. A local map of the bottom of chromosome 1 is shown at top. The recombination rate at each marker position is shown in parentheses. Recombination events were fewest at the bottom of chromosome 1, and no recombination was detected at the markers located between T13D8 and F1N21. Black spears indicate the positions and directions of BACs used as probes in the genomic DNA gel blot analysis. The white spear indicates the F8A5 BAC described in the text.
(B) Overview of chromosome rearrangements in rev3-1. (Top) Original structures and positions of two genes disrupted in the rev3-1 mutant. In the wild-type plant, the F8A5.6 gene, consisting of six exons (gray rectangles), is located on F8A5. The AtREV3 gene, consisting of 22 exons (black rectangles), is located on T1F15. (Center) Predicted chromosome rearrangements by ion beam irradiation. The chromosome was broken at three points (vertical arrows), and then fragment 2 was inverted and fragment 1 was translocated. (Bottom) Chromosome and gene structures in the rev3-1 mutant. The 12th exon of the AtREV3 gene is rejoined to the first intron of the F8A5.6 gene in the opposite direction. The 3′ part of AtREV3 gene is rejoined to the noncoding region in T2K10, and the 3′ part of the F8A5.6 gene is rejoined to the other broken end in T2K10.
Chromosome Rearrangement Occurs in rev3-1, Leading to a Disruption of Two Genes
Given that ion beam irradiation often induces large chromosomal rearrangements in plant cells (Shikazono et al., 2001), it is possible that the unusual distribution of recombinations in rev3-1 might be caused by a chromosome inversion induced by ion beam irradiation. To test this possibility and to identify the mutation responsible for the rev3-1 phenotype, we searched for the ends of the inverted region. The sequences of seven BACs located around the boundaries of the recombining and nonrecombining regions were used as probes to detect the restriction enzyme–digested patterns of rev3-1 DNA (see supplemental data online). As a result, the probes prepared from BACs T2K10, F12B7, and T1F15 showed altered band patterns when hybridized to rev3-1 DNA. A 9.2-kb fragment corresponding to F12B7 and T1F15 and a 6.6-kb fragment corresponding to T2K10 were missing from the pattern of rev3-1 DNA.
To narrow the possible mutated DNA region, the 9.2-kb (right, distal region to centromere in Figure 3A) and 6.6-kb (left, proximal region to centromere) regions were examined by PCR. As a result, a 1.6-kb subfragment in the 9.2-kb region and a 0.8-kb subfragment in the 6.6-kb region were not amplified. The failure to amplify these subfragments might be the result of rearrangements within them. To elucidate the rearrangement in the rev3-1 chromosome, thermal asymmetric interlaced (TAIL)–PCR was performed using arbitrary degenerated primers with the specific primers designed for the upstream region of the 1.6-kb subfragment and the downstream region of the 0.8-kb subfragment. Sequencing of the TAIL-PCR products revealed that the sequence of T1F15 was broken and rejoined to the sequence of T2K10. However, the other half of T2K10 was rejoined to a fragment other than T1F15. A search for homologous sequences in the Arabidopsis DNA database revealed that this fragment corresponded to F8A5, which was located ∼200 kb downstream of T2K10 (Figure 3A).
These results suggest that there are at least three breakpoints on chromosome 1. To clarify the three breaks and their rejoining sites, TAIL-PCR was performed with specific primers designed from the upstream and downstream sequences of the breakpoints in F8A5 and these PCR products were sequenced and assembled. The results indicate that the rev3-1 mutant has a large chromosomal rearrangement, including an inversion and a translocation (Figure 3B). In summary, chromosome 1 was broken at a minimum of three points, on T2K10, F8A5, and T1F15. One fragment from the first breakpoint (on T2K10) to the second breakpoint (on F8A5) was translocated and rejoined to the right side of the third breakpoint (on T1F15). Another fragment of ∼2 Mb from the second breakpoint to the third breakpoint was inverted and then rejoined to the left side of the first breakpoint and the translocated fragment.
Because of the chromosomal rearrangements, at least three breaks appeared to disrupt two presumptive genes in the rev3-1 mutant (Figure 3B). One gene (F8A5.6) encodes a protein that is homologous with the members of the large GTP binding protein family. Another gene encodes the catalytic subunit of DNA polymerase ζ (AtREV3). Analysis of the nucleotide sequence flanking the three breakpoints revealed that the 12th exon of the AtREV3 gene was rejoined to the first intron of the F8A5.6 gene in the opposite direction. This rearrangement disrupted the open reading frames of both the AtREV3 and F8A5.6 genes (Figure 3B). The 3′ part of the AtREV3 gene was rejoined to the one broken end of T2K10, where no gene is present. Similarly, the 3′ part of the F8A5.6 gene was rejoined to the other broken end of T2K10. Comparison of the nucleotide sequences flanking the rejoined sites and the corresponding sequences of the wild type demonstrated that the first, second, and third breakpoints had deletions of 20, 28, and 17 bp, respectively. Moreover, a few nucleotides of T or GTA at the first and second breaks were present at both rejoining sites, suggesting that these short homologous sequences were used to rejoin the ends. At the third breakpoint, however, no homologous sequence was found on the rejoined site. These findings indicate that the rejoinings occurred by nonhomologous end joining, as reported previously (Shikazono et al., 2001).
The rev3-1 Mutant Phenotype Is Attributable to a Disruption of the AtREV3 Gene
Because two genes are disrupted in the rev3-1 mutant, either or both mutations might be responsible for the mutant's sensitivity to UV light. First, we focused on the F8A5.6 gene. However, this gene does not seem to be transcribed in wild-type plants, because no transcripts were detected by reverse transcriptase–mediated PCR (data not shown). Therefore, we speculate that this gene is a pseudogene and cannot be responsible for the rev3-1 mutant phenotype. To confirm this speculation, we analyzed a T-DNA–inserted F8A5.6-disrupted line and found that its sensitivity to UV light was the same as that observed in wild-type plants even when both F8A5.6 alleles were broken (data not shown). Together, these results led us to hypothesize that the AtREV3 gene, rather than the F8A5.6 gene, is responsible for the phenotype of the rev3-1 mutant.
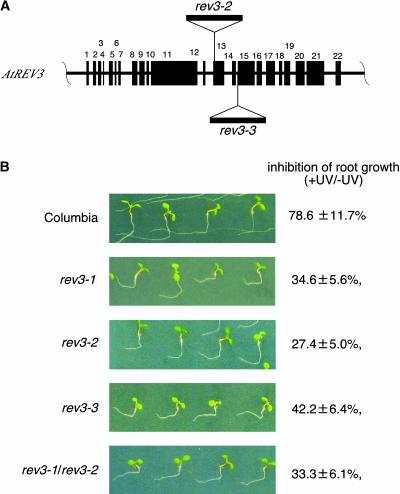
To test this hypothesis, we searched for other rev3 alleles. A search of the T-DNA insertion sequence databases of the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu) for sequences homologous with the AtREV3 gene found two lines, SALK_029237 and SALK_067237, which we named rev3-2 and rev3-3. rev3-2 has a T-DNA inserted at the13th exon, whereas rev3-3 has a T-DNA inserted at the border of the 14th intron and the 15th exon (Figure 4A). These two mutant lines and the progeny obtained by crossing them with rev3-1 were examined using the root-bending assay. Without UV irradiation, the root length of the wild-type, rev3-1, rev3-2, rev3-3, and rev3-1/rev3-2 plants were almost the same. When irradiated with 2 kJ m−2 UV-B light and incubated under the light condition, the root growth of wild-type seedlings was reduced slightly (78.6 ± 11.7% of the control value), whereas the root growth of rev3-1 seedlings was only 34.6 ± 5.6% of that of the control rev3-1 seedlings without UV light treatment (Figure 4B).
Figure 4.
UV-B Light Sensitivities of rev3-2 and rev3-3 Mutants Generated by T-DNA Insertion.
(A) Disruption of AtREV3 by T-DNA insertion. Black rectangles represent 22 exons of the AtREV3 gene. The T-DNA is inserted in the 13th exon in rev3-2 and at the border of the 14th intron and the 15th exon in rev3-3.
(B) UV light sensitivities of rev3-1, rev3-2, rev3-3, and rev3-1/rev3-3 seedlings. Seedlings were exposed to 2 kJ m−2 UV-B light and then incubated under continuous white light for 3 days. Root growth is expressed as a percentage of the average length of nonirradiated roots ± sd.
The inhibition of root growth of the rev3-2 and rev3-3 seedlings (27.4 ± 5.0% and 42.2 ± 6.4% of the root growth of the controls, respectively) was almost the same as that of the rev3-1 plants. Similarly, when the plants were irradiated with 0.6 kJ m−2 UV-B light and incubated under the dark condition, the root growth of wild-type seedlings was 82.0 ± 9.8% of the root growth of the control, whereas the root growth of the rev3-1, rev3-2, and rev3-3 seedlings was 51.7 ± 12.1%, 51.7 ± 9.2%, and 55.1 ± 9.9% of the root growth of the controls, respectively. The root growth of rev3-1/rev3-2 plants was 33.3 ± 6.1% of the root growth of the control under the light condition (Figure 4B) and 54.0 ± 8.5% of the root growth of the control under the dark condition, suggesting that rev3-1 and rev3-2 are allelic. Inhibition of the root growth of rev3-1/rev3-3 plants by UV-B irradiation in both light and dark conditions appeared to be similar to that of rev3-1, although the data were insufficient to show this statistically (data not shown). These results suggest that rev3-1 and rev3-3 also are allelic.
When we analyzed the genetic trait of the rev3-2 mutant, the offspring of rev3-2/+ plants were segregated at a ratio of 1:3 ( UV light sensitive:UV light resistant) (P < 0.05, n = 276), indicating a single recessive trait. In addition, all examined UV light–sensitive offspring (n = 66) contained T-DNA insertions but no intact AtREV3 gene (data not shown). In conclusion, these results suggest that the UV light hypersensitivity of the rev3-1 mutant is caused by a disruption of the AtREV3 gene.
Features of the AtREV3 Gene
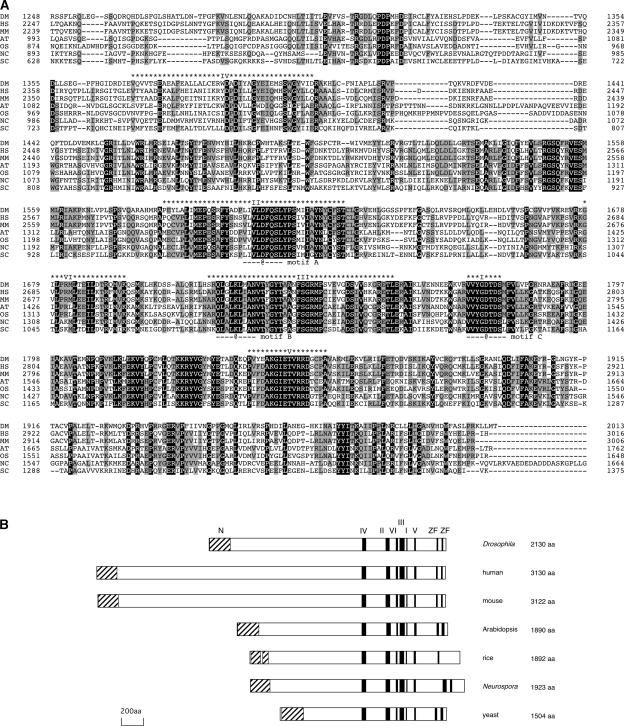
According to the annotation by The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org/), the AtREV3 gene consists of 22 exons and contains an open reading frame of 5685 bp (gene identifier, At1g67500). After sequencing of the reverse transcriptase–mediated PCR products, we found that all predicted exon/intron splicing sites are correct except for the prediction of intron 5. Based on this result, it is likely that the correct length of the open reading frame is 5673 bp, which encodes a protein of 1890 amino acids. AtREV3 is highly similar to other REV3s ( Figure 5). In particular, the C-terminal half of the protein contains all six polymerase domains that are conserved in the B-type DNA polymerase family (Figure 5A). Three residues that are known to interact with the template DNA in other B-type polymerases also are found in AtREV3 and other REV3 sequences. In addition, AtREV3 has an N-terminal homologous region and two repeats of zinc finger domains that are conserved in almost all other REV3s (Figure 5B). These facts strongly suggest that AtREV3 is involved in DNA synthesis. A search for a sequence homologous with AtREV3 revealed a possible REV3 homolog (43% identity) in the rice genome shown in TIGR (http://www.tigr.org/). This finding suggests that REV3 proteins are present in both monocotyledonous and dicotyledonous plants.
Figure 5.
Sequence Alignments of REV3 Proteins.
(A) Alignment of the C-terminal parts of the conserved domains in REV3 proteins from fruit fly (DM), human (HS), mouse (MM), Arabidopsis (AT), rice (OS), N. crassa (NC), and yeast (SC). Amino acid sequences were aligned using CLUSTAL X. The positions of the amino acids in each protein are shown at left and right. The residues conserved in all sequences are shown in reverse. The residues conserved in more than four sequences are shown in gray. Asterisks above the sequences indicate the six conserved polymerase domains. The “at” signs (@) under the sequences indicate residues shown to form the active center in other B-type polymerases.
(B) Primary structures of eukaryotic REV3 proteins. Black boxes indicate the B-type polymerase domains (I to IV) and zinc finger domains (ZF). Hatched boxes indicate the N-terminal homologous region (N). aa, amino acids.
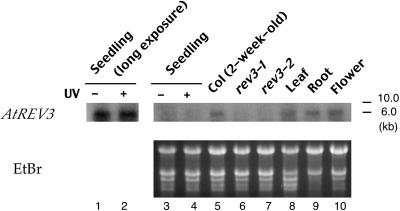
RNA gel blot analysis showed that the AtREV3 transcript is ∼6 kb, which is consistent with the predicted open reading frame of 5673 bp. The AtREV3 transcript in the wild-type plants was more abundant in 2-week-old plants than in 1-week-old seedlings (Figure 6, lanes 3 and 5) and was expressed ubiquitously in different tissues, such as leaf, root, and flower, of 4-week-old plants (lanes 8 to 10). However, the transcript was not detected in the rev3-1 and rev3-2 mutants (lanes 6 and 7). If the AtREV3 gene is involved in UV sensitivity in the wild-type plant and therefore the rev3-1 or rev3-2 mutant becomes hypersensitive to UV light, induction of the AtREV3 transcript would be expected to depend on the UV light treatment. However, when the seedlings were irradiated with 3 kJ/m−2 UV-B light, the expression level was not changed at 6 h after the end of the exposure (Figure 6, lanes 1 and 2). The lack of induction of AtREV3 by UV light is not too different from the weak induction of the REV3 gene of yeast by UV light (Singhal et al., 1992). However, the REV3 gene of Neurospora crassa is inducible by UV light (Sakai et al., 2002).
Figure 6.
Expression Patterns of the AtREV3 Gene.
One-week-old wild-type Arabidopsis seedlings were irradiated with 2 kJ m−2 UV-B light (lanes 2 and 4) or not irradiated (lanes 1 and 3). After a 6-h incubation, RNA was extracted from the seedlings. RNA also was extracted from 2-week-old plants of the wild type (lane 5), rev3-1 (lane 6), or rev3-2 (lane 7) and from different tissues of 4-week-old wild-type plants (lanes 8 to 10). Ten micrograms of total RNA was blotted on a membrane and hybridized with the probe prepared from AtREV3 cDNA. EtBr, ethidium bromide.
The AtREV3 Gene Seems to Be Involved in TLS
If AtREV3 is involved in TLS in Arabidopsis, as is reported in yeast and mammal cells (Lawrence et al., 2000), then the deletion of AtREV3 should stall DNA replication after DNA damage. In preliminary observations, rev3-1 plants treated with UV-B light had a twisted root in which the row of epidermal cells was disturbed severely (data not shown). We speculate that this phenomenon is attributable to nonuniform cell division: in the root cells, UV light–induced damage might inhibit DNA replication and affect cell division patterns in the root. If this is correct, DNA synthesis should be disturbed in the rev3-1 plants treated with UV-B light.
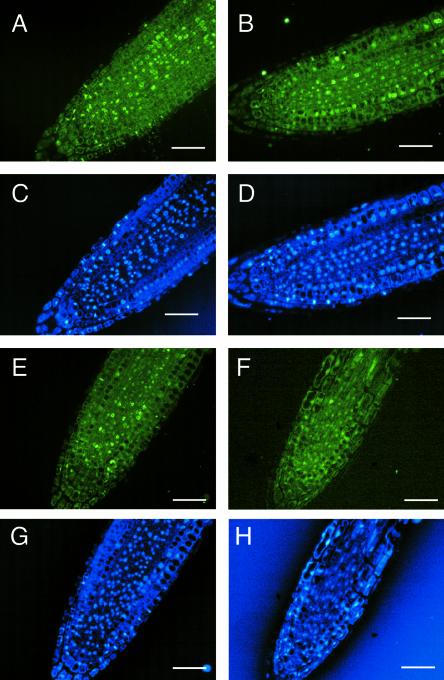
To test this hypothesis, we observed the efficiency of incorporation of bromodeoxyuridine (BrdU), an analog of thymidine, into the root tip cells (Figure 7). Incorporation of BrdU often is used as an index of DNA replication and cell proliferation (Gratzner, 1982; Nagata et al., 2000). Although BrdU is known to be incorporated in DNA during dark repair and TLS (DiGiuseppe and Dresler, 1989; Srivastava et al., 1993), the amount that is incorporated by these processes is quite small compared with the amount incorporated during replication. Without UV-B light treatment, the tissues of wild-type and rev3-1 roots were well stained with the anti-BrdU antibody (Figures 7A and 7E), indicating that the vigorously proliferating root tip cells incorporated BrdU. The wild-type roots treated with 2 kJ m−2 UV-B light could not be distinguished from the nontreated roots ( Figure 7B). However, in the UV-B light–treated rev3-1 roots, the number of immunostained cells was reduced greatly (Figure 7F). This finding indicates that DNA replication in the rev3-1 roots was inhibited even at 36 to 60 h after UV-B irradiation, at which time DNA replication in the UV light–treated wild-type plants was recovered completely.
Figure 7.
BrdU Incorporation in the Root Tip Cells of the Wild Type and rev3-1.
(A) to (D) Wild-type cells.
(E) to (H) rev3-1 cells.
Three-day-old seedlings were irradiated with 2 kJ m−2 UV-B light ([B], [D], [F], and [H]) or not irradiated ([A], [C], [E], and [G]). After ∼36 h, the seedlings were soaked in BrdU solution for 24 h. BrdU incorporation was detected by anti-BrdU ([A], [B], [E], and [F]). The same sections were stained with 4′,6-diamidino-2-phenylindole ([C], [D], [G], and [H]). Bars = 50 μm.
Some nuclei of UV-B light–treated rev3-1 roots showed considerable BrdU incorporation (data not shown). However, very few cells incorporated BrdU, and incorporation was not synchronous, in contrast to what was observed in the nontreated root or in wild-type roots. It is likely that this asynchronous DNA replication caused an irregular cell division in the UV light–treated rev3-1 roots. Another possibility is that BrdU incorporation in the UV-B light–treated rev3-1 roots was caused by other DNA repair activities that are used to complement the loss of the AtREV3-dependent pathway. In summary, AtREV3 is required for DNA replication. The loss of AtREV3 activity leads to the UV light–hypersensitive phenotype in the rev3-1 mutant.
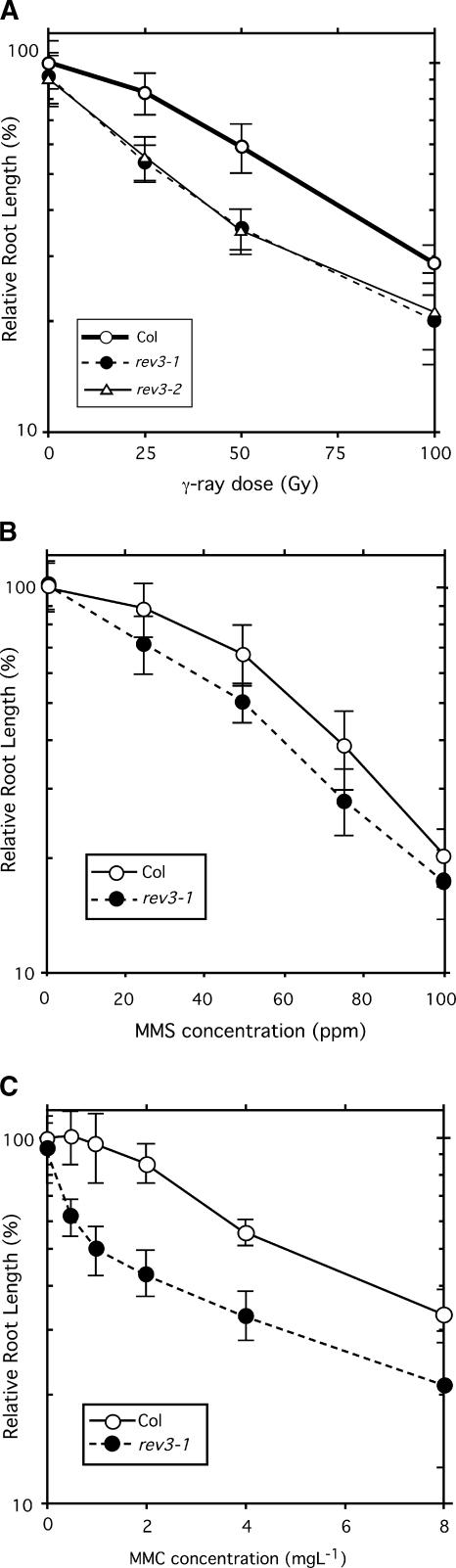
In other organisms, REV3 is reported to be involved not only in the replication of UV light–damaged DNA but also in other types of DNA damage, such as the formation of apurine/apyrimidine (AP) sites (Lawrence et al., 2000). If AtREV3 also is involved in TLS of several kinds of DNA damage, rev3-1 plants that have suffered such DNA damage should show root growth defects similar to those caused by UV treatments. To test this prediction, wild-type, rev3-1, and rev3-2 seedlings were challenged with several agents that cause different types of DNA damage (Figure 8). When exposed to γ-rays, the rev3-1 and rev3-2 seedlings became hypersensitive (i.e., root growth was inhibited compared with that of the wild type) (Figure 8A). When the seedlings were explanted to agar plates supplemented with 20 to 100 ppm of methyl methanesulfonate (MMS), an alkylating agent that provokes base modification, the root growth of rev3-1 plants was only slightly shorter than that of the wild type (Figure 8B). On the other hand, the growth of rev3-1 seedlings was inhibited on plates supplemented with 2 to 8 mg/L mitomycin C (MMC), which is thought to cause intrastrand or interstrand cross-links on double-stranded DNA. This finding suggests that AtREV3 has diverse functions in Arabidopsis. Together, these results indicate that AtREV3 is required for the tolerance of various DNA-damaging agents, including UV light, γ-rays, and MMC. These results are consistent with our hypothesis that AtREV3 is involved in the TLS mechanisms in Arabidopsis.
Figure 8.
Responses of Wild-Type, rev3-1, and rev3-2 Seedlings to DNA-Damaging Agents.
Three-day-old seedlings were irradiated with γ-rays (A) or explanted to MMS-containing (B) or MMC-containing (C) agar plates and incubated for another 3 days. Root growth after the treatments was measured using NIH Image. Each value represents an average of 16 to 25 measurements. Error bars indicate 1 sd. Col, Columbia wild type; Gy, Gray units.
DISCUSSION
AtREV3 Seems to Be Involved in the Damage-Tolerance Mechanisms of Higher Plants
The results described above show that the disruption of the AtREV3 gene, the Arabidopsis homolog of yeast REV3, resulted in hypersensitivity to UV-B light under both light and dark conditions. Under photoreactivating conditions, most UV light–induced damage in higher plants is repaired by the activities of photolyases (Britt, 1999). Elimination of any of the photolyase genes drastically reduced the DNA-repair activity in the light (Jiang et al., 1997; Landry et al., 1997). Under nonphotoreactivating conditions, UV light–induced DNA damage is repaired by a dark repair pathway that includes NER (Britt, 1999). Disruption of a particular subunit of the NER complex reduces repair activity in the dark condition (Liu et al., 2000). However, in the rev3-1 mutant, the major forms of UV light–induced damage, CPDs and (6-4) photoproducts, seem to be repaired normally under both conditions (Figure 2). Thus, AtREV3 does not seem to be involved in the photorepair or dark repair pathways. Instead, we propose a novel mechanism that allows plants to tolerate DNA damage. If AtREV3 is disrupted, this mechanism does not work and DNA synthesis is blocked for some time, resulting in a delay of the restart of the cell cycle. In fact, we observed that DNA replication was inhibited in UV light–treated roots of AtREV3-disrupted plants (Figure 7), which probably was responsible for the growth arrest of these roots.
Even with a normal damage-repair system, plants would have difficulty repairing all DNA damage. For example, rice plants grown in the field are known to accumulate as much as 2 to 3 CPDs per Mb throughout the day (Hidema et al., 1999). In Arabidopsis, our ELISA experiments showed that >50% of UV-B light–induced CPDs and (6-4) photoproducts were removed rapidly within 5 h in the light (Tanaka et al., 2002) (Figure 2). This finding suggests that by 24 h after UV-B light exposure, the amount of DNA damage in the UV light–treated plants would become indistinguishable from that of untreated plants. Even under such conditions, however, DNA synthesis was inhibited in rev3-1 plants (Figure 7). This finding suggests that in the absence of a functional AtREV3, a small amount of DNA damage that was not repaired by the major DNA-repair activities would be sufficient to prevent DNA replication. Thus, our results raise the possibility that Arabidopsis has a TLS pathway that involves AtREV3 and that AtREV3 is needed to allow DNA replication to proceed in the presence of minor DNA damage. However, direct evidence of a TLS pathway is lacking. Further studies will be needed to confirm that such a pathway is present in plants.
REV3 Proteins Are Involved in the Damage-Tolerance Pathway in Eukaryotic Cells and Play a Mutagenic Role
The damage-tolerance mechanism was investigated in detail in yeast using a series of RAD6 epistatic mutants (Friedberg et al., 1991). This pathway is independent of the RAD3 (excision repair) and RAD50 (recombination repair) pathways and does not repair the damage itself. The main functions of this pathway are (1) to find the stalled replication fork, (2) to resolve the replication complex, and (3) to continue the replication without being interrupted by any repair process. TLS is the most important activity among the damage-tolerance pathways. TLS accomplishes the replication of damaged DNA using a specialized DNA polymerase instead of the replication-type polymerase.
If the replication complex encounters a damaged site on the template DNA, it will stop (Figure 9A). This stalled complex is detected by RAD5 gene products and then removed by the protein degradation system involving RAD6 gene products (Friedberg et al., 1995). After the template is cleared of the replication complex, TLS-type polymerase, which is more tolerant of a distorted template structure, binds to the template and extends the daughter strand by inserting nucleotides on the opposite side of the lesion (Figure 9B). Because the TLS-type polymerase has low processivity, it does not extend the strand very long and is replaced by the replication-type polymerase to continue the replication (Figure 9C). DNA polymerase ζ (pol ζ) is one of the TLS-type polymerases; it consists of the catalytic subunit REV3 and the regulatory subunit REV7 and is known to bypass several DNA lesions, such as pyrimidine dimers or AP sites (Figure 9B). In the case of AP sites, another protein, REV1, works together with pol ζ. REV1 is a member of the Y-family of polymerases, which have deoxycytidyl transferase activity (Nelson et al., 1996b). Thus, REV1 inserts a dCTP opposite an AP site, which then is followed by pol ζ to extend the daughter strand.
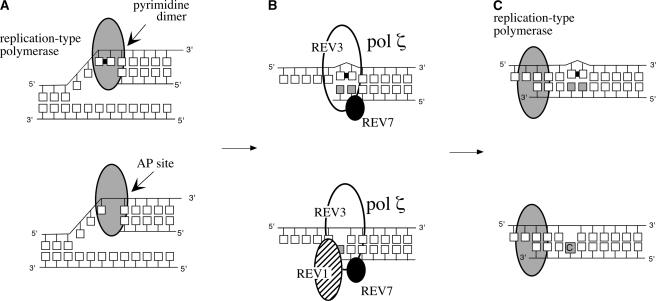
Figure 9.
Scheme of TLS.
(A) DNA damage, such as pyrimidine dimers or AP sites, distorts the template structure and prevents replication by the replication-type polymerase.
(B) The stalled replication complex is removed, and a TLS-type polymerase, such as pol ζ, bypasses the damaged site. pol ζ consists of a large catalytic subunit of REV3 and a regulatory subunit of REV7. Opposite the AP site, REV1 inserts a deoxycytidine. This TLS step causes a replication error.
(C) Because of its low processivity, the TLS-type polymerase is replaced by the replication-type polymerase and replication is continued.
Based on a genetic analysis of yeast mutants, two TLS pathways were identified, an error-free TLS pathway and an error-prone TLS pathway. The former is accomplished by DNA polymerase η (pol η) (McDonald et al., 1997), and the latter is believed to be accomplished by pol ζ (Lawrence and Christensen, 1976). Because purified pol ζ does not have proofreading ability (Nelson et al., 1996a), it is regarded as a faulty polymerase. A subsequent study demonstrated that pol ζ is unable to bypass a pyrimidine dimer by itself and that an additional polymerase is required to complete the TLS (Prakash and Prakash, 2002). It also was found that pol ζ has a relatively high fidelity on a normal template and is able to extend a strand from the mismatched primer termini (Lawrence et al., 2000). Therefore, it is hypothesized that TLS often is accomplished by the sequential action of two polymerases. If the first polymerase incorporates a wrong nucleotide opposite the lesion, the second polymerase, such as pol ζ, would extend the strand from the mismatched nucleotide pair and keep the mutation. For example, in the replication of (6-4) photoproducts, pol η predominantly inserts a G opposite the 3′ nucleotide, and then the strand is extended by pol ζ (Johnson et al., 2001). Because (6-4) photoproducts most frequently occur in the 5′-TC-3′ sequence, the insertion of a G opposite the C retains the original sequence, which is why pol η is regarded as an error-free polymerase. However, if a (6-4) photoproduct is formed on the 5′-TT-3′ sequence, the insertion of a G opposite the T and extension from this mismatched pair by pol ζ would result in a T-to-C substitution. Therefore, pol ζ would cause a mutation, which is why it is regarded as an error-prone polymerase.
AtREV3 Is Essential for the Tolerance of DNA Damage
REV3-deficient mutants of several organisms have been shown to be sensitive to various DNA-damaging agents. In yeast, for example, the rev3 mutant is sensitive not only to UV light but also to x-rays and MMS treatments (Haynes and Kunz, 1981). In general, low linear energy transfer ionizing radiation, such as that from γ-rays and x-rays, or monofunctional alkylating agents, such as MMS, induce base damage in DNA. These kinds of damage are removed mainly by the base excision repair pathway and result in the AP sites. To bypass the AP sites, cooperation between REV1 and pol ζ is necessary, as described previously (Figure 9), which explains the sensitivity of the yeast rev3 mutant to x-rays and MMS in addition to UV light. If this cooperation is necessary, the hypersensitivity of the rev3-1 mutant to γ-ray irradiation (Figure 8A) suggests that AtREV3 is involved in bypassing MMS-induced damage. However, the AtREV3-disrupted mutant did not appear to be sensitive to MMS (Figure 8B). Similarly, in N. crassa, a REV3-deficient strain was reported to be sensitive to UV light but less sensitive to γ-rays and MMS (Sakai et al., 2002). These discrepancies may be attributable to the different assay systems used in yeast, N. crassa, and Arabidopsis. Another possibility is that each organism has an alternative, non-pol ζ–mediated pathway for bypassing the DNA damage induced by γ-rays and MMS.
Although MMC severely inhibited the root growth of the rev3-1 mutant (Figure 8C), it only slightly inhibited the growth of the N. crassa REV3-deficient strain (Sakai et al., 2002). On the other hand, the yeast rev3 mutant is sensitive to cisplatin and 8-methoxysporalen (Grossmann et al., 2000), which, like MMC, cause interstrand cross-links (ICLs) in double-stranded DNA. Because ICL prevents DNA strand separation, the DNA cannot be repaired by the usual excision repair pathway. It is unclear why several REV3-deficient mutants are sensitive to agents that produce ICLs. However, one possibility is that ICLs require some specific DNA-repair process that involves REV3. Grossmann et al. (2000) proposed an ICL repair pathway in which ICLs are excised by an NER and then the resultant double-strand break is repaired by the recombination repair pathway or by the nonhomologous end-joining pathway. In addition, Holbeck and Strathern (1997) reported that in yeast, the high mutation frequency induced in the repair process of double-strand breaks was abolished by elimination of the REV3 gene. This fact implies that REV3 is involved in the repair of double-strand breaks. Therefore, it is possible that AtREV3 also is involved in other DNA maintenance processes.
REV3 Is Nonessential for Survival
Based on its primary structure, DNA polymerase ζ is classified as a member of the B-type DNA polymerase family (Filée et al., 2002). The B-type polymerase family also includes polymerases α, δ, and ɛ, all of which are involved in the replication of chromosomal DNA and are essential for survival in many organisms (Burgers, 1998). By contrast, the survival of REV3-deficient mutants of yeast (Friedberg et al., 1995), N. crassa (Sakai et al., 2002), fruit fly (Eeken et al., 2001), and Arabidopsis (this work) indicates that pol ζ is not essential for survival, at least in these organisms. Moreover, the finding that cultured human cells expressing an antisense RNA of REV3 are viable and grow normally (Gibbs et al., 1998), and the fact that the nematode genome does not have a gene homologous with REV3 (Lawrence et al., 2000), provide further evidence that REV3 is dispensable. On the other hand, three independent groups have reported that a disruption of REV3 resulted in embryonic lethality in knockout mice (Bemark et al., 2000; Esposito et al., 2000; Wittschieben et al., 2000). It is possible that, although the conserved REV3 function involving TLS is not essential for the survival of cells, some additional function of mammalian REV3s are required for the developmental process in mammals. In support of this hypothesis, mammalian REV3s have a large domain in the N-terminal half that is not present in the REV3s of lower organisms or in any other polymerases (Figure 5B).
Why Is the Mutagenic Repair Pathway Conserved?
As mentioned above, the REV3 gene is the largest factor controlling mutagenesis in yeast and animal systems. In yeast, REV3 is responsible for 50 to 75% of spontaneous mutations (Quah et al., 1980), for >98% of UV light–induced base substitutions (Lawrence and Christensen, 1979), and for >90% of UV light–induced frameshift mutations (Lawrence et al., 1984). The presence of a REV3 homolog protein in many eukaryotes raises a question: why is such a risky, mutagenic pathway conserved?
For unicellular organisms, the inhibition of cell division is lethal. Thus, it is easy to imagine that unicellular organisms passively use a mutagenic repair system rather than stop replication. Alternatively, unicellular organisms may benefit from a mutagenic pathway to modify their genomes to adapt to hostile environments. By contrast, in higher animals, mutagenic repair may serve a different purpose. Because higher animals are multicellular organisms, the inhibition of cell division does not necessarily mean death. Thus, it is not essential for higher animals to replicate damaged DNA by error-prone TLS with pol ζ. At the same time, mutations in a small number of cells caused by the error-prone TLS may not be so toxic for the whole individual. There are three reasons for this. First, because the genomes of higher animals consist largely of noncoding sequences, the probability that a mutation will occur in the coding region is small. Second, higher animals have an apoptotic pathway to remove mutated cells from the tissues. Third, in higher animals, somatic cells and reproductive cells are different, so mutations in the somatic cells do not affect the next generation.
Like higher animals, higher plants also contain large noncoding sequences in their genomes. Many plants are polyploid and seem to be hardly affected by mutations. However, it is not clear whether plant cells have an apoptosis-like system for removing mutated cells. Another difference is that plants are totipotent and do not possess a special reproductive cell lineage until the very late stage of development. Therefore, mutations that occur in somatic cells can be transmitted to the next generation. This was shown by Ries et al. (2000), who exposed Arabidopsis plants to modest doses of UV-B light. These plants produced one to four germinal recombinations in 250,000 seeds. This fact also suggests that the plant genome can be disturbed by mild environmental stresses, although the frequency with which this occurs is low. In recent preliminary experiments (our unpublished data), we found that the frequency of several kinds of base substitutions was reduced in AtREV3-disrupted plants. This finding suggests that AtREV3, like the REV3s of other organisms, is responsible for some mutations. Another indication of the importance of AtREV3 is the relatively uniform expression of the AtREV3 gene in adult plants, suggesting that mutagenic TLS may occur routinely in Arabidopsis. If this is true, it seems unavoidable that a few mutations would be transmitted to the next generation. Further studies are needed to determine whether mutations by the error-prone TLS have any adverse effects in plants or whether they are even beneficial.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia was the wild-type plant used in this study. The rev3-2 line (SALK_029237), the rev3-3 line (SALK_067237), and the F8A5.6 disruption line (SALK_060920) were provided by the Salk Institute Genomic Analysis Laboratory. Plants were grown at 23°C in a growth chamber (LH200RD; NK System, Osaka, Japan) under continuous white light from fluorescent lamps (FL40SS-W/37 [Toshiba Lighting and Technology, Tokyo, Japan] or FLR40SEX D-HG [NEC Lighting, Tokyo, Japan]; ∼40 μE·m−2·s−1) filtered through a glass plate. This light source emits wavelengths no shorter than 310 nm.
Mutant Isolation
Seeds were irradiated with 220-megaelectron volt carbon ions from an azimuthally varying field cyclotron at a dose of 150 Gray units, as described previously (Tanaka et al., 1997). Approximately 750 M1 seeds were grown and self-pollinated. The obtained seeds (M2) were grown, and the seeds of each M2 plant were harvested, resulting in the establishment of 3000 M2 lines. The UV-B light sensitivity of all M2 lines was examined by a root-bending assay.
UV-B Light Source
UV-B light was supplied by a UV lamp (CSL-15B; COSMO BIO, Tokyo, Japan) that radiates at wavelengths of >280 nm with a high peak at 312 nm (manufactured and measured by Vilber Lourmat, Cedex, France). UV-B light doses were measured with a UV-B light radiometer (CSV-312; COSMO BIO) whose filter transmits UV-B irradiation with a peak at 313 nm.
Root-Bending Assay and Analysis of the Root Growth Rate
The root-bending assay was performed as described previously (Britt et al., 1993) with modifications. Seedlings were grown vertically on nutritive agar plates (2% sucrose and 0.1% [v/v] commercial nutrient; Hyponex, Osaka, Japan) under continuous white light for 3 days. To isolate mutants, seedlings were exposed to 0.5 kJ m−2 UV-B light at a rate of 0.25 kJ m−2 min−1 and then incubated for another 3 days under the dark condition. For analysis of UV-B light sensitivity, seedlings were exposed to 0.2 to 0.8 kJ m−2 (for the dark condition) or 2 to 4 kJ m−2 (for the light condition) UV-B light and then incubated in the dark or under continuous white light (∼40 μE·m−2·s−1) for 3 days. The length of root growth after UV-B irradiation was measured using NIH Image software (version 1.62) and expressed as a percentage of the average length of nonirradiated wild-type roots. To plot root growth, wild-type and rev3-1 seeds were sown on a nutritive agar plate and incubated under continuous white light at 23°C. After the seeds germinated, the positions of the root tip ends were marked at the back of the plate at ∼24-h intervals to calculate the growth rate.
Long-Term UV-B Irradiation
Plants were grown in a pot containing soil (Metro-Mix 350; Scotts-Sierra Horticultural Products, Marysville, OH) under a 16-h photoperiod in a photochamber (BIOTRON; NK System). Ten-day-old seedlings were irradiated with UV-B light at 7.6 to 14 kJ m−2 for 7 h/day. The UV-B light dose was adjusted by varying the distance from the UV-B lamp to the plants. After 14 days of UV-B light treatment, the fresh weight of each plant was measured. More than 20 plants were used for each data point.
ELISA
Five-day-old seedlings grown on agar plates were irradiated with UV-B light and then kept in the dark or under light from fluorescent lamps (FL40SS-W/37 [Toshiba Lighting and Technology] or FLR40SEX D-HG [NEC Lighting]; ∼40 μE·m−2·s−1) filtered through a glass plate. The plants were harvested at various time points, and the DNA was extracted with a Plant Mini Kit (Qiagen, Hilden, Germany). The amounts of cyclobutane pyrimidine dimers and (6-4) photoproducts were determined by ELISA using specific antibodies, as described previously (Tanaka et al., 2002). Values were expressed as percentages of the amount of DNA damage immediately after the UV-B light treatment in each line.
Mapping Analysis
The rev3-1 line was crossed with the Landsberg erecta ecotype. F2 plants from this cross were grown, and one leaf was removed from each plant and stored for DNA preparation. Then, the plants were allowed to self-pollinate to prepare F2 lines. Approximately 600 F2 lines were screened for UV-B light sensitivity using the root-bending assay. DNA was extracted from the leaf stocks of homozygous UV-B light–sensitive lines and analyzed by the simple sequence length polymorphism method (Bell and Ecker, 1994) or the cleaved amplified polymorphic sequence method (Konieczny and Ausubel, 1993). The sequences of PCR-based markers are listed in the TAIR database (http://www.Arabidopsis.org/) or in the supplemental data online.
Measurement of Plant Sensitivities to γ-Rays, Methyl Methanesulfonate, and Mitomycin C
Seeds were set on nutritive agar plates and grown vertically under continuous white light for 3 days. To test γ-ray sensitivity, the seedlings were irradiated with γ-rays from a 60Co irradiation facility (JAERI, Takasaki, Japan). To test methyl methanesulfonate (MMS) or mitomycin C (MMC) sensitivity, 3-day-old seedlings were transplanted to the surface of nutritive agar plates supplemented with 25 to 100 ppm MMS or 0.5 to 8 mg L−1 MMC. The plates were placed vertically so that the new root would grow at a right angle to the previous root. After a 3-day incubation under continuous white light, new root growth was measured as described above.
Multiple Alignment of REV3 Proteins
The REV3 sequences of yeast, N. crassa, human, mouse, and fruit fly were obtained from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/). The rice BAC sequence encoding a protein homologous with REV3 was obtained by a BLAST (Basic Local Alignment Search Tool) search of the rice genome at TIGR (http://www.tigr.org/). The original predicted open reading frame in the database is missing an exon that we found in the genomic sequence and included in this alignment to make the best alignment with the AtREV3 sequence. The amino acid sequences of the REV3 proteins were aligned with the CLUSTAL X program (Thompson et al., 1997). The amino acid sequence identity between AtREV3 and OsREV3 was calculated using the BLAST2 program at NCBI.
Genetic Analysis of the rev3-1 Mutant
The rev3-1 plant was backcrossed with the parental Columbia line, and ∼100 F2 seeds derived from this cross were placed on soil, grown, and self-pollinated, and the seeds were pooled to generate F2 lines. More than 20 F3 seedlings were examined for each F2 line using the root-bending assay, and the lines in which all seedlings showed UV light sensitivity were identified as homologous UV light–sensitive lines. The number of UV light–sensitive lines and other lines was examined using the χ2 test.
Detection of DNA Synthesis
DNA synthesis was detected with a bromodeoxyuridine (BrdU) labeling and detection kit (Roche Diagnostics, Mannheim, Germany). Root tissues were incubated in the labeling solution containing 10 μM BrdU and 1 μM 5-fluoro-2′-deoxyuridine (ICN, Irvine, CA) at room temperature for 24 h. After fixing with 4% paraformaldehyde-phosphate buffer (Wako Chemicals, Osaka, Japan), tissues were dehydrated and embedded in Technovit 7100 resin (Hereaus Kulzer, Wehrheim, Germany) as described previously (Fujie et al., 1993). The detection of BrdU was performed according to the manufacturer's instructions. Fluorescence images were obtained with an Axioscope fluorescence microscope (Zeiss, Jena, Germany).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact A. Sakamoto, sakamoto@taka.jaeri.go.jp.
Accession Numbers
Accession numbers for the sequences mentioned in this article are as follows: AAC18785 (Arabidopsis REV3), AB114052 (Arabidopsis REV3 cDNA based on the revised open reading frame, P14284 (yeast REV3), BAB83627 (N. crassa REV3), O60673 (human REV3), XP_125533 (mouse REV3), AAG30224 (fruit fly REV3), and AP003982 (a BAC that includes a rice REV3-like gene).
Supplementary Material
Acknowledgments
We thank Chihiro Suzuki, Shinya Takahashi, Tomohiro Baba, Satoshi Goto, Masanori Hatashita, and Masaaki Mizobuchi for their technical assistance, Yutaka Oono and Issay Narumi for critical readings of the manuscript, and Shigemitsu Tano, Hiroshi Watanabe, and Tamaki Hirose for their helpful comments. We acknowledge the ABRC and the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants. This work was supported in part by a grant-in-aid (15201010) for scientific research from the Ministry of Education, Science, Sports and Culture of Japan.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.012369.
Footnotes
Online version contains Web-only data.
References
- Ahmad, M., Jarillo, J.A., Klimczak, L.J., Landry, L.G., Peng, T., Last, R.L., and Cashmore, A.R. (1997). An enzyme similar to animal type II photolyases mediates photoreactivation in Arabidopsis. Plant Cell 9, 199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada, K. (1999). The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 601–639. [DOI] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Bemark, M., Khamlichi, A.A., Davies, S.L., and Neuberger, M.S. (2000). Disruption of mouse polymerase ζ (Rev3) leads to embryonic lethality and impairs blastocyst development in vitro. Curr. Biol. 10, 1213–1216. [DOI] [PubMed] [Google Scholar]
- Britt, A.B. (1999). Molecular genetics of DNA repair in higher plants. Trends Plant Sci. 1, 20–25. [DOI] [PubMed] [Google Scholar]
- Britt, A.B., Chen, J.-J., Wykoff, D., and Mitchell, D. (1993). A UV-sensitive mutant of Arabidopsis defective in the repair of pyrimidine-pyrimidinone(6-4) dimers. Science 261, 1571–1574. [DOI] [PubMed] [Google Scholar]
- Burgers, O.M.J. (1998). Eukaryotic DNA polymerases in DNA replication and DNA repair. Chromosoma 107, 218–227. [DOI] [PubMed] [Google Scholar]
- DiGiuseppe, J.A., and Dresler, S.L. (1989). Bleomycin-induced DNA repair synthesis in permeable human fibroblasts: Mediation of long-patch and short-patch repair by distinct DNA polymerases. Biochemistry 28, 9515–9520. [DOI] [PubMed] [Google Scholar]
- Eeken, J.C.J., Romeijn, R.J., de Jong, A.W.M., Pastink, A., and Lohman, P.H.M. (2001). Isolation and genetic characterisation of the Drosophila homologue of (SCE)REV3, encoding the catalytic subunit of DNA polymerase ζ. Mutat. Res. 485, 237–253. [DOI] [PubMed] [Google Scholar]
- Esposito, G., Godin, I., Klein, U., Yaspo, M.-L., Cumano, A., and Rajewsky, K. (2000). Disruption of the Rev3l-encoded catalytic subunit in mice of polymerase ζ results in early embryonic lethality. Curr. Biol. 10, 1221–1224. [DOI] [PubMed] [Google Scholar]
- Filée, J., Forterre, P., Sen-Lin, T., and Laurent, J. (2002). Evolution of DNA polymerase families: Evidence for multiple gene exchange between cellular and viral proteins. J. Mol. Biol. 54, 763–773. [DOI] [PubMed] [Google Scholar]
- Friedberg, E.C., Siede, W., and Cooper, A.J. (1991). Cellular responses to DNA damage in yeast. In The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis, and Engineering, J.R. Broach, J.R. Pringle, and E.W. Jones, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 147–192.
- Friedberg, E.C., Walker, G.C., and Siede, W. (1995). DNA Repair and Mutagenesis. (Washington, DC: American Society of Microbiology Press).
- Fujie, M., Kuroiwa, H., Suzuki, T., Kawano, S., and Kuroiwa, T. (1993). Organelle DNA synthesis in the quiescent center of Arabidopsis thaliana (Col.). J. Exp. Bot. 44, 689–693. [Google Scholar]
- Gallego, F., Fleck, O., Li, A., Wyrzykowska, J., and Tinland, B. (2000). AtRAD1, a plant homologue of human and yeast nucleotide excision repair endonucleases, is involved in dark repair of UV damages and recombination. Plant J. 21, 507–518. [DOI] [PubMed] [Google Scholar]
- Gibbs, P.E.M., McGregor, W.G., Maher, V.M., and Nisson, P. (1998). A human homolog of the Saccharomyces cerevisiae REV3 gene which encodes the catalytic subunit of DNA polymerase ζ. Proc. Natl. Acad. Sci. USA 95, 6876–6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratzner, H.G. (1982). Monoclonal antibody to 5-bromo- and 5-indodeoxyuridine: A new reagent for detection of DNA replication. Science 218, 474–475. [DOI] [PubMed] [Google Scholar]
- Grossmann, K.F., Ward, A.M., and Moses, R.E. (2000). Saccharomyces cerevisiae lacking Smn1, Rev3 or Rad51 have a normal S-phase but arrest permanently in G2 after cisplatin treatment. Mutat. Res. 461, 1–13. [DOI] [PubMed] [Google Scholar]
- Harborne, J.B., and Williams, C.A. (2000). Advances in flavonoid research since 1992. Phytochemistry 55, 481–504. [DOI] [PubMed] [Google Scholar]
- Haynes, R.H., and Kunz, B.A. (1981). DNA repair and mutagenesis in yeast. In The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance, J.N. Strathern, E.W. Jones, and J.R. Broach, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 371–414.
- Hidema, J., Kang, S.-H., and Kumagai, T. (1999). Changes in cyclobutyl pyrimidine dimer levels in rice (Oryza sativa L.) growing indoors and outdoors with or without supplemental UV-B radiation. J. Photochem. Photobiol. 52, 1–3. [Google Scholar]
- Holbeck, S.L., and Strathern, J.N. (1997). A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics 147, 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins, M.E., Harlow, G.R., Liu, Z., Shotwell, M.A., Ma, J., and Mount, D.W. (1995). Radiation-sensitive mutant of Arabidopsis thaliana. Genetics 140, 725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C.Z., Yee, J., Mitchell, D.L., and Britt, A.B. (1997). Photorepair mutants of Arabidopsis. Proc. Natl. Acad. Sci. USA 94, 7441–7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R.E., Haracska, L., Prakash, S., and Prakash, L. (2001). Role of DNA polymerase η in the bypass of a (6-4) TT photoproduct. Mol. Cell. Biol. 21, 3559–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R.E., Prakash, S., and Prakash, L. (1999). Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, pol η. Science 283, 1001–1004. [DOI] [PubMed] [Google Scholar]
- Kimura, S., et al. (2002). A novel DNA polymerase homologous to Escherichia coli DNA polymerase I from a higher plant, rice (Oryza sativa L.). Nucleic Acids Res. 30, 1585–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Landry, L.G., Stapleton, A.E., Lim, J., Hoffman, P., Hays, J.B., Walbot, V., and Last, R. (1997). An Arabidopsis photolyase mutant is hypersensitive to ultraviolet-B radiation. Proc. Natl. Acad. Sci. USA 94, 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C.W., and Christensen, R. (1976). UV mutagenesis in radiation-sensitive strains of yeast. Genetics 82, 207–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C.W., and Christensen, R.B. (1979). Ultraviolet-induced reversion of cyc1 alleles in radiation-sensitive strains of yeast. III. rev3 mutant strains. Genetics 92, 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C.W., Gibbs, P.E.M., Murante, R.S., Wang, X.-D., Li, Z., McManus, T.P., McGregor, W.G., Nelson, J.R., Hinkle, D.C., and Maher, V.M. (2000). Roles of DNA polymerase ζ and REV1 protein in eukaryotic mutagenesis and translesion replication. Cold Spring Harbor Symp. Quant. Biol. LXV, 61–69. [DOI] [PubMed]
- Lawrence, C.W., O'Brien, T., and Bond, J. (1984). UV-induced reversion of his4 frameshift mutations in rad6, rev1 and rev3 mutants of yeast. Mol. Gen. Genet. 195, 487–490. [DOI] [PubMed] [Google Scholar]
- Lin, W., Wu, X., and Wang, Z. (1999). A full-length cDNA of hREV3 is predicted to encode DNA polymerase ζ for damage-induced mutagenesis in humans. Mutat. Res. 433, 89–98. [DOI] [PubMed] [Google Scholar]
- Liu, Z., Hall, J.D., and Mount, D.W. (2001). Arabidopsis UVH3 gene is a homolog of the Saccharomyces cerevisiae RAD2 and human XPG DNA repair genes. Plant J. 26, 329–338. [DOI] [PubMed] [Google Scholar]
- Liu, Z., Hossain, G.S., Islas-Osuna, M.A., Mitchell, D.L., and Mount, D.W. (2000). Repair of UV damage in plants by nucleotide excision repair: Arabidopsis UVH1 DNA repair gene is a homolog of Saccharomyces cerevisiae Rad1. Plant J. 21, 519–528. [DOI] [PubMed] [Google Scholar]
- Matsuda, T., Bebenek, K., Masutani, C., Hanaoka, F., and Kunkel, T.A. (2000). Low fidelity DNA synthesis by human DNA polymerase-η. Nature 404, 1011–1013. [DOI] [PubMed] [Google Scholar]
- McDonald, J.P., Levine, A.S., and Woodgate, R. (1997). The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics 147, 1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, A., Christensen, R.B., Alley, J., Beck, A.K., Bernstine, E.G., Lemontt, J.F., and Lawrence, C.W. (1989). REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J. Bacteriol. 171, 5659–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata, Y., Min, T.K., Nakano, T., Asami, T., and Yoshida, S. (2000). Treatment of dark-grown Arabidopsis thaliana with a brassinosteroid-biosynthesis inhibitor, brassinozole, induces some characteristics of light-grown plants. Planta 211, 781–790. [DOI] [PubMed] [Google Scholar]
- Nakajima, S., Sugiyama, M., Iwai, S., Hitomi, K., Otoshi, E., Kim, S.-T., Jiang, C.-Z., Todo, T., Britt, A.B., and Yamamoto, K. (1998). Cloning and characterization of a gene (UVR3) required for photorepair of 6-4 photoproducts in Arabidopsis thaliana. Nucleic Acids Res. 26, 638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, J.R., Lawrence, C.W., and Hinkle, D.C. (1996. a). Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science 272, 1646–1649. [DOI] [PubMed] [Google Scholar]
- Nelson, J.R., Lawrence, C.W., and Hinkle, D.C. (1996. b). Deoxycytidyl transferase activity of yeast REV1 protein. Nature 382, 729–731. [DOI] [PubMed] [Google Scholar]
- Prakash, S., and Prakash, L. (2002). Translesion DNA synthesis in eukaryotes: A one- or two-polymerase affair. Genes Dev. 16, 1872–1883. [DOI] [PubMed] [Google Scholar]
- Quah, S.-K., von Borstel, R.C., and Hastings, P.J. (1980). The origin of spontaneous mutation in Saccharomyces cerevisiae. Genetics 96, 819–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries, G., Heller, W., Puchta, H., Sandermann, H., Seidlitz, H.K., and Hohn, B. (2000). Elevated UV-B radiation reduces genome stability in plants. Nature 406, 98–101. [DOI] [PubMed] [Google Scholar]
- Reuven, N.B., Arad, G., Maor-Shoshani, A., and Livneh, Z. (1999). The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA, and SSB and is specialized for translesion replication. J. Biol. Chem. 274, 31763–31766. [DOI] [PubMed] [Google Scholar]
- Rozema, J., van de Staaij, J., Björn, L.O., and Caldwell, M. (1997). UV-B as an environmental factor in plant life: Stress and regulation. Tree 12, 22–28. [DOI] [PubMed] [Google Scholar]
- Sakai, W., Ishi, C., and Inoue, H. (2002). The upr-1 gene encodes a catalytic subunit of the DNA polymerase ζ which is involved in damage-induced mutagenesis in Neurospora crassa. Mol. Genet. Genomics 267, 401–408. [DOI] [PubMed] [Google Scholar]
- Shikazono, N., Tanaka, A., Watanabe, H., and Tano, S. (2001). Rearrangements of the DNA in carbon ion-induced mutants of Arabidopsis thaliana. Genetics 157, 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal, R.K., Hinkle, D.C., and Lawrence, C.W. (1992). The REV3 gene of Saccharomyces cerevisiae is transcriptionally regulated more like a repair gene than one encoding a DNA polymerase. Mol. Gen. Genet. 236, 17–24. [DOI] [PubMed] [Google Scholar]
- Srivastava, V., Miller, S., and Busbee, D. (1993). Immunofluorescent evaluation of DNA repair synthesis using interactive laser cytometry. Cytometry 14, 144–153. [DOI] [PubMed] [Google Scholar]
- Tanaka, A., Sakamoto, A., Ishigaki, Y., Nikaido, O., Sun, G., Hase, Y., Shikazono, N., Tano, S., and Watanabe, H. (2002). An ultraviolet-B-resistant mutant with enhanced DNA repair in Arabidopsis. Plant Physiol. 129, 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, A., Shikazono, N., Yokota, Y., Watanabe, H., and Tano, S. (1997). Effects of heavy ions on the germination and survival of Arabidopsis thaliana. Int. J. Radiat. Biol. 72, 121–127. [DOI] [PubMed] [Google Scholar]
- Tang, M., Pham, P., Shen, X., Taylor, J.-S., O'Donnell, M., Woodgate, R., and Goodman, M.F. (2000). Roles of E. coli DNA polymerase IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature 404, 1014–1018. [DOI] [PubMed] [Google Scholar]
- Tang, M., Shen, X., Frank, E.G., O'Donnell, M., Woodgate, R., and Goodman, M.F. (1999). UmuD′2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl. Acad. Sci. USA 96, 8919–8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sloun, P.P.H., Romeijin, R.J., and Eeken, J.C.J. (1999). Molecular cloning, expression and chromosomal localisation of the mouse Rev3l gene, encoding the catalytic subunit of polymerase ζ. Mutat. Res. 433, 109–116. [DOI] [PubMed] [Google Scholar]
- Wagner, J., Gruz, P., Kim, S.-R., Yamada, M., Matsui, K., Fuchs, R.P.P., and Nohmi, T. (1999). The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol. Cell 4, 281–286. [DOI] [PubMed] [Google Scholar]
- Wittschieben, J., Shivji, M.K.K., Lalani, E., Jacobs, M.A., Marini, F., Gearhart, P.J., Rosewell, I., Stamp, G., and Wood, R.D. (2000). Disruption of developmentally regulated Rev3l gene causes embryonic lethality. Curr. Biol. 10, 1217–1220. [DOI] [PubMed] [Google Scholar]
- Xu, H., Swoboda, I., Bhalla, P.L., Sijbers, A.M., Zhao, C., Ong, E.-K., Hoeijmakers, J.H.J., and Singh, M.B. (1998). Plant homologue of human excision repair gene ERCC1 points to conservation of DNA repair mechanisms. Plant J. 13, 823–829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.