Abstract
SURE (sugar responsive) is a cis element in plant sugar signaling. The SURE element was reported first for potato, in which it confers sugar responsiveness to the patatin promoter. A SURE binding transcription factor has not been isolated. We have isolated a transcription factor cDNA from barley and purified the corresponding protein. The transcription factor, SUSIBA2 (sugar signaling in barley), belongs to the WRKY proteins and was shown to bind to SURE and W-box elements but not to the SP8a element in the iso1 promoter. Nuclear localization of SUSIBA2 was demonstrated in a transient assay system with a SUSIBA2:green fluorescent protein fusion protein. Exploiting the novel transcription factor oligodeoxynucleotide decoy strategy with transformed barley endosperm provided experimental evidence for the importance of the SURE elements in iso1 transcription. Antibodies against SUSIBA2 were produced, and the expression pattern for susiba2 was determined at the RNA and protein levels. It was found that susiba2 is expressed in endosperm but not in leaves. Transcription of susiba2 is sugar inducible, and ectopic susiba2 expression was obtained in sugar-treated leaves. Likewise, binding to SURE elements was observed for nuclear extracts from sugar-treated but not from control barley leaves. The temporal expression of susiba2 in barley endosperm followed that of iso1 and endogenous sucrose levels, with a peak at ∼12 days after pollination. Our data indicate that SUSIBA2 binds to the SURE elements in the barley iso1 promoter as an activator. Furthermore, they show that SUSIBA2 is a regulatory transcription factor in starch synthesis and demonstrate the involvement of a WRKY protein in carbohydrate anabolism. Orthologs to SUSIBA2 were isolated from rice and wheat endosperm.
INTRODUCTION
Development of the cereal seed is orchestrated by the coordinated activities of a large number of genes that encode metabolic and regulatory enzymes as well as other proteins (Olsen, 2001). This results in a triploid endosperm, the embryo, pericarp, seed coat, and other tissues of the mature grain. The endosperm structure consists of two tissues, the interior starch-filled endosperm and the outer epidermal layer called the aleurone. Starch, which is a mixture of amylopectin (a heavily branched polyglucan) and amylose (a mostly linear polyglucan), is deposited in the endosperm as granules. The synthesis and deposition of starch in the endosperm depend on enzymes such as ADP–glucose pyrophosphorylase, starch synthases, starch-branching enzymes (SBEs), and starch-debranching enzymes. Most, if not all, of these enzymes exist in two or more isoforms (for recent reviews on starch biosynthesis, see Ball et al., 1998; Buléon et al., 1998; Myers et al., 2000; Smith, 2001; Nakamura, 2002). Starch-debranching enzymes are grouped into two distinct classes, isoamylase and pullulanase, each with different isoforms (Nakamura, 1996). Isoamylase (EC 3.2.1.68) is an essential enzyme in amylopectin synthesis. However, the precise role of the enzyme in this process is not clear, and different models have been proposed (Ball et al., 1998; Smith, 2001; Nakamura, 2002).
It has been reported that the expression of starch synthesis genes, such as starch synthase in potato (Visser et al., 1991); ADP–glucose pyrophosphorylase in potato (Müller-Röber et al., 1990), sweet potato (Bae and Liu, 1997), Arabidopsis (Rook et al., 2001), and tomato (Li et al., 2002); SBE in potato (Kossman et al., 1991), maize (Kim and Guiltinan, 1999), and Arabidopsis (Khoshnoodi et al., 1998); and isoamylase in barley (Sun et al., 1999), is sugar inducible. In contrast to the situation in bacteria, yeast, and mammals, in which sugar signaling cascades have been studied extensively, the sugar signaling transduction pathways in plants are poorly understood (Rolland et al., 2002). Generally, in higher plants, high sugar levels stimulate the expression of genes involved in sink function, such as growth, storage of proteins, and the biosynthesis of starch and other carbohydrates, whereas low sugar levels promote the photosynthesis and mobilization of energy reserves, such as the breakdown of storage starch or lipids.
Sugar signaling can be dissected into three steps: sugar sensing, signal transduction, and target gene expression. However, this division is confused by the dual function of sugars as nutrients and signaling molecules, and by the interaction (in plants and animals) between sugar signaling and hormonal networks. In plants, this complexity is increased further by the vital role of sugar production through photosynthesis. Hexoses, sucrose, and trehalose might serve as elicitors of plant sugar signaling (Goddijn and Smeekens, 1998; Rolland et al., 2002). Hexokinase, sucrose, glucose transporters, and various sugar receptors have been proposed as components of the sugar-sensing machinery (Sheen et al., 1999; Smeekens, 2000; Rolland et al., 2002).
The Ser/Thr protein kinase Snf1 is a central participant in yeast sugar signaling (Carlson, 1999). Snf1 phosphorylates downstream components and also is itself activated by phosphorylation. Snf-related protein kinases (SnRKs) are found in yeast, mammals, and plants, in which they participate in a large number of regulatory functions (Halford and Hardie, 1998; Hardie et al., 1998). There is evidence that some plant SnRKs are functionally homologous with Snf1 in plant sugar signaling, although the exact nature of their responses to sugars remains to be clarified (Rolland et al., 2002). Other players implicated in sugar signaling transduction pathways in plants are sugar metabolites, 14-3-3 proteins, trehalose-6-phosphate, and Ca2+ (Rolland et al., 2002).
Little is known about the cis and trans factors that mediate the final steps in plant sugar signaling. To date, five different types of cis elements have been identified in sugar-regulated plant promoters: the SURE (sugar-responsive) (Grierson et al., 1994), SP8 (Ishiguro and Nakamura, 1994), TGGACGG (Maeo et al., 2001), G-box (Giuliano et al., 1988), and B-box (Grierson et al., 1994; Zourelidou et al., 2002) elements. Only two putative transcription factors, SPF1 and STK, with relevance to plant sugar signaling have been isolated. SPF1 binds to the SP8 sequence, in which it functions as a repressor (Ishiguro and Nakamura, 1994), and STK binds to the B-box as an activator (Zourelidou et al., 2002).
We are interested in the endosperm-specific expression of the sbeIIb (encoding SBEIIb) and iso1 (encoding isoamylase1) genes during barley seed development. We have shown previously that barley iso1 harbors an SP8a element and that it contributes to the endosperm specificity of iso1 expression by recruiting a repressor in nonexpressing tissues (Sun et al., 1999). Similarly, the endosperm-specific expression of barley sbeIIb (Sun et al., 1998) is determined partly by a repressor binding to the B-box-like (Bbl) element in the second intron of the gene (Ahlandsberg et al., 2002b). In the work described here, we have isolated a transcription factor, SUSIBA2 (sugar signaling in barley), and examined its interaction with the iso1 promoter. We show that SUSIBA2 represents a transcription factor involved in the regulation of starch synthesis.
RESULTS
Isolation and Characterization of the susiba2 cDNA
The sugar responsiveness of the sporamin and β-amylase genes in sweet potato was reported more than a decade ago (Hattori et al., 1990), and two promoter sequences, SP8a and SP8b, were implicated as cis elements in the coordinated regulation of the sporamin multigene family and the β-amylase gene (Ishiguro and Nakamura, 1992). Subsequently, a cDNA that encodes a transcription factor named SPF1 was isolated from sweet potato, and it was reported that SPF1 binds to the SP8a and SP8b promoter elements of the β-amylase and sporamin genes of tuberous roots (Ishiguro and Nakamura, 1994). Later, clones for SPF1-like proteins were isolated from cucumber (Kim et al., 1997) and parsley (Rushton et al., 1996).
To continue our studies of the sugar-mediated regulation of the barley iso1 gene and the involvement of the SP8a element (Sun et al., 1999), we set out to isolate a cDNA clone for the barley SPF1 ortholog. Based on the alignment of the sweet potato, cucumber, and parsley SPF1 cDNA sequences, primers were designed for reverse transcriptase–mediated (RT) PCR amplification of barley cDNA using total RNA isolated from developing barley endosperm at 9 days after pollination. Only one PCR product with the expected size was obtained (data not shown). The amplified product was used as a probe to screen an endosperm cDNA library from 10 days after pollination and a leaf library. From screening of 106 plaque-forming units, 14 positive clones were recovered. All positive clones were subjected to subcloning, restriction mapping, and sequence analysis. Restriction mapping and sequence analysis revealed that the clones grouped into three distinct classes. They were designated susiba1, susiba2, and susiba3, respectively.
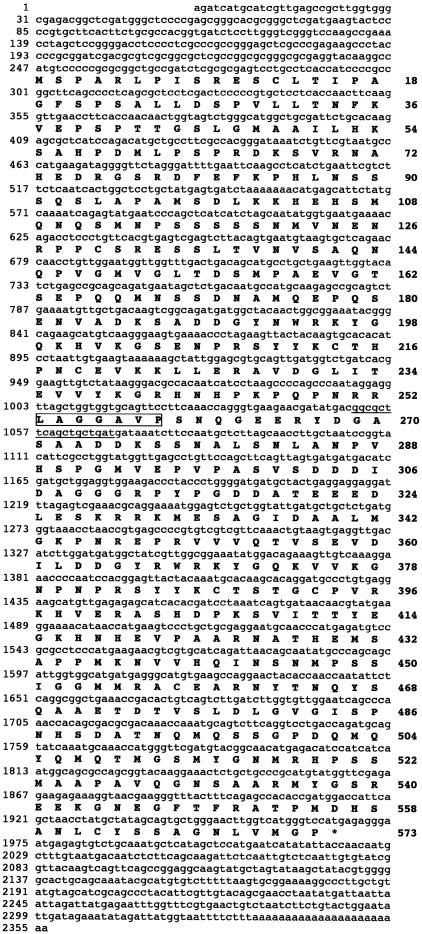
From group 2, one representative clone was sequenced completely on both DNA strands. The length of the susiba2 cDNA is 2355 nucleotides, and the open reading frame starts at position 247 and ends at position 1966 (Figure 1). The cDNA encodes a 573–amino acid polypeptide, SUSIBA2, with a calculated molecular mass of 62.2 kD, and it has 5′ and 3′ untranslated regions and a poly(A+) tail sequence. These findings, together with the results from RNA gel blot analyses (see below), suggest that the susiba2 cDNA is a full-length clone. One interesting feature of susiba2 is that an 18-nucleotide sequence between nucleotides 1051 and 1068 (Figure 1) is 100% complementary to the gene for the human insulin-like growth factor1 (IGF1) receptor (nucleotides 173 to 190).
Figure 1.
Structure of the susiba2 cDNA.
The 18-nucleotide sequence complementary to the human IGF1 receptor gene is underlined. The amino acid sequence of the tryptic fragment obtained by microsequencing from the overproduced SUSIBA2 (see Figure 3) is indicated with an open box.
SUSIBA2 Belongs to the WRKY Superfamily of Transcription Factors
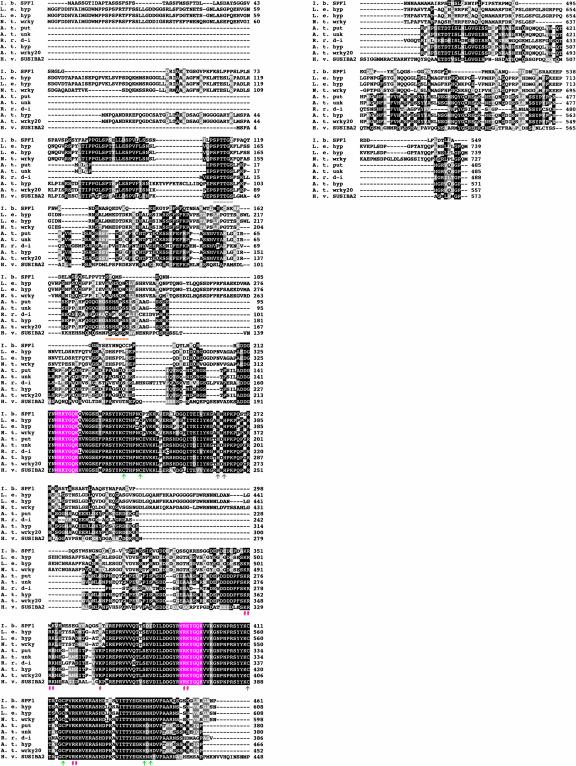
The deduced amino acid sequence of SUSIBA2 was used in a BLAST (Basic Local Alignment Search Tool) search of the National Center for Biotechnology Information (NCBI) databases. Alignment of the 9 top-scoring matches with the SUSIBA2 sequence demonstrated that all 10 proteins are highly similar and that SUSIBA2 belongs to group 1 of the WRKY superfamily of plant transcription factors (Figure 2). The two WRKY domains (Eulgem et al., 2000) with their zinc finger motifs are highly conserved in SUSIBA2. Also, the N-terminal Ser- and Thr-rich regions that are present in many WRKY proteins are found in SUSIBA2. The function of these regions is not known but likely involves regulation. Ser- and Thr-rich regions are characteristic of activation domains in transcription factors.
Figure 2.
Analysis of the SUSIBA2 Primary Structure.
The amino acid sequence of SUSIBA2 was aligned with those of the nine closest matching proteins from a BLAST search. Identical amino acids are shown in black boxes, and similar amino acids are shown in gray boxes. The WRKYGQK peptide stretch is shown in pink. Putative nuclear localization signals are indicated with orange number signs (#) under the sequences. Ser- and Thr-rich regions are underlined in orange. The zinc finger–like
motifs in the two WRKY domains are indicated with green vertical arrows under the sequences. A. t. hyp, Arabidopsis hypothetical protein; A. t. put, Arabidopsis putative protein; A. t. unk, Arabidopsis unknown protein; A. t. wrky20, Arabidopsis WRKY transcription factor 20; H. v. SUSIBA2, barley SUSIBA2; I. b. SPF1, sweet potato SPF1; L. e. hyp, tomato hypothetical proteins; N. t. wrky, tobacco WRKY protein; R. r. d-i, white broom drought-induced protein.
As would be expected from transcription factors, WRKY proteins contain nuclear localization signals (NLSs). The two most common NLSs are monopartite signals, which are short stretches enriched in basic amino acids, and bipartite signals, which are composed of two short basic stretches separated by a spacer (Merkle, 2001). The monopartite NLS depicted for WRKY proteins is conserved in SUSIBA2 (amino acids 328 to 331; Figure 2). However, closer inspection of the sequences suggests that the monopartite NLS in SUSIBA2 and other WRKY proteins also might constitute a bipartite NLS close to the consensus motif, KR-(24-74)-R..RK, for early auxin-inducible proteins (Abel and Theologis, 1995). One putative bipartite NLS in SUSIBA2 is KR-(66)-RK (amino acids 328 to 397). The other possibility is KR-(37)-R.RK (amino acids 328 to 370) and involves the WRKYGQK sequence itself in the C-terminal WRKY domain. The function of the WRKYGQK heptapeptide is unknown, and we cannot exclude the possibility that it participates in the nuclear localization process, although a bipartite NLS arrangement for the N-terminal WRKYGQK sequence was not found.
A different kind of NLS reported for the Arabidopsis WRKY protein, AtWRKY6 (Robatzek and Somssich, 2001), is not present in SUSIBA2, although one of the Lys residues implicated in this motif is conserved in SUSIBA2 (corresponding to Lys-344) and some other WRKY proteins (Figure 2). The most distinguishing feature of SUSIBA2 relative to other WRKY proteins is the 37–amino acid insertion after the C-terminal WRKY domain (amino acids 435 to 471; Figure 2). A BLAST search with the insertion sequence did not show any matches in the NCBI databases (see below), suggesting that SUSIBA2 is a novel type of WRKY protein.
Overproduction of SUSIBA2 in Escherichia coli
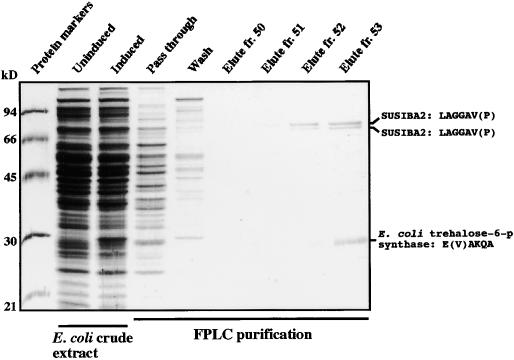
The susiba2 cDNA was inserted into a pET expression vector and overexpressed in E. coli after isopropylthio-β-galactoside induction. The overproduced protein was purified by one-step fast protein liquid chromatography on a His tag affinity column. Results from a typical overproduction and purification procedure are shown in Figure 3. The purified polypeptides were used for microsequencing, activity characterization, and antibody production.
Figure 3.
Purification of SUSIBA2.
His-tagged SUSIBA2 was subjected to fast protein liquid chromatography (FPLC) purification on a Ni affinity column after overexpression of the susiba2 cDNA construct in E. coli. Amino acid sequences obtained by microsequencing of tryptic fragments are shown at right. Molecular masses (in kilodaltons) are indicated at left. fr., fraction.
The purified protein fraction contained three polypeptides. They were separated by SDS-PAGE and subjected to in situ trypsin digestion. The digested fragments from each polypeptide were separated by HPLC and applied to mass spectrometry analysis. The upper two bands corresponded to polypeptides with molecular masses of ∼70 and 68 kD and with the identical sequence LAGGAVP. This sequence is found in the deduced amino acid sequence of SUSIBA2 (Figure 1; amino acids 235 to 241). The lower band corresponded to a 28-kD polypeptide with the sequence EIAKQA. This sequence matches E. coli trehalose-6-phosphate synthase (amino acids 234 to 239). The two SUSIBA2 polypeptides were extracted and used for the production of polyclonal antibodies.
The DNA Binding Activity of SUSIBA2
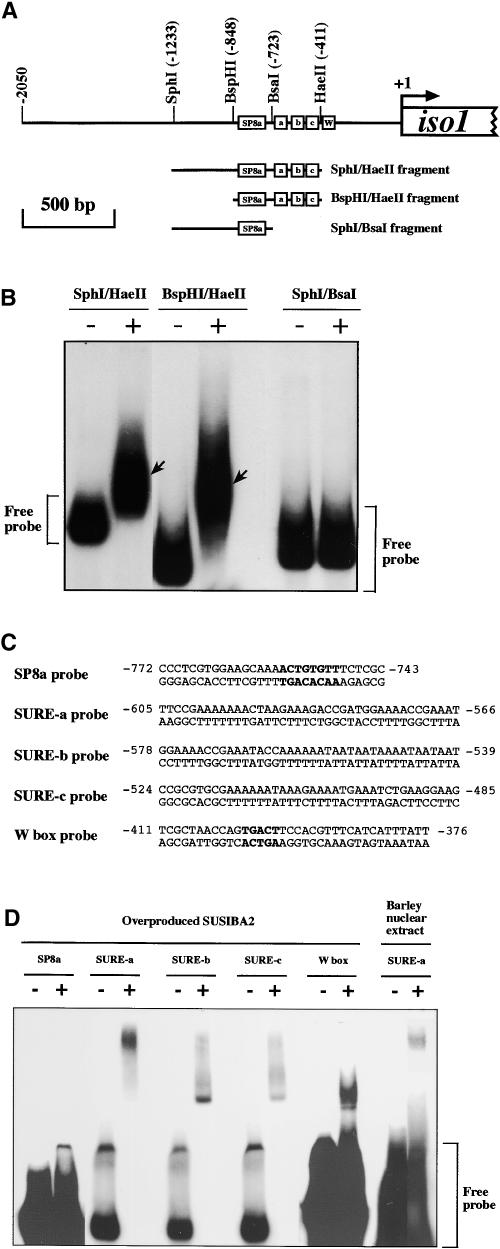
As an initial test of the DNA binding activity of SUSIBA2, we performed electrophoretic mobility shift assays (EMSAs) with three overlapping nucleotide fragments from the barley iso1 promoter, all containing the SP8a sequence (Figures 4A and 4B). The 822-bp SphI-HaeII fragment, encompassing SP8a and flanking sequences, and the 437-bp BspHI-HaeII fragment, containing SP8a and downstream flanking sequence, formed prominent DNA-protein complexes with the purified SUSIBA2 fraction. To our surprise, however, the 510-bp SphI-BsaI fragment, containing SP8a and upstream flanking sequence, bound very poorly to SUSIBA2. We concluded that the target for SUSIBA2 in the iso1 promoter is not SP8a but one or more sites downstream of the SP8a element. From further analysis of the segment downstream of SP8a in the BspHI-HaeII fragment, we found that it harbors three A-rich sequence segments, 5′-AAAACTAAGAAAGACCGATGGAAAA-3′, 5′-AATACCAAAAAA- TAATAATAAAA-3′, and 5′-CGAAAAAATAAAGAAAATGAAAT-3′ (nucleotides −597 to −573, −568 to −546, and −517 to −495 relative to the transcriptional start site, respectively), each of which shares a high degree of identity with the SURE element, as first reported by Grierson et al. (1994) from work on the potato class-1 patatin promoter.
Figure 4.
Binding of SUSIBA2 to DNA Sequences Containing the SP8a, SURE, and W-Box Elements.
(A) A portion of the iso1 promoter and restriction fragments spanning the SP8a element and flanking sequences. The positions of the three SURE elements (a, b, and c) and the W-box (W) are indicated.
(B) EMSA using the different SP8a-containing restriction fragments incubated in the presence (+) or absence (−) of SUSIBA2. The positions of the DNA-SUSIBA2 complexes and free probes are indicated.
(C) The sequences of five oligonucleotides containing the SP8a, SURE-a, SURE-b, SURE-c, and W-box sequences. The SP8a and W-box sequences are shown in boldface.
(D) EMSA using the different oligonucleotides incubated in the presence (+) or absence (−) of overproduced SUSIBA2 or barley nuclear extract. The position of the free probe is indicated.
To follow up on this observation, we performed EMSAs with oligonucleotides consisting of the iso1 SURE sequences, referred to as SURE-a, SURE-b, and SURE-c for the upstream, middle, and downstream sequences, respectively (Figures 4C and 4D). As is evident in Figure 4D, SUSIBA2 bound to all three SURE probes, whereas, again, binding to the SP8a probe was negligible. The strongest binding activity was observed for the SURE-a sequence, which yielded a major DNA-protein complex of a low relative mobility (Mr). The same complex was obtained with the SURE-b and SURE-c probes, although only as a minor component; the major complex with the SURE-b and SURE-c probes had a higher Mr. This high Mr complex also could be discerned as a faint band with the SURE-a probe. With the SURE-b and SURE-c probes, additional complexes of intermediate Mr were seen. When the SURE-a probe was incubated with a nuclear extract from barley endosperm, a complex was formed with the same Mr as with the overproduced SUSIBA2, suggesting that the complex produced with the endogenous SUSIBA2 was the same as that produced with the purified protein. Because SUSIBA2 is a WRKY protein, it also would be expected to bind to the W-box, the highly conserved binding site for WRKY proteins (Eulgem et al., 2000). A sequence identical to the consensus sequence for the W-box, TGACT (nucleotides −400 to −396), was located in the iso1 promoter, and the iso1 W-box sequence was included in the binding assay (Figures 4C and 4D). With the W-box probe, two DNA-SUSIBA2 complexes could be discerned. Thus, the results support the conclusion that SUSIBA2 does not bind to the SP8a element and show that, instead, it interacts with the SURE and W-box elements.
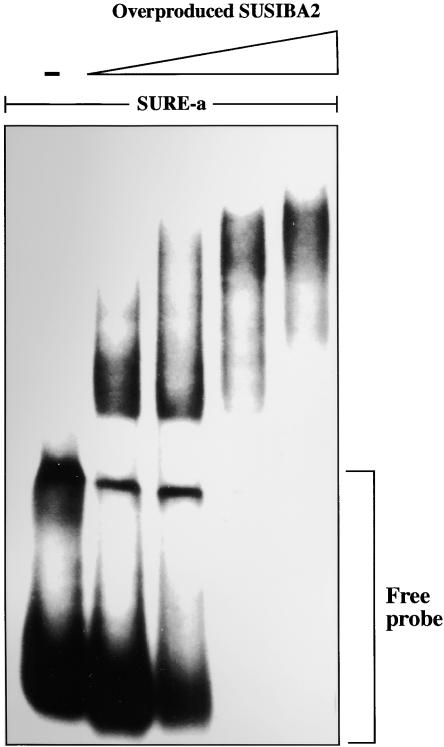
The observation that the SURE-SUSIBA2 complexes appeared with different Mr indicated that SUSIBA2 might exist in two or more oligomeric states. This possibility was illustrated further by a titration EMSA in which the SURE-a probe was incubated with increasing amounts of SUSIBA2 protein (Figure 5). As the concentration of SUSIBA2 increased from 0.01 to 0.08 μM, the migration of the formed complex shifted from high to low Mr. The same phenomenon was observed for the SURE-b and SURE-c probes (data not shown).
Figure 5.
Titration of SUSIBA2 Oligomerization.
The SURE-a probe was used for EMSA in the absence (−) or presence of different concentrations (0.035, 0.07, 0.14, and 0.28 μg, in 50-μL reactions, corresponding to 0.01, 0.02, 0.04 and 0.08 μM proteins, respectively) of overproduced SUSIBA2. The position of the free probe is indicated.
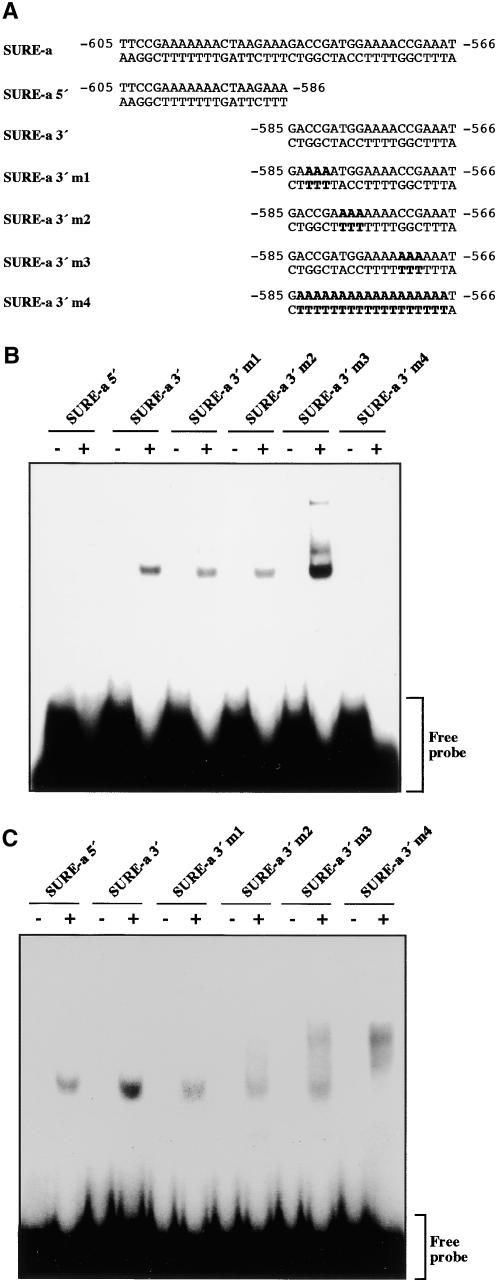
Having demonstrated that SUSIBA2 binds to the SURE element, we were interested in further exploring the interaction between SUSIBA2 and the SURE sequence. To this end, we constructed engineered versions of the SURE-a element in the barley iso1 promoter and tested their affinity for the SUSIBA2 protein (Figures 6A and 6B). Our first observation was that the binding activity could be localized to the 20-nucleotide 3′ end of the SURE-a sequence (SURE-a 3′). Subsequent modifications of the 3′ end revealed that, although the SURE element is an A-rich sequence, increasing the number of consecutive A nucleotides in the upstream portion of SURE-a 3′ (SURE-a 3′ m1 and SURE-a 3′ m2) decreased its affinity for SUSIBA2. Converting most of SURE-a 3′ to a poly(A) stretch (SURE-a 3′ m4) resulted in the complete loss of binding activity. Interestingly, when the number of consecutive A nucleotides in the downstream region of SURE-a 3′ was increased (SURE-a 3′ m3), the binding activity was enhanced significantly and additional complexes of higher Mr were produced. When the EMSA was performed with nuclear extracts from barley endosperm (Figures 6A and 6C), the effects of the m1, m2, and m3 mutations in the SURE-a 3′ probe were similar to those with the purified SUSIBA2. One difference was that, with the nuclear extract, the SURE-a 3′ m3 probe also exhibited decreased binding activity compared with SURE-a 3′, albeit to a lower extent than did the SURE-a 3′ m1 and SURE-a 3′ m2 probes. Contrary to the situation with purified SUSIBA2, the nuclear extract displayed binding activity to the 5′ end of the SURE-a sequence and to the SURE-a 3′ m4 probe.
Figure 6.
EMSA with Engineered SURE-a Oligonucleotides and Overproduced SUSIBA2.
(A) Sequences of SURE-a, the SURE-a 5′ probe (SURE-a 5′), the SURE-a 3′ probe (SURE-a 3′), and the engineered SURE-a 3′ probes (SURE-a 3′ m1 to m4). Engineered nucleotides are shown in boldface.
(B) EMSA with the different SURE-a probes in the presence (+) or absence (−) of overproduced SUSIBA2. The position of the free probe is indicated.
(C) EMSA with the different SURE-a probes in the presence (+) or absence (−) of nuclear extracts from barley endosperm. The position of the free probe is indicated.
To further demonstrate that the complex formed between the nuclear extracts and the SURE element contained SUSIBA2, we attempted an antibody supershift analysis. However, our antiserum was raised against a denatured SUSIBA2 protein, and the antibodies reacted poorly with native SUSIBA2. As an alternative means to address this issue, we examined the composition of the DNA-protein complex after SDS-PAGE. A narrow strip of the labeled complex between SURE-a 5′ and a nuclear extract on the polyacrylamide gel was excised. The proteins were eluted and separated on an SDS gel, followed by protein gel blot analysis with the SUSIBA2 antiserum and preimmune serum. Because the overproduced SUSIBA2 protein used for antibody production contained a His tag, we also included a control in which the protein gel blot was probed with His tag antibodies. The SUSIBA2 antibodies produced a double band corresponding to 68 to 66 kD, the expected molecular mass for endogenous SUSIBA2 (Figure 7). No bands were detected with the preimmune serum, whereas the His tag antibodies reacted with a polypeptide of ∼96 kD.
Figure 7.
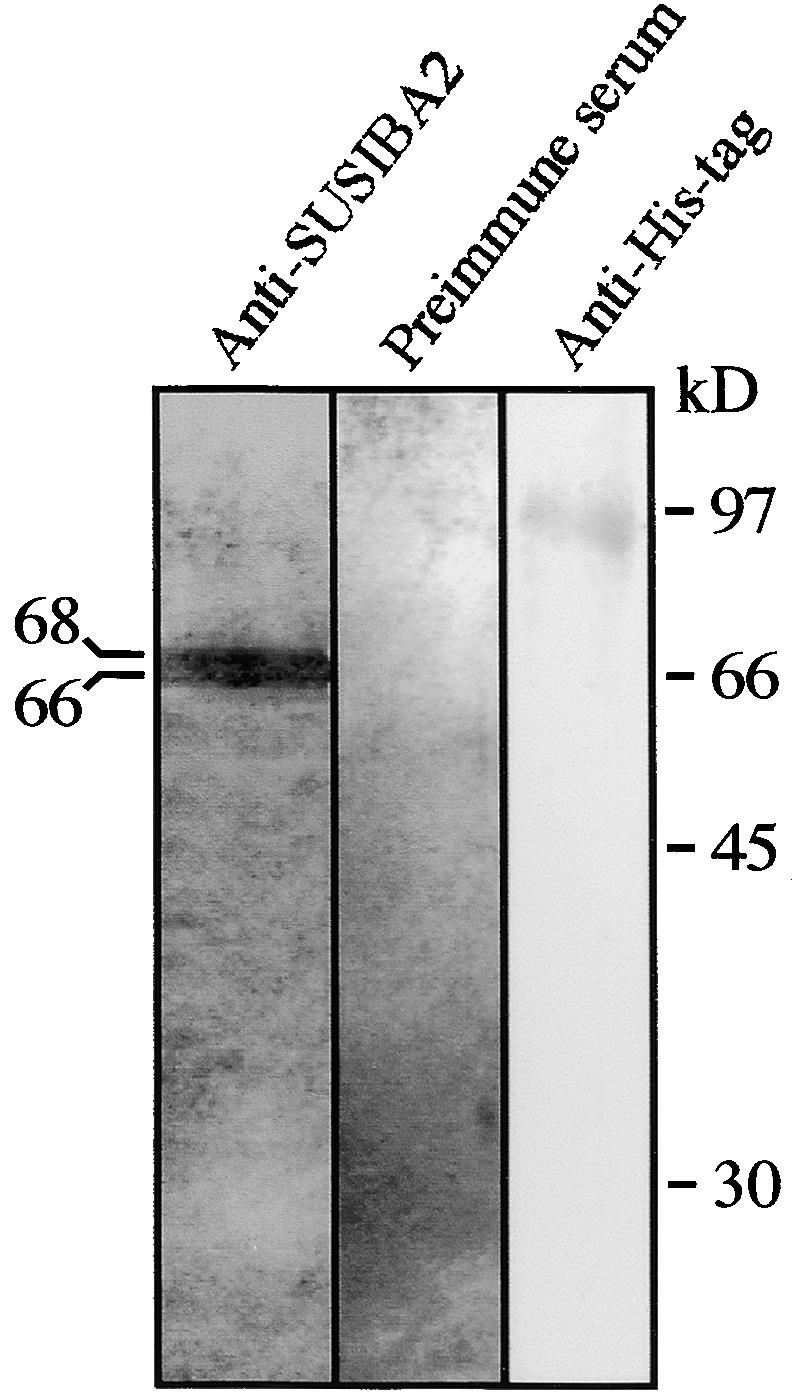
Protein Analysis of the Complex Formed between the SURE Element and Nuclear Extracts.
EMSA with the SURE-a 5′ probe and a nuclear extract from barley endosperm was performed, and the complex produced was subjected to SDS-PAGE and protein gel blot analysis with antibodies against SUSIBA2 (anti-SUSIBA2) serum before immunization with SUSIBA2 (preimmune serum) or antibodies against a 6-His tag (anti-His tag). Molecular masses (in kilodaltons) are indicated.
Expression of susiba2 and iso1 Correlates with Sucrose Levels
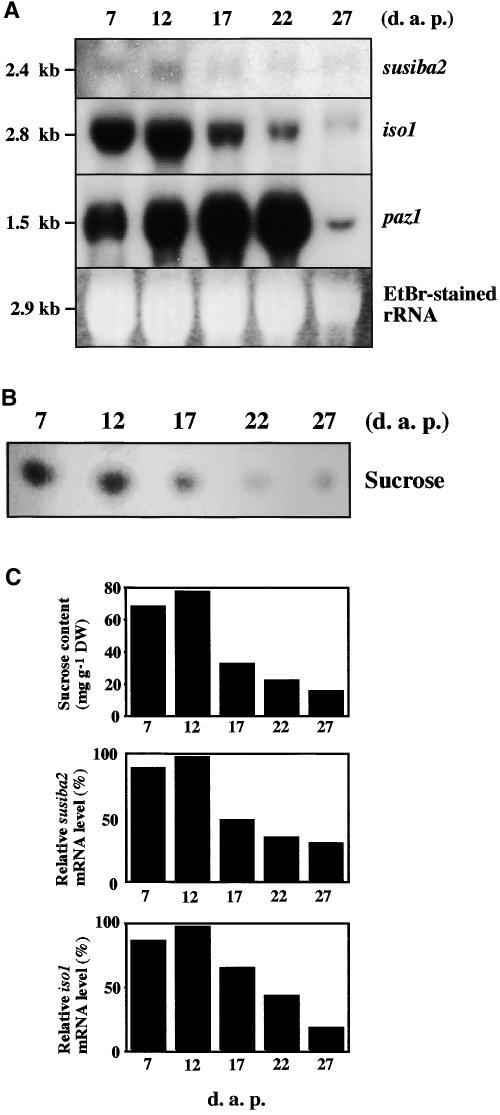
In previous investigations, we showed that the expression of the barley iso1 gene is sugar inducible (Sun et al., 1999). In light of different reports suggesting the involvement of the SURE element in sugar signaling (Kim et al., 1991; Rhee and Staswick, 1992; Grierson et al., 1994; Fu et al., 1995; Mita et al., 1995; Kim and Guiltinan, 1999; Li et al., 2002) and the results presented here showing the SURE binding activity of the SUSIBA2 transcription factor, it was of interest to study the expression pattern of the susiba2 gene. For the examination of temporal expression, barley endosperms were collected at 7 to 27 days after pollination and total RNA was isolated. RNA gel blot analy- sis was performed with susiba2- or iso1-specific probes or with a PCR-amplified probe for the paz1 gene, which encodes the storage protein Z (Sørensen et al., 1989) and which was included for comparison. The expression level for susiba2 was low compared with that for iso1, but the patterns were similar, with a peak at ∼12 days after pollination (Figure 8A). A similar time course was obtained for the barley sbeIIa and sbeIIb genes (Sun et al., 1998). Expression of the paz1 gene increased up to or beyond 22 days after pollination and then declined sharply, consistent with the results reported by Sørensen et al. (1989). To study the correlation between sucrose and susiba2 expression, we monitored the endogenous sucrose concentrations during endosperm development using an enzymatic assay and thin layer chromatography. The sucrose concentrations were determined in seeds from the same batches used for transcript analysis. A comparison between the temporal expression patterns for susiba2 and iso1 transcript accumulation and sucrose levels showed a good correlation (Figures 8B and 8C).
Figure 8.
Temporal Expression Profiles for susiba2 and iso1.
(A) RNA gel blot analysis of susiba2 and iso1 gene activity during barley endosperm development. Total RNA was isolated from barley endosperm on different days after pollination (d.a.p.) and used for RNA gel blot analysis. The blots were hybridized with probes specific for susiba2, iso1, or paz1. The sizes for the hybridizing fragments and ethidium bromide (EtBr)–stained rRNA are shown.
(B) Thin layer chromatography analysis of endogenous sucrose levels in developing barley endosperm from different days after pollination. Extracts from 0.2 mg dry weight of developing endosperm from each time point were applied to thin layer chromatography analysis.
(C) Correlation between endogenous sucrose content and relative susiba2 and iso1 mRNA levels in developing endosperm. Sucrose content was determined by an enzymatic assay and is presented in milligrams per gram dry weight of tissue (DW). Relative mRNA levels (in percentage of the levels at 12 days after pollination) were determined from the RNA gel blots in (A) using NIH Image software.
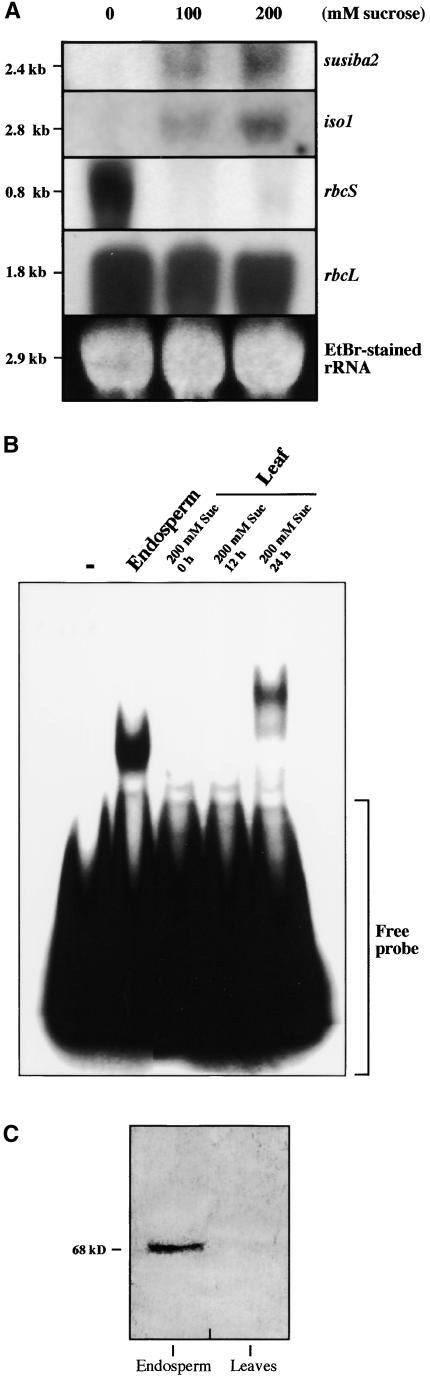
No susiba2 transcripts were detected in untreated barley leaves. However, ectopic susiba2 expression in leaves was achieved after supplying exogenous sugar (Figure 9A). The same was true for iso1, in agreement with previous results (Sun et al., 1999). As a comparison, the photosynthesis genes rbcS and rbcL, which encode the small and large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase, respectively, were expressed abundantly in control leaves (0 mM sucrose). The nuclear rbcS was downregulated dramatically by sucrose, whereas the plastidic rbcL was not affected notably. The sugar-induced expression levels for susiba2 and iso1 in leaves were quite low compared with those observed in endosperm and were detected only after prolonged exposure. The expression studies were extended to the protein level, using both EMSA with nuclear extracts (Figure 9B) and protein gel blot analyses with the SUSIBA2 antibody (Figure 9C). Consistent with the transcript assessments, nuclear extracts from barley leaves gave rise to a complex with the SURE-a probe after but not before sugar treatment. The immunological examination showed that SUSIBA2 was detected in endosperm but not in leaves. We failed to obtain satisfactory protein gel blot results with extracts from sugar-treated leaves.
Figure 9.
Tissue-Specific and Ectopic Expression of susiba2 and iso1 in Barley Leaves.
(A) Barley leaves were treated with exogenous sucrose at the concentrations indicated. Total RNA was isolated and hybridized to probes specific for susiba2, iso1, rbcS, and rbcL. The sizes for the hybridizing molecules and ethidium bromide (EtBr)–stained rRNA are shown.
(B) EMSA was performed with the SURE-a 3′ probe in the absence (−) or presence of barley nuclear extracts isolated from endosperm or leaves. The leaves were treated with sucrose and harvested at the times indicated. The position of the free probe is indicated.
(C) Protein gel blot analysis of susiba2 expression in barley endosperm and leaves.
In Vivo Analysis of Transformed Barley Endosperm Demonstrates the Relevance of the SURE Element for iso1 Promoter Activity
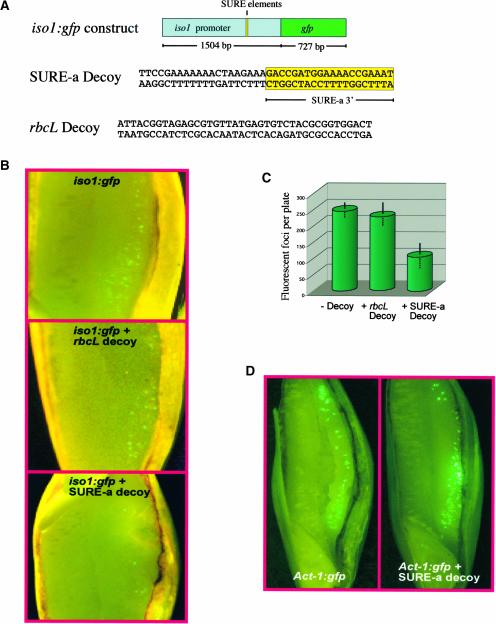
To test the function of the SURE element in the regulation of iso1 expression, we performed transient assays in which the transcription of a chimeric iso1 reporter construct was studied in transformed barley endosperm. In this experiment, the entire (1.5 kb) iso1 promoter was fused to the green fluorescent protein (gfp) reporter gene. The exploitation of the GFP reporter system for the analysis of promoter activity has been used successfully in transformed barley embryos (Ahlandsberg et al., 1999, 2002b). Because the iso1 gene is not expressed in embryos (Sun et al., 1999), we needed first to establish a protocol for the transformation of barley endosperm. This was accomplished recently (S. Palmqvist, S. Ahlandsberg, C. Sun, and C. Jansson, unpublished results), allowing us to perform transient assays with gfp reporter constructs also in transformed barley endosperm.
The activity of the iso1 promoter, manifested as GFP fluorescence, was followed in transformed barley endosperm in the presence or absence of a transcription factor oligodeoxynucleotide (ODN) decoy containing the SURE-a sequence (Figure 10A). The ODN decoy approach involves flooding the cells with enough double-stranded decoy to compete for binding of transcription factors with their consensus sequences in target genes. If present in high enough concentrations, these decoys can negate the ability of the transcription factor to regulate gene expression. The transcription factor ODN technology is used widely in medical investigations (Mann and Dzau, 2000). Here, we chose to adopt the strategy for our studies on barley endosperm. As a negative control, we constructed a decoy containing a sequence from the rbcL gene. As demonstrated in Figure 10B, the presence of the SURE-a decoy in the endosperm cells blocked most, although not all, iso1 promoter activity, whereas the rbcL decoy had no obvious effect. The results of nine independent experiments are summarized in Figure 10C. To affirm that the SURE-a decoy did not interfere nonspecifically with the transcriptional machinery, we fused the gfp gene downstream of the constitutive intronic actin promoter (Act-1) from rice (McElroy et al., 1990, 1991; Ahlandsberg et al., 1999) and monitored its activity in transformed barley endosperm (Figure 10D). No inhibitory effect on GFP fluorescence was detected in the presence of the SURE-a decoy.
Figure 10.
Effects of the SURE-a ODN Decoy on iso1 Expression in Transformed Barley Endosperm.
(A) Scheme of the iso1:gfp construct and sequences of the SURE-a and rbcL ODN decoys.
(B) Transient expression assays of GFP fluorescence in transformed barley endosperm cells. Barley endosperm cells were transformed by microprojectile bombardment with the iso1:gfp construct in the absence (top) or presence of the rbcL (middle) or SURE-a (bottom) ODN decoy. For cotransformation, the iso1:gfp construct and the ODN decoys were mixed at a molar ratio of 1:100.
(C) Average number (means ± se) of fluorescent foci per plate from nine independent transformation events. An F test indicated significant difference between means for the absence and presence of the SURE-a decoy (P = 0.0017) and no significant difference between means for the absence and presence of the rbcL decoy (P = 0.62).
(D) Transient expression assays of GFP fluorescence in transformed barley endosperm cells. Barley endosperm cells were transformed by microprojectile bombardment with the Act-1:gfp construct in the absence (left) or presence (right) of the SURE-a ODN decoy. For cotransformation, the Act-1:gfp construct and the ODN decoy were mixed at a molar ratio of 1:100.
Nuclear Localization of SUSIBA2
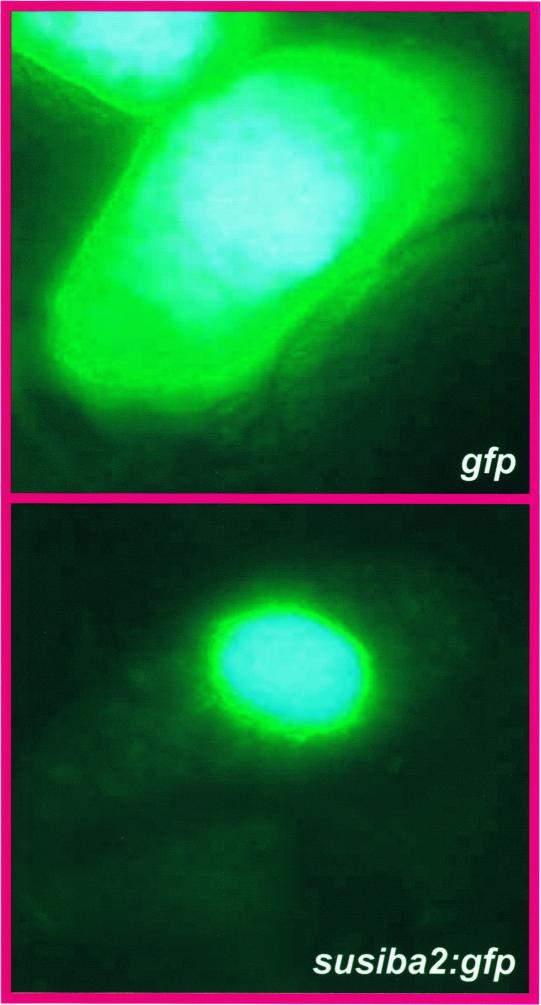
The subcellular localization of SUSIBA2 was determined in a transient assay system using transformed barley endosperm cells (Figure 11). For this investigation, the gfp reporter gene was fused in frame C terminal to the susiba2 open reading frame. A construct that encodes GFP alone served as a control. Endosperm cells transformed with the control construct showed GFP fluorescence throughout the entire cytosol and the nucleus. By contrast, cells transformed with the SUSIBA2:GFP fusion protein showed GFP fluorescence exclusively in the nucleus.
Figure 11.
Nuclear Localization of the SUSIBA2:GFP Fusion Protein.
Transient expression assays of GFP fluorescence in transformed barley endosperm cells. Barley endosperm cells were transformed by microprojectile bombardment with a construct encoding GFP only (top) or with a construct encoding the SUSIBA2:GFP fusion protein (bottom).
Isolation of cDNA for SUSABA2 Orthologs in Rice and Wheat
The involvement of the SUSIBA2 transcription factor in the regulation of iso1 expression in barley endosperm suggested that SUSIBA2 orthologs also should be found in sink tissues in other plants. The same primers used for cloning of the susiba2 cDNA were used for RT-PCR amplification of total RNA isolated from rice and wheat endosperm. For both species, only one major RT-PCR product was obtained (data not shown). The PCR products were subcloned and sequenced. The deduced amino acid sequences from the amplified regions in the rice and wheat clones share a high degree of identity with the corresponding segment in SUSIBA2 (see supplemental data online). By contrast, in this region between the conserved WRKY domains, SUSIBA2 and its rice and wheat orthologs all are significantly different from the SP8a binding SPF1 protein. The entire sequence for the rice ortholog could be retrieved from GenBank with the temporary name WRKY20. SUSIBA2 and the rice ortholog share a high degree of amino acid identity (80%) along their entire lengths.
DISCUSSION
We have identified a class of transcription factors, the SUSIBAs, and are studying the influence of the SUSIBA isomers on the regulation of starch synthesis in barley. To date, we have isolated cDNA clones for three members of the SUSIBA family: SUSIBA1, SUSIBA2, and SUSIBA3. The PCR product used in the barley library screening that resulted in the isolation of the susiba family was generated based on sequence similarities between cDNAs for SPF1 and SPF1-like proteins. Thus, the weak interaction between SUSIBA2 and the SP8a element might seem surprising. However, as is evident in Figure 2, the PCR primers used correspond to regions in the conserved WRKY domains, and, apart from these two domains, the overall degree of identity between SUSIBA2 and SPF1 is rather low (28%; see supplemental data online). Furthermore, as has been suggested (Eulgem et al., 2000), it has not been shown conclusively that SPF1 binds to the reported SP8a element, because the oligonucleotide used in the binding assay also contained a W-box. Sequence analysis of SUSIBA1, SUSIBA2, and SUSIBA3 shows that they are closely related and belong to the WRKY superfamily of plant transcription factors. The major difference between SUSIBA1 and SUSIBA2 relates to the number of WRKY domains (see supplemental data online). SUSIBA2 contains two WRKY domains and belongs to group 1 of the WRKY proteins, whereas SUSIBA1 contains one WRKY domain and belongs to group 2. SUSIBA3 has not been characterized in detail. In the work described here, we focus on SUSIBA2 and its communication with the iso1 promoter.
The SUSIBA2 Transcription Factor
Purified SUSIBA2 appeared in two forms, SUSIBA2-70 and SUSIBA2-68, of ∼70 and 68 kD, respectively (Figure 3). It is possible that SUSIBA2-68 is a truncated version of SUSIBA2-70. This notion is supported by preliminary results, which indicate that the level of SUSIBA2-70 increases at the expense of SUSIBA2-68 with increasing concentrations of protease inhibitors. Whether the truncation is at the N or C terminus, and if it is of biological significance in vivo, remain to be elucidated. Because the purified SUSIBA2 still contains the N-terminal His tag, it seems less likely that the truncation should be at the N terminus, because that would leave too few His residues on SUSIBA2-68 to bind to the Ni column. Alternatively, the size difference between SUSIBA2-70 and SUSIBA2-68 is an effect of post-translational modification.
The SUSIBA2 preparation also contained the E. coli enzyme trehalose 6-phosphate synthase (Figure 3). This enzyme also copurified with the SUSIBA2 polypeptides during repeated runs on the Ni column. There do not seem to be enough His residues in trehalose 6-phosphate synthase to generate high affinity to the Ni resin. Instead, it is conceivable that the copurification was attributable to the association between SUSIBA2 and trehalose 6-phosphate synthase. Although such an association is likely to be nonspecific, we must consider the possibility that it reflects an interaction of biological significance between SUSIBA2 and barley trehalose 6-phosphate synthase. A role for trehalose and trehalose-6-phosphate in plant sugar signaling has been proposed (Goddijn and Smeekens, 1998).
As has been demonstrated here, purified SUSIBA2 binds strongly to the SURE element and the W-box but not to the SP8a motif (Figures 4 to 6). Binding to the W-box is consistent with SUSIBA2 being a WRKY protein, whereas binding to the SURE element displays a novel feature for a WRKY protein. The WRKY proteins seem to be specific to the plant kingdom and are best known for their participation in various stress responses and senescence (Eulgem et al., 2000). Our results demonstrate that their sphere of activities has to be expanded to include carbohydrate anabolism. We show that SUSIBA2 is a regulatory transcription factor in starch synthesis and that it binds to the SURE element. Although the binding of nuclear protein fractions to the SURE element in the potato patatin (Grierson et al., 1994) and maize sbeI (Kim and Guiltinan, 1999) promoters, to the SP8a sequence in the barley iso1 promoter (Sun et al., 1999), and to the Bbl element in the second intron of the barley sbeIIb gene (Ahlandsberg et al., 2002b) has been documented previously, the interacting trans factors have not been isolated or identified.
Analysis of the SUSIBA2 amino acid sequence shows that it conforms well to the general features of group 1 of the WRKY transcription family (Figure 2). Our understanding of structure-function relationships in WRKY proteins is poor. It is believed generally that the C-terminal WRKY domain mediates sequence-specific binding to the cognate DNA elements, whereas the N-terminal WRKY domain facilitates DNA binding or engages in protein–protein interactions (Eulgem et al., 2000). The zinc fingers in each WRKY domain might be involved in binding to either DNA or proteins. What aspects of SUSIBA2 that are responsible for binding to the SURE element are not known. Site-directed mutagenesis in combination with binding assays is under way in our laboratory to address this question. It is probable that the unique properties of SUSIBA2, compared with other, previously known WRKY proteins, are attributable to regions outside of the two WRKY domains, where the amino acid sequences are relatively divergent (Figure 2). Another region that merits attention is the insertion in SUSIBA2 right after the C-terminal WRKY domain (Figure 2).
Using RT-PCR, we found orthologs to SUSIBA2 in rice and wheat (see supplemental data online). The deduced amino acid sequences from the PCR products include the region between the two WRKY domains, and analysis of these segments from the rice and wheat sequences shows that they share a high degree of identity between themselves and with SUSIBA2. Because the open reading frame for the rice clone could be obtained in its entirety, we were able to compare the complete primary sequence between SUSIBA2 and the rice ortholog (see supplemental data online). The two proteins are very similar in the extreme C and N termini. Notably, the SUSIBA2-specific C-terminal insertion (Figure 2) is present in the rice sequence. Preliminary data suggest that we have identified SUSIBA2 orthologs in potato and Arabidopsis (data not shown).
Given the large number (>70) of WRKY genes in Arabidopsis and the conservation of the WRKY domains, it is an apparently unexpected outcome that the initial RT-PCR amplification of barley, rice, and wheat RNA yielded only one major product. One likely explanation for this finding is that the RNA was isolated from developing endosperms, in which only a subset of WRKY genes is expressed.
As demonstrated using a SUSIBA2:GFP chimeric protein, SUSIBA2 is targeted to the nucleus. Nuclear import can occur by diffusion or carrier-mediated active transport (Görlich and Mattaj, 1996). As a result of its predicted molecular mass of 62 kD, translocation of SUSIBA2 across the nuclear pore is likely to require active transport via NLS-dependent receptor binding. Several NLSs were found in SUSIBA2, one of which involved a WRKY motif (Figure 2). Whether one or more of these NLSs is responsible for the nuclear import of SUSIBA2 remains to be established. As shown by Robatzek and Somssich (2001), a classic NLS is not always a prerequisite for nuclear targeting.
Transcriptional Analyses of the susiba2 Gene
The expression pattern of the susiba2 gene mimicked that of barley iso1. The temporal expression of both genes followed changes in the endogenous sucrose levels and peaked at 12 days after pollination (Figure 8). Both genes are expressed in the endosperm but not in leaves. However, they can be expressed ectopically in leaves after sugar treatment (Figure 9A). Again, the expression levels followed the sucrose concentration. The spatial expression of susiba2 was confirmed at the protein level; SUSIBA2 was present in the endosperm but not in leaves (Figure 9C). That SUSIBA2 was produced in sugar-treated leaves is suggested by the sugar-induced SURE binding activity in nuclear extracts from barley leaves (Figure 9B). We did not get good immunological signals from sugar-treated barley leaves using our SUSIBA2 antibody. A probable reason is that the SUSIBA2 protein was present at concentrations too low to be detected by protein gel blot analysis, which is less sensitive than EMSA. The data from the expression analyses are consistent with a role for SUSIBA2 as a positive regulatory transcription factor for iso1 activity. Experimental evidence for this view was obtained recently in antisense inhibition studies (data not shown). They also suggest that the expression of the susiba2 gene itself is controlled via sugar signaling.
The coding region of susiba2 contains an 18-nucleotide sequence that is exactly complementary to the human IGF1 receptor gene (Figure 1). We present this observation without any attempt at explanation. The sequence is not conserved in any of the susiba2 orthologs except for that of wheat. It also is not present in the recently deposited partial EST sequences for barley WRKY proteins implicated in drought tolerance (Ozturk et al., 2002). However, the 18-mer sequence is conserved in susiba1, susiba2, and susiba3, and it might serve as a signature sequence for susiba genes.
We noted that the expression of iso1 in transformed barley endosperm was confined mainly to the periphery of the endosperm, toward the aleurone layer (Figure 10B). The significance of this finding is unclear. The endosperms used for transformation were from seeds 20 days after pollination, which is near maturity. Because programmed cell death of the starchy endosperm tissue in barley progresses from the center, the uneven distribution of iso1 activity might indicate that at 20 days after pollination, only endosperm cells along the aleurone layer are metabolically viable (for a recent review of endosperm development, see Olsen, 2001).
The Nature and Relevance of the SURE Element
The importance of the SURE cis element for the sucrose induction of gene activity in plants was first demonstrated by Grierson et al. (1994) from work on the potato patatin promoter and has been corroborated by a large number of studies in different plants (Kim et al., 1991; Rhee and Staswick, 1992; Fu et al., 1995; Mita et al., 1995; Kim and Guiltinan, 1999; Li et al., 2002). In some of these studies, the assignment of the SURE sequence as a regulatory element was inferred from experimental data (i.e., binding assays), whereas in other cases, it was based primarily on computational analyses. Here, we show that three SURE elements, SURE-a, SURE-b, and SURE-c, also are located in the barley iso1 promoter and that they provide binding sites for the SUSIBA2 transcription factor (Figures 4 to 7).
Most of our attention in this study was directed toward SURE-a, because it appeared to have the strongest SUSIBA2 binding activity (Figure 4). The fact that SURE-a produced a similar binding pattern with purified SUSIBA2 and nuclear extracts (Figure 4) indicates that the in vivo assembly of the SUSIBA2 complex on the iso1 promoter does not involve regulatory proteins other than SUSIBA2 itself. The binding assays also suggest that SUSIBA2 can oligomerize in a concentration-dependent manner (Figures 4 and 5). The structural features of the SURE elements in the barley iso1 promoter need to be assessed. As judged by mutation analyses of the SURE-a sequence (Figure 6), a large number of A nucleotides does not in itself constitute a SUSIBA2 binding element. In fact, the presence of long poly(A) stretches in the upstream region (SURE-a 3′ m1), the middle region (SURE-a 5′ and SURE-a 3′ m2), or the entirety (SURE-a 3′ m4) of SURE-a diminished or abolished its SUSIBA2 binding activity (Figure 6).
Given the large number of DNA binding proteins in nuclei, it is plausible that the complex formations observed for SURE-a 5′ and SURE-a 3′ m4 with nuclear extracts are attributable to proteins other than SUSIBA2 (Figures 6A and 6C). Notably, extending the poly(A) stretch in the downstream half of SURE-a (SURE-a 3′ m3) significantly augmented the affinity for the purified SUSIBA2 protein, although not for nuclear extracts, and promoted the assembly of high Mr complexes (Figure 6). The apparent lower SURE-a 3′ m3 binding capacity for nuclear extracts compared with that for purified SUSIBA2 is not understood. It might reflect the fact that the amount of purified SUSIBA2 used in the binding assays exceeded that present in the added nuclear extracts.
The complex formed between the SURE-a 5′ element and the nuclear extract harbored two polypeptides of 68 and 66 kD that reacted with the SUSIBA2 antibodies (Figure 7). These data demonstrate that the SURE binding activity in the nuclear extracts was attributable to SUSIBA2. Whether the SUSIBA2 duplex observed in Figure 7 corresponds to the overproduced SUSIBA2-70 and SUSIBA2-68 is not known. Occasionally, two additional faint bands in the 56- to 54-kD region were discerned on the protein gel blot (data not shown), possibly as a result of degradation during the extraction procedure. The nature of the 96-kD polypeptide recognized by the His tag antibodies is unknown.
Its presence in the barley iso1 and maize sbe1 promoters suggests that the SURE element also should be found in other starch synthesis genes subject to sugar induction. Indeed, a database search of the proximal and distal promoter regions of sequences revealed that SURE-like sequences are present in promoters of several genes that encode enzymes involved in starch synthesis. Among those are barley sbeIIb and the barley genes for starch synthase I (ssI) (Gubler et al., 2000) and the small subunit of ADP–glucose pyrophosphorylase (agpaseS) (Thorbjørnsen et al., 2000). Although all three SURE sequences in the barley iso1 promoter, as well as those identified in other genes, bear a good resemblance to the patatin SURE sequence in pair-wise comparisons, it is difficult to arrive at a consensus sequence for the SURE element. An alignment of a subset of reported SURE sequences is shown in the supplemental data online. The compilation also includes the SURE sequences from barley ssI and agpaseS.
As can be seen, the SURE element is best described as an A-rich sequence with the consensus core AA/TAA. Other A-rich or A/T-rich promoter elements with demonstrated or implied protein binding activity are known from both plants and other organisms, such as the A-box in gbssI from wheat (Kluth et al., 2002) and developmentally regulated genes from Drosophila (Gasser and Laemmli, 1986), the A-stretch in psbD from tobacco (Allison and Maliga, 1995), the A-rich sequence in the Npg1 gene from tobacco (Tebbutt et al., 1994), the A/T-rich region in URA3 from yeast (Roy et al., 1990), the A-rich site in the α-MHC gene from rat (Molkentin and Markham, 1994), the A-rich region in the Igκ gene from mouse (Costa and Atchison, 1996), and the A/T-rich AT hook binding (Dragan et al., 2003) and SPKK binding (Churchill and Suzuki, 1989) regions in eukaryotic DNA. A/T-rich regions also might be expected in some promoters, because they facilitate DNA unwinding.
We noted that, in contrast to the barley sbeIIb promoter, which contains three SURE elements, no SURE-like sequence was found in the promoter of the barley sbeIIa gene. Both sbeIIb and sbeIIa contain a W-box (data not shown). We reported previously (Sun et al., 1998) that the expression of barley sbeIIb is endosperm specific, whereas that of sbeIIa is not. We suggest that the W-box in the iso1, sbeIIb, and sbeIIa promoters serves as a general activating element, whereas the sugar responsiveness conferred to iso1 and sbeIIb by the SURE elements contributes to their endosperm specificity (by the higher sugar levels in sink tissues compared with embryos and vegetative tissues). Further control of endosperm-specific expression probably is exerted via the binding of repressor proteins in nonexpressing tissues. From earlier work, we concluded that repressors are recruited to the SP8a element in the barley iso1 promoter (Sun et al., 1999) and to the Bbl element in the second intron of barley sbeIIb (Ahlandsberg et al., 2002b).
The importance of the SURE element for iso1 expression is demonstrated by the decoy experiment (Figure 10). The activity remaining in the presence of the SURE-a decoy could indicate that the decoy did not efficiently trap the SURE binding activity. More likely, it demonstrates that elements other than SURE are sufficient to maintain a basic level of iso1 activity. The involvement of the SURE element in the regulation of iso1 expression is in agreement with the ectopic expression of iso1 in sugar-treated barley leaves (Sun et al., 1999). The emerging transcription factor ODN technology offers a powerful and convenient means to assess transcription factor function and the involvement of cis elements both in vitro and in vivo. The strategy has gained rapid popularity in animal sciences in studies of gene therapy (Morishita et al., 1998; Mann and Dzau, 2000). The present report demonstrates the applicability of the transcription factor ODN technology also in plant biology.
The enrichment of W-boxes in the promoters of defense-related wrky genes suggests that these genes are subject to autoregulation and/or controlled by other members of the WRKY superfamily (Dong et al., 2003). It is worth considering whether the barley susiba2 gene participates in such a self-regulatory network. We have not completed the sequencing of the susiba2 genomic clone and do not know if the susiba2 promoter contains W-boxes or SURE elements. However, thanks to the rice genome project, we were able to analyze the entire locus for the orthologous gene, WRKY20, in rice. Inspection of the 1.2-kb region upstream of the predicted transcription start site revealed that the WRKY20 gene harbors one W-box sequence (TGACT; nucleotides −796 to −792) and one SURE sequence (see supplemental data online). Whether these putative elements are present in barley susiba2, and to what extent they would contribute to the regulation of susiba2 activity and starch synthesis, are questions that remain to be addressed.
METHODS
Plant Material
Barley (Hordeum vulgare cv Pongo), rice (Oryza sativa), and wheat (Triticum aestivum) were grown in soil in a climate chamber under a 16-h-light/8-h-dark photoperiod as described by Sun et al. (1998)(1999).
Oligonucleotides
The following oligonucleotides were used: 1, 5′-CCAAGAAGTTATTACAAGTG-3′; 2, 5′-TGGTTATGTTTTCCTTCGTA-3′; 3, 5′-GGAATTCCATATGTCCCCCGCGCGGCTGCC-3′; 4, 5′-CGGATCCGGCTGAACTGACTTGTAAC-3′; 5, 5′-CCCTCGTGGAAGCAAAACTGTGTTTCTCGC-3′; 6, 5′-GCGAGAAACACAGTTTTGCTTCCACGAGGG-3′; 7, 5′-TTCCGAAAAAAACTAAGAAAGACCGATGGAAAACCGAAAT-3′; 8, 5′-ATTTCGGTTTTCCATCGGTCTTTCTTAGTTTTTTTCGGAA-3′; 9, 5′-GGAAAACCGAAATACCAAAAAATAATAATAAAATAATAAT-3′; 10, 5′-ATTATTATTTTATTATTATTTTTTGGTATTTCGGTTTTCC-3′; 11, 5′-CCGCGTGCGAAAAAATAAAGAAAATGAAATCTGAAGGAAG-3′; 12, 5′-CTTCCTTCAGATTTCATTTTCTTTATTTTTTCGCACGCGG-3′; 13, 5′-TCGCTAACCAGTGACTTCCACGTTTCATCATTTATT-3′; 14, 5′-AATAAATGATGAAACGTGGAAGTCACTGGTTAGCGA-3′; 15, 5′-TTCCGAAAAAAACTAAGAAA-3′; 16, 5′-TTTCTTAGTTTTTTTCGGAA-3′; 17, 5′-GACCGATGGAAAACCGAAAT-3′; 18, 5′-ATTTCGGTTTTCCATCGGTC-3′; 19, 5′-GAAAAATGGAAAACCGAAAT-3′; 20, 5′-ATTTCGGTTTTCCATTTTTC-3′; 21, 5′-GACCGAAAAAAAACCGAAAT-3′; 22, 5′-ATTTCGGTTTTTTTTCGGTC-3′; 23, 5′-GAAAAAAAAAAAAAAAAAAT-3′; 24, 5′-ATTTTTTTTTTTTTTTTTTC-3′; 25, 5′-ATGACTCGAGCAGATTTTGGATTGCTAATGA-3′; 26, 5′-ATGACCATGGGCCACCTCGTGTTGGTTCTTCGT-3′; 27, 5′-ATTACGGTAGAGCGTGTTATGAGTGTCTACGCGGTGGACT-3′; and 28, 5′-AGTCCACCGCGTAGACACTCATAACACGCTCTACCGTAAT-3′.
Computational Analyses
Searches of the NCBI databases were performed with the Basic Local Alignment Search Tool (BLAST) service (http://www.ncbi.nlm.nih.gov/BLAST/). Alignments of nucleotide and amino acid sequences were performed using the CLUSTAL W service at the European Bioinformatics Institute (http://www.ebi.ac.uk/clustalw), except for the alignment of sweet potato, cucumber, and parsley SPF1 cDNA sequences, which was performed with the MacVector program (Accelrys, Paris, France), and alignment of the SURE element sequences, which were run on the T-Coffee server (http:// www.ch.embnet.org/software/TCoffee.html). For presentation purposes, the ALN output from the CLUSTAL W program was fed into the BOXSHADE server (http://www.ch.embnet.org/software/BOX_form.html).
Isolation of cDNA Clones
Oligonucleotides 1 and 2 were constructed according to the consensus sequences for the SPF1 and SPF1-like cDNA sequences. Total RNA was isolated from developing barley, rice, and wheat endosperm according to Sun et al. (1999). First-strand cDNAs were produced as described (Frohman et al., 1988) using oligonucleotide 2. Reverse transcriptase–mediated (RT) PCR was performed according to standard protocols (Sambrook et al., 1989) using oligonucleotides 1 and 2. The PCR products were subcloned and sequenced. The RT-PCR product from barley was used as a probe to screen a barley cDNA library from developing endosperm 10 days after pollination. Library construction and screening, subcloning, DNA gel blot analysis, and sequence analysis were as described (Sun et al., 1998, 1999).
Overproduction of SUSIBA2 in Escherichia coli, Microsequencing, Antibody Production, and Protein Gel Blot Analysis
Two primers were designed: oligonucleotide 3, with an NdeI restriction site, and oligonucleotide 4, with a BamHI restriction site. These primers were used for PCR amplification of the susiba2 cDNA from the isolated full-length cDNA clone. The amplified cDNA was inserted into the expression vector pET-15b (Novagen, Madison, WI) between the NdeI and BamHI sites. The constructed plasmid was transformed into E. coli strain BL21(DE3). Overexpression was performed according to the manual provided by the manufacturer, except that the isopropylthio-β-galactoside induction was performed at 37°C for 2 h. The overproduced protein was purified by fast protein liquid chromatography on a column with a Ni+ chelating resin. SDS-PAGE was run as described previously (Sun et al., 1997).
Trypsin digestion and microsequencing were performed in collaboration with Amersham Pharmacia Biotech. Antisera were produced by AgriSera (Vännäs, Sweden) after immunizing rabbits with the Coomassie Brilliant Blue R 250–stained SUSIBA2-70 and SUSIBA2-68 polypeptide bands excised from the SDS gel. Protein gel blot analysis was performed as described (Sun et al., 1997). Polyclonal His tag antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Electrophoretic Mobility Shift Assay
Electrophoretic mobility shift assays (EMSAs) were performed essentially as described by Sun et al. (1999). For protein gel blot analysis of the EMSA complex formed between the nuclear protein extract and the SURE element, a polyacrylamide gel with a SURE-a 5′ protein complex was positioned over the x-ray film and a <1-cm-tall section of the labeled complex was sliced out from the gel. The gel pieces were homogenized in loading buffer and applied directly to an SDS gel.
RNA Gel Blot Analysis
RNA isolation and RNA gel blot analysis were performed as described previously (Sun et al., 1998, 1999).
Sucrose Isolation and Analysis, Exogenous Sucrose Induction, and Nuclear Protein Isolation
Sucrose isolation, enzymatic sucrose analysis, thin layer chromatography, and sucrose induction of ectopic gene expression in barley leaves were performed as described (Sun et al., 1998, 1999). Nuclear proteins were isolated from leaves and endosperm as described by Sun et al. (1999).
Transformation and GFP Assay of Barley Endosperm
The 1504-bp promoter region of the iso1 gene was amplified by PCR using oligonucleotide primers 25 and 26 and fused to the gfp plasmid pN1473GFP as described by Ahlandsberg et al. (1999)(2002a, 2002b), yielding the construct p48. The SURE-a and rbcL oligodeoxynucleotide decoys were generated by annealing oligonucleotides 7 and 8, and 27 and 28, respectively, in 14 mM Tris-HCl, pH 8.0, and 7 mM MgCl2. The Act-1:gfp construct was as described (Ahlandsberg et al., 2002b). The SUSIBA2:GFP fusion protein was constructed by ligating the NcoI-SacI fragment of the susiba2 cDNA into NcoI-SacI–digested pN1473GFP. Transformation of barley endosperm by microprojectile bombardment using the DuPont PDS 1000 He Biolistic Delivery System (Bio-Rad Laboratories, Hercules, CA) and transient assay of GFP fluorescence were performed as described by Ahlandsberg et al. (2002a) with the following exceptions. Barley caryopses at 19 to 22 days after pollination were bisected longitudinally and placed endosperm side up on DG3B medium containing Murashige and Skoog (1962) medium supplemented with 100 g/L sucrose, 1.0 mg/L thiamine-HCl, 0.25 g/L myo-inositol, 1.0 g/L casein hydrolase, and 0.69 g/L Pro solidified by 5.0 g/L Gelrite (Duchefa, Haarlem, The Netherlands) for 2 h before bombardment. Six bisected caryopses were placed in the center of a 20- × 90-mm Petri dish with DG3B medium, 13 cm below the stopping screen, and bombarded once using a 7.6-MPa rupture disc. After bombardment, the caryopses were kept for 48 h in darkness on plates before GFP fluorescence assays.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Christer Jansson, christer.jansson@vbsg.slu.se.
Accession Numbers
The susiba2 cDNA sequence has been deposited in GenBank with the accession number AY323206. The cDNA sequences for the rice and wheat susiba2 orthologs have been deposited with accession numbers AY324392 and AY324393, respectively. The GenBank accession numbers for the other sequences referred to in this work are as follows: Arabidopsis gene for β-amylase, S77076; Arabidopsis hypothetical protein, T08930; Arabidopsis WRKY transcription factor 20, Q93WV0; Arabidopsis putative protein, NP567752; Arabidopsis unknown protein, AAK76566; barley agpaseS, AJ239130; barley iso1, AF142589; barley sbeIIb, AF064563; barley ssI, AF234163; barley WRKY proteins implicated in drought tolerance, BM816210 and BM816211; E. coli trehalose-6-phosphate synthase, S33584; human IGF1 receptor, NM_000875; maize sbeI, AF072724; potato patatin class-I gene, M18880; potato gene for proteinase inhibitor II, X04118; potato gene for sucrose synthase, U24087; soybean gene for vegetative storage protein, M76980; sweet potato SPF1, S51529; tobacco WRKY protein, BAB61056; tomato hypothetical proteins, CAC36397 and CAC36402; white broom drought-induced protein, AAL32033; and rice putative WRKY transcription factor 20, BAC55609.
Supplementary Material
Acknowledgments
We thank the reviewers for thorough work and constructive criticism. This work was supported by grants from the Swedish Research Council for Environment, Agricultural Sciences, and Spatial Planning, the Nordic Joint Committee for Agricultural Research, the Foundation for Strategic Research, and Helge Ax:son Johnsons Stiftelse.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.014597.
Footnotes
Online version contains Web-only data.
References
- Abel, S., and Theologis, A. (1995). A polymorphic bipartite motif signals nuclear targeting of early auxin-inducible proteins related to PS-IAA4 from pea (Pisum sativum). Plant J. 8, 87–96. [DOI] [PubMed] [Google Scholar]
- Ahlandsberg, S., Sathish, P., Sun, C., and Jansson, C. (1999). Green fluorescent protein as a reporter system in the transformation of barley cultivars. Physiol. Plant. 107, 194–200. [Google Scholar]
- Ahlandsberg, S., Sathish, P., Sun, C., and Jansson, C. (2002. a). A set of useful monocotyledon transformation vectors. Biotechnol. Lett. 23, 1871–1875. [Google Scholar]
- Ahlandsberg, S., Sun, C., and Jansson, C. (2002. b). An intronic element directs endosperm-specific expression of the sbeIIb gene during barley seed development. Plant Cell Rep. 20, 864–868. [Google Scholar]
- Allison, L.A., and Maliga, P. (1995). Light-responsive and transcription-enhancing elements regulate the plastid psbD core promoter. EMBO J. 14, 3721–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae, J.M., and Liu, J.R. (1997). Molecular cloning and characterization of two novel isoforms of the small subunit of ADP-glucose pyrophosphorylase from sweet potato. Mol. Gen. Genet. 254, 179–185. [DOI] [PubMed] [Google Scholar]
- Ball, S., van de Wal, M.H.B.J., and Visser, R.G.F. (1998). Progress in understanding the synthesis of amylose. Trends Plant Sci. 3, 462–467. [Google Scholar]
- Buléon, A., Colonna, P., Planchot, V., and Ball, S. (1998). Starch granules: Structure and biosynthesis. Int. J. Biol. Macromol. 23, 85–112. [DOI] [PubMed] [Google Scholar]
- Carlson, M. (1999). Glucose repression in yeast. Curr. Opin. Microbiol. 2, 202–207. [DOI] [PubMed] [Google Scholar]
- Churchill, M.E.A., and Suzuki, M. (1989). ‘SPKK’ motifs prefer to bind to DNA at A/T-rich sites. EMBO J. 8, 4189–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, M.W., and Atchison, M.L. (1996). Identification of an Sp1-like element within the immunoglobulin κ 3′ enhancer necessary for maximal enhancer activity. Biochemistry 35, 8662–8669. [DOI] [PubMed] [Google Scholar]
- Dong, J., Chen, C., and Chen, Z. (2003). Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 51, 21–37. [DOI] [PubMed] [Google Scholar]
- Dragan, A.I., Liggins, J.R., Crane-Robinson, C., and Privalov, P.L. (2003). The energetics of specific binding of AT-hooks from HMGA1 to target DNA. J. Mol. Biol. 327, 393–411. [DOI] [PubMed] [Google Scholar]
- Eulgem, T., Rushton, P.J., Robatzek, S., and Somssich, I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Frohman, M.A., Dush, M.K., and Martin, G.R. (1988). Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85, 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H., Kim, S.Y., and Park, W.D. (1995). High-level tuber expression and sucrose inducibility of a potato sus4 sucrose synthase gene require 5′ and 3′ flanking sequences and the leader intron. Plant Cell 7, 1387–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser, S.M., and Laemmli, U.K. (1986). Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell 46, 521–530. [DOI] [PubMed] [Google Scholar]
- Giuliano, G., Pichersky, E., Malik, V.S., Timko, M.P., Scolnic, P.A., and Cashmore, A.R. (1988). An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc. Natl. Acad. Sci. USA 85, 7089–7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddijn, O., and Smeekens, S. (1998). Sensing trehalose biosynthesis in plants. Plant J. 14, 143–146. [DOI] [PubMed] [Google Scholar]
- Görlich, D., and Mattaj, I.W. (1996). Nucleocytoplasmic transport. Science 271, 1513–1518. [DOI] [PubMed] [Google Scholar]
- Grierson, C., Du, J.-S., de Torres Zabala, M., Beggs, K., Smith, C., Holdsworth, M., and Bevan, M.W. (1994). Separate cis sequences and trans factors direct metabolic and developmental regulation of a potato tuber storage protein gene. Plant J. 5, 815–826. [DOI] [PubMed] [Google Scholar]
- Gubler, F., Li, Z., Fieg, S., Jacobsen, J.V., and Morell, M.K. (2000). Cloning and characterization of a starch synthase I gene (accession No. AF234163) from barley. Plant Physiol. 122, 1459.
- Halford, N.G., and Hardie, D.G. (1998). SNF1-related protein kinases: Global regulators of carbon metabolism in plants? Plant Mol. Biol. 37, 735–748. [DOI] [PubMed] [Google Scholar]
- Hardie, D.G., Carling, D., and Carlson, M. (1998). The AMP-activated/SNF1 protein kinase subfamily: Metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67, 821–855. [DOI] [PubMed] [Google Scholar]
- Hattori, T., Nakagawa, S., and Nakamura, K. (1990). High-level expression of tuberous root storage protein genes of sweet potato in stems of plantlets grown in vitro on sucrose medium. Plant Mol. Biol. 14, 595–604. [DOI] [PubMed] [Google Scholar]
- Ishiguro, S., and Nakamura, K. (1992). The nuclear factor SP8BF binds to the 5′-upstream regions of three different genes coding for major proteins of sweet potato tuberous roots. Plant Mol. Biol. 18, 97–108. [DOI] [PubMed] [Google Scholar]
- Ishiguro, S., and Nakamura, K. (1994). Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol. Gen. Genet. 244, 563–571. [DOI] [PubMed] [Google Scholar]
- Khoshnoodi, J., Larsson, C.-T., Larsson, H., and Rask, L. (1998). Differential accumulation of Arabidopsis thaliana Sbe2.1 and Sbe2.2 transcripts in response to light. Plant Sci. 135, 183–193. [Google Scholar]
- Kim, D.-J., Smith, S.M., and Leaver, C.J. (1997). A cDNA encoding a putative SPF1-type DNA-binding protein from cucumber. Gene 185, 265–269. [DOI] [PubMed] [Google Scholar]
- Kim, K.-N., and Guiltinan, M.J. (1999). Identification of cis-acting elements important for expression of the starch-branching enzyme I gene in maize endosperm. Plant Physiol. 121, 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.-R., Costa, M.A., and An, G. (1991). Sugar response element enhances wound response of potato proteinase inhibitor II promoter in transgenic tobacco. Plant Mol. Biol. 17, 973–983. [DOI] [PubMed] [Google Scholar]
- Kluth, A., Sprunck, S., Becker, D., Lörz, H., and Lütticke, S. (2002). 5′ deletion of a gbsss1 promoter region from wheat leads to changes in tissue and developmental specificities. Plant Mol. Biol. 49, 669–682. [DOI] [PubMed] [Google Scholar]
- Kossman, J., Visser, R.G.F., Müller-Röber, B., Willmitzer, L., and Sonnewald, U. (1991). Cloning and expression analysis of a potato cDNA that encodes branching enzyme: Evidence for coexpression of starch biosynthetic genes. Mol. Gen. Genet. 230, 39–44. [DOI] [PubMed] [Google Scholar]
- Li, X., Xing, J., Gianfagna, T.J., and Janes, H.W. (2002). Sucrose regulation of ADP-glucose pyrophosphorylase subunit gene transcript levels in leaves and fruits. Plant Sci. 162, 239–244. [DOI] [PubMed] [Google Scholar]
- Mann, M.J., and Dzau, V.J. (2000). Therapeutic applications of transcription factor decoy oligonucleotides. J. Clin. Invest. 106, 1071–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeo, K., Tomiya, T., Hayashi, K., Akaike, M., Morikami, A., Ishiguro, S., and Nakamura, K. (2001). Sugar-responsible elements in the promoter of a gene for β-amylase of sweet potato. Plant Mol. Biol. 46, 627–637. [DOI] [PubMed] [Google Scholar]
- McElroy, D., Blowers, A.D., Jenes, B., and Wu, R. (1991). Construction of expression vectors based on the rice actin 1 (Act1) 5′ region for use in monocot transformation. Mol. Gen. Genet. 231, 150–160. [DOI] [PubMed] [Google Scholar]
- McElroy, D., Zhang, W., Cao, J., and Wu, R. (1990). Isolation of an efficient promoter for use in rice transformation. Plant Cell 2, 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle, T. (2001). Nuclear import and export of proteins in plants: A tool for the regulation of signaling. Planta 213, 499–517. [DOI] [PubMed] [Google Scholar]
- Mita, S., Suzuki-Fujii, K., and Nakamura, K. (1995). Sugar-inducible expression of a gene for β-amylase in Arabidopsis thaliana. Plant Physiol. 107, 895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin, J.D., and Markham, B.E. (1994). An M-CAT binding factor and an RSRF-related A-rich binding factor positively regulate expression of the α-cardiac myosin heavy-chain gene in vivo. Mol. Cell. Biol. 14, 5056–5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita, R., Higaki, J., Tomita, N., and Ogihara, T. (1998). Application of transcription factor “decoy” strategy as means of gene therapy and study of gene expression in cardiovascular disease. Circ. Res. 82, 1023–1028. [DOI] [PubMed] [Google Scholar]
- Müller-Röber, B.T., Kossman, J., Hannah, L.C., and Willmitzer, L. (1990). One of two different ADP-glucose pyrophosphorylase genes from potato responds strongly to elevated levels of sucrose. Mol. Gen. Genet. 224, 136–146. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Myers, A.M., Morell, M.K., James, M.G., and Ball, S.G. (2000). Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol. 122, 989–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, Y. (1996). Some properties of starch debranching enzymes and their possible role in amylopectin biosynthesis. Plant Sci. 121, 1–18. [Google Scholar]
- Nakamura, Y. (2002). Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: Rice endosperm as a model tissue. Plant Cell Physiol. 43, 718–725. [DOI] [PubMed] [Google Scholar]
- Olsen, O.-A. (2001). Endosperm development: Cellularization and cell fate specification. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 233–267. [DOI] [PubMed] [Google Scholar]
- Ozturk, Z.N., Talamé, V., Deyholos, M., Michalowski, C.B., Galbraith, D.W., Gozukirmizi, N., Tuberosa, R., and Bohnert, H.J. (2002). Monitoring large-scale changes in transcript abundance in drought- and salt-stressed barley. Plant Mol. Biol. 48, 551–573. [DOI] [PubMed] [Google Scholar]
- Rhee, Y., and Staswick, P.E. (1992). Nucleotide sequence of a soybean vegetative storage protein vspB gene. Plant Physiol. 98, 794–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek, S., and Somssich, I.E. (2001). A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence-and defence-related processes. Plant J. 28, 123–133. [DOI] [PubMed] [Google Scholar]
- Rolland, F., Moore, B., and Sheen, J. (2002). Sugar sensing and signaling in plants. Plant Cell 14 (suppl.), S185.–S205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook, F., Corke, F., Card, R., Munz, G., Smith, C., and Bevan, M.W. (2001). Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic expression by abscisic acid signaling. Plant J. 26, 421–433. [DOI] [PubMed] [Google Scholar]
- Roy, A., Exinger, F., and Losson, R. (1990). cis- and trans-acting regulatory elements of the yeast URA3 promoter. Mol. Cell. Biol. 10, 5257–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton, P.J., Torres, J.T., Parniske, M., Wernert, P., Hahlbrock, K., and Somssich, I.E. (1996). Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 15, 5690–5700. [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sheen, J., Zhou, L., and Jang, J.C. (1999). Sugars as signaling molecules. Curr. Opin. Plant Biol. 2, 410–418. [DOI] [PubMed] [Google Scholar]
- Smeekens, S. (2000). Sugar-induced signal transduction in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 49–81. [DOI] [PubMed] [Google Scholar]
- Smith, A.M. (2001). The biosynthesis of the starch granule. Biomacromolecules 2, 335–341. [DOI] [PubMed] [Google Scholar]
- Sørensen, M.B., Cameron-Mills, V., and Brandt, A. (1989). Transcriptional and post-transcriptional regulation of gene expression in developing barley endosperm. Mol. Gen. Genet. 217, 195–201. [Google Scholar]
- Sun, C., Sathish, P., Ahlandsberg, S., Deiber, A., and Jansson, C. (1997). Identification of four starch-branching enzymes in barley endosperm: Partial purification of forms I, IIa and IIb. New Phytol. 137, 215–222. [DOI] [PubMed] [Google Scholar]
- Sun, C., Sathish, P., Ahlandsberg, S., and Jansson, C. (1998). The two genes encoding starch-branching enzyme IIa and IIb are differentially expressed in barley. Plant Physiol. 118, 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, C., Sathish, P., Ahlandsberg, S., and Jansson, C. (1999). Analyses of isoamylase gene activity in wild-type barley indicate its involvement in starch synthesis. Plant Mol. Biol. 40, 431–443. [DOI] [PubMed] [Google Scholar]
- Tebbutt, S.J., Rogers, H.J., and Lonsdale, D.M. (1994). Characterization of a tobacco gene encoding a pollen-specific polygalacturonase. Plant Mol. Biol. 25, 283–297. [DOI] [PubMed] [Google Scholar]
- Thorbjørnsen, T., Villand, P., Ramstad, V.E., Kleczkowski, L.A., Olsen, O.-A., and Opsahl-Ferstad, H.-G. (2000). Nucleotide sequence of the ADP-glucose pyrophosphorylase promoter (accession No. AF239130) of barley, a gene involved in endosperm starch formation. Plant Physiol. 122, 1459.
- Visser, R.G.F., Stolte, A., and Jacobsen, E. (1991). Expression of a chimaeric granule-bound starch synthase-GUS gene in transgenic potato plants. Plant Mol. Biol. 17, 691–699. [DOI] [PubMed] [Google Scholar]
- Zourelidou, M., de Torres-Zabala, M., Smith, C., and Bevan, M.W. (2002). Storekeeper defines a new class of plant-specific DNA-binding proteins and is a putative regulator of patatin expression. Plant J. 30, 489–497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.