Abstract
Mitochondria fulfill a wide range of metabolic functions in addition to the synthesis of ATP and contain a diverse array of proteins to perform these functions. Here, we present the unexpected discovery of the presence of the enzymes of glycolysis in a mitochondrial fraction of Arabidopsis cells. Proteomic analyses of this mitochondrial fraction revealed the presence of 7 of the 10 enzymes that constitute the glycolytic pathway. Four of these enzymes (glyceraldehyde-3-P dehydrogenase, aldolase, phosphoglycerate mutase, and enolase) were also identified in an intermembrane space/outer mitochondrial membrane fraction. Enzyme activity assays confirmed that the entire glycolytic pathway was present in preparations of isolated Arabidopsis mitochondria, and the sensitivity of these activities to protease treatments indicated that the glycolytic enzymes are present on the outside of the mitochondrion. The association of glycolytic enzymes with mitochondria was confirmed in vivo by the expression of enolase– and aldolase–yellow fluorescent protein fusions in Arabidopsis protoplasts. The yellow fluorescent protein fluorescence signal showed that these two fusion proteins are present throughout the cytosol but are also concentrated in punctate regions that colocalized with the mitochondrion-specific probe Mitotracker Red. Furthermore, when supplied with appropriate cofactors, isolated, intact mitochondria were capable of the metabolism of 13C-glucose to 13C-labeled intermediates of the trichloroacetic acid cycle, suggesting that the complete glycolytic sequence is present and active in this subcellular fraction. On the basis of these data, we propose that the entire glycolytic pathway is associated with plant mitochondria by attachment to the cytosolic face of the outer mitochondrial membrane and that this microcompartmentation of glycolysis allows pyruvate to be provided directly to the mitochondrion, where it is used as a respiratory substrate.
INTRODUCTION
The role of the mitochondrion in the synthesis of cellular ATP is both well established and well studied. However, it is becoming increasingly apparent that the mitochondrion also is responsible for a wide range of other metabolic processes. This is particularly the case in autotrophic organisms such as plants, in which mitochondria provide precursors for a number of essential biosynthetic processes such as nitrogen fixation and the biosynthesis of amino acids, tetrapyrroles, and vitamin cofactors (Douce, 1985; Douce and Neuberger, 1989; Mackenzie and McIntosh, 1999). The unique demands placed on the mitochondrion in the plant cell are reflected by the presence of additional respiratory chain components that are not seen in animal mitochondria (Moller, 1986; Vanlerberghe and McIntosh, 1997; Moller, 2001) as well as a number of plant-specific metabolite exchanges between the mitochondrion and the cytosol (Douce and Neuberger, 1989). Plant mitochondria also have been shown to be a site of synthesis for fatty acids (Gueguen et al., 2000) and vitamin cofactors such as folic acid (Mouillon et al., 2002) and ascorbic acid (Bartoli et al., 2000) and have been implicated in the synthesis and export of iron-sulfur clusters (Kushnir et al., 2001). The pivotal importance of mitochondrial function in the physiology and development of higher plants is demonstrated by the fact that mutations of the mitochondrial genome frequently lead to cytoplasmic male sterility (Hanson, 1991; Schnable and Wise, 1998) and by the recent implication of plant mitochondria in programmed cell death (Balk and Leaver, 2001; Robson and Vanlerberghe, 2002). The breadth of mitochondrial functions is such that relatively few of these functions have been studied in great detail, and in most cases, the genes and proteins that support these processes remain unidentified. An important starting point in increasing our understanding of the manifold roles of mitochondria in plants would be the establishment of a complete catalog of the identities of all of the proteins present in plant mitochondria.
One way to achieve this goal is to identify mitochondrial targeting sequences present in nuclear genes that encode proteins that are destined for the mitochondrion. The complete sequencing of the genomes of model plant species (Arabidopsis Genome Initiative, 2000; Goff et al., 2002; Yu et al., 2002) means that this now can be done in a systematic and comprehensive manner, and several bioinformatic tools have been developed that attempt to assign cellular localization by analysis of N-terminal targeting sequences (Emanuelsson et al., 2000). However, there is considerable disagreement between these methods regarding which proteins are mitochondrially targeted, and all of the programs mispredict a small proportion of proteins (Heazlewood et al., 2003). Furthermore, because these methods concentrate on the analysis of N-terminal targeting sequences, they do not correctly predict the localization of proteins such as the mitochondrial carrier family, which are encoded by genes that do not contain N-terminal targeting sequences (Laloi, 1999). Thus, although these bioinformatic approaches give some insight into which nuclear genes encode mitochondrial proteins, the catalogs they generate will not be comprehensive and will require additional experimental validation.
Therefore, several groups have used a more empirical approach to identifying mitochondrial proteins that involves the isolation of pure mitochondria from plant tissues and the identification of proteins with a combination of two-dimensional electrophoretic fractionation and mass spectrometry. This approach has been taken with Arabidopsis cells (Kruft et al., 2001; Millar et al., 2001), rice shoots (Heazlewood et al., 2003), and a variety of pea tissues (Bardel et al., 2002). These studies report the identification of between 40 and 135 plant mitochondrial proteins, and collectively they provide a useful starting point for the establishment of a catalog of the mitochondrial proteome. However, there are a number of problems with this approach that will ultimately prevent full coverage of the proteome. Many of these problems relate to the electrophoretic techniques used to fractionate the protein sample and result from the insolubility of hydrophobic proteins during isoelectric focusing (Santoni et al., 1999), the lack of resolution of proteins of similar mass and charge, and the inability to readily resolve proteins outside the mass range of 10 to 100 kD (Link et al., 1999). An additional problem is that low-abundance proteins are significantly underrepresented in such separations, particularly when a relatively small number of very abundant proteins predominate. This is a major issue, because it is the low-abundance proteins that remain the least well characterized, and some of these proteins may fulfill key mitochondrial functions. For example, the proteins of the intermembrane space fall into this low-abundance category, and some intermembrane space proteins are crucially important in the execution of the cell death program (Green and Reed, 1998; Balk et al., 1999). Similarly, the outer mitochondrial membrane is essential to mitochondrial function (Vander Heiden et al., 2000), but its protein complement remains almost completely uncharacterized as a result of the predominance of porin (Werhahn et al., 2001).
Given the potential importance of low-abundance mitochondrial proteins, a proteomic strategy was designed specifically to identify low-abundance mitochondrial proteins. We took two different approaches. In the first, liquid chromatography rather than electrophoresis was used to fractionate proteins before identification by mass spectrometry. This method can dramatically increase the number of proteins that can be identified in a single analysis, is highly sensitive, and overcomes many of the aforementioned limitations of electrophoretic separations (Washburn et al., 2001; Wolters et al., 2001; Koller et al., 2002). The second approach was to prefractionate the mitochondrial proteome to isolate low-abundance intermembrane space and outer membrane proteins. This prefractionation reduces the complexity of the protein sample and increases the representation of low-abundance proteins relative to total protein in the fraction. Proteins present in these fractions were further resolved using two-dimensional gel electrophoresis and identified by tandem mass spectrometry (MS/MS). An unexpected finding from these two complementary approaches was that the enzymes of glycolysis are present at low abundance in mitochondrial fractions from Arabidopsis cells. The presence of these enzymes was confirmed by enzyme assays, and protease treatments of intact mitochondria indicated that the enzymes were attached to the outside of the mitochondrion. Furthermore, an analysis of the fate of 13C-fructose-1,6-bisphosphate or 13C-glucose supplied to isolated mitochondria demonstrated that these enzymes retain the capacity to function as an intact glycolytic sequence in vitro. We hypothesize that the glycolytic sequence may be attached loosely to the outside of mitochondria and may function to ensure the provision of a localized supply of pyruvate to support mitochondrial respiration.
RESULTS
A Purified Arabidopsis Mitochondrial Fraction Contains Glycolytic Proteins
As part of an ongoing project to increase our knowledge of the plant mitochondrial proteome, we have identified mitochondrial proteins purified from a dark-grown Arabidopsis cell suspension culture (Millar et al., 2001). Mitochondria were purified using two sequential Percoll gradients to yield mitochondria that are free of contaminating cytosol and peroxisomes and that contain only a slight contamination of plastids (plastid contamination of the mitochondrial fraction was <0.2% [Millar et al., 2001]). To increase sensitivity and overcome the limitations of two-dimensional gel electrophoresis, proteins were identified by separating tryptic peptides of total mitochondrial protein with reverse-phase HPLC and direct elution of the separated peptides into an Applied Biosystems QSTAR mass spectrometer (LC-MS). Collision-induced dissociation spectra of selected doubly or triply charged peptides were used to identify proteins by matching to the TIGR Arabidopsis protein set (ATH1a.pep, release 3.0, July 2002) using ProID software (Applied Biosystems). Among the several hundred mitochondrial proteins that were identified (data not shown) were a number of the enzymes of glycolysis (Table 1).
Table 1.
Identification of Glycolytic Proteins by LC-MS/MS Analysis of Purified Arabidopsis Mitochondria
| Enzyme | AGI Number | Matched Sequence | Experiments |
|---|---|---|---|
| Hexokinase | At1g50460 | MLLTFVDDLPTGR | 3 |
| ELAFTFSFPVK | 1 | ||
| At2g19860 | VGLDMLVAALVNDTIGTLAGGR | 2 | |
| GARLSAAGIYGILK | 1 | ||
| IISGMYLGEILR | 1 | ||
| ELGFTFSFPVK | 1 | ||
| Aldolase | At2g36460 | LGEGAAESLHVK | 1 |
| NLNAMNQLK | 1 | ||
| At3g52930 | NLNAMNQLK | 1 | |
| Triosephosphate isomerase | At3g55440 | ELGGQADVDGFLVGGASLKPEFIDIIK | 1 |
| AILNESSEFVGDK | 1 | ||
| Glyceraldehyde-3-P dehydrogenase | At3g04120 | TLLFGEKPVTVFGIR | 1 |
| At1g13440 | AASFNIIPSSTGAAK | 1 | |
| Phosphoglycerate mutase | At1g09780 | RGWDAQVLGEAPHK | 2 |
| Enolase | At2g36530 | AVGNVNNIIGPALIGK | 1 |
| Pyruvate kinase | At5g52920 | AVIVATNMLESMIVHPTPTR | 1 |
| At3g55650 | AGMNVARFNFSHGSHSYHQETLDNLR | 1 | |
| GDLGMEIPIEKMFLAQK | 1 |
AGI number is the locus identifier given by the Arabidopsis Genome Initiative. Experiments indicate the number of independent experiments in which the same peptide was identified. Total mitochondrial protein from Arabidopsis was digested with trypsin, and the resulting peptides were separated by reverse-phase HPLC with direct elution into an Electrospray Ionisation-Quadrupole time of flight mass spectrometer. Spectra obtained after collision-induced dissociation were matched to translated Arabidopsis open reading frames.
In total, we identified 7 of the 10 enzymes that constitute the glycolytic pathway. With the exception of pyruvate kinase, none of the identified enzymes contained N-terminal extensions typical of organellar targeting signals, indicating that these proteins are the cytosolic rather than the plastidic isoforms of the glycolytic enzymes (Target P; TIGR [http://www.tigr.org]; data not shown). Two isoforms of pyruvate kinase were identified. One (At5g52920) contains a predicted organellar targeting presequence and is predicted by Target P to be targeted to the plastid (P = 0.885). The second (At3g55650) does not contain the N-terminal extension typical of an organellar targeting sequence and probably represents a cytosolic isoform. Two isoforms of hexokinase were identified, representing the products of a putative hexokinase (At1g50460) and AtHX2 (At2g19860) (Jang et al., 1997). Two isoforms of cytosolic aldolase also were identified. Two peptides matching glyceraldehyde-3-P dehydrogenase were not discriminatory between the products of genes At3g04120 and At1g13440. For triosephosphate isomerase, phosphoglycerate mutase, and enolase, only single gene products were identified.
A Purified Subfraction of Arabidopsis Mitochondria Consisting of Proteins of the Intermembrane Space and Outer Mitochondrial Membrane Contains Glycolytic Proteins
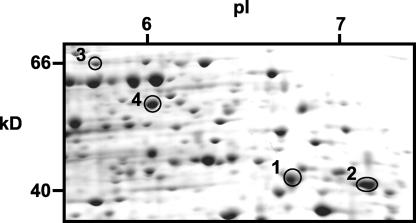
To establish the submitochondrial location of mitochondrial proteins, we undertook a proteomic investigation of specific mitochondrial subfractions. Subfractionation of mitochondria was achieved via osmotic shock to rupture the outer membrane and by subsequent separation of mitoplasts from released intermembrane space (IMS) and outer mitochondrial membrane (OMM) by centrifugation (Sweetlove et al., 2001). A further centrifugation step was used to pellet OMM fragments and leave a supernatant containing IMS proteins. However, this supernatant also is likely to contain OMM proteins released into the solution by the rupturing of the outer membrane, and we consider this an IMS/OMM fraction. This IMS/OMM protein fraction was fractionated further by two-dimensional gel electrophoresis (Figure 1). Selected protein spots were excised and identified by MS/MS. An initial survey of 50 protein spots revealed the presence of four glycolytic enzymes in this protein fraction (Table 2). These were aldolase, glyceraldehyde-3-P dehydrogenase, phosphoglycerate mutase, and enolase. These enzymes matched gene products found in the LC-MS survey of total mitochondrial proteins (Table 1).
Figure 1.
Proteins of the IMS and OMM Separated by Two-Dimensional Gel Electrophoresis.
An IMS/OMM fraction was prepared from isolated Arabidopsis mitochondria by osmotic shock and centrifugation (for details, see Methods). Proteins were separated by isoelectric focusing on 3 to 10 nonlinear immobilized pH gradients for the first dimension and by SDS-PAGE for the second dimension. For clarity, only the relevant section of the gel is shown. The numbered spots are those that have been identified by MS/MS (see Table 2).
Table 2.
Identification of Glycolytic Proteins in an IMS/OMM Fraction of Arabidopsis Mitochondria
| Enzyme | Spot No. | AGI Number | No. MP (P<0.05) | Match MM/pI | Gel MM/pI |
|---|---|---|---|---|---|
| Aldolase | 1 | At3g52930 | 8 | 38,858/6.05 | 40,000/6.8 |
| Glyceraldehyde-3-P dehydrogenase | 2 | At3g04120 | 3 | 37,077/6.34 | 40,000/7.3 |
| Phosphoglycerate mutase | 3 | At1g09780 | 4 | 60,857/5.32 | 66,000/5.7 |
| Enolase | 4 | At2g36530 | 13 | 47,947/5.54 | 57,000/6.0 |
Spot number refers to the protein spot excised from the gel shown in Figure 1. AGI number is the locus identifier given by the Arabidopsis Genome Initiative. No. MP refers to the number of peptides matching the same protein (at P < 0.05). Match MM/pI indicates the predicted molecular mass/pI of the translated open reading frame. Gel MM/pI indicates the molecular mass/pI observed on the gel shown in Figure 1. IMS/OMM was isolated from isolated Arabidopsis mitochondria by osmotic shock and centrifugation. A total of 500 μg of IMS/OMM protein was fractionated by isoelectric focusing and SDS-PAGE. The indicated protein spots were excised from the gel, digested with trypsin, and analyzed by MS/MS. Spectra obtained after collision-induced dissociation were matched to translated Arabidopsis open reading frames.
Glycolytic Proteins Are Associated with Mitochondria in Vivo
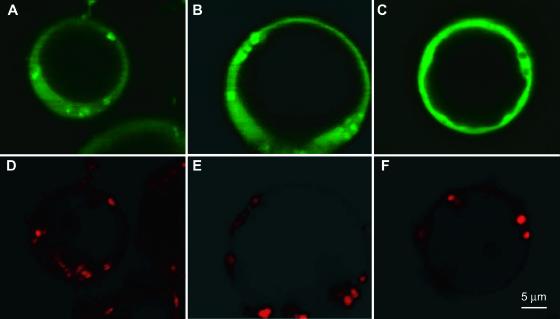
To determine whether the in vitro association of glycolytic enzymes with mitochondria also occurs in vivo, we constructed protein–yellow fluorescent protein (YFP) fusions for two of the identified glycolytic proteins, enolase and aldolase. Full-length cDNAs encoding enolase (At2g36530) and aldolase (At3g52930) were cloned into the plant expression vector pGHiB between the 35S promotor of Cauliflower mosaic virus and the 3′ terminus with a cDNA encoding YFP. These constructs were introduced into Arabidopsis protoplasts by polyethylene glycol–mediated transformation, and after 48 h, transformed protoplasts were loaded with the mitochondria-selective probe Mitotracker Red (Molecular Probes). Fluorescence from Mitotracker Red and YFP was visualized by confocal microscopy. The YFP signal for both aldolase- and enolase-YFP was present throughout the cytosol (Figures 2A and 2B). However, we also found a number of concentrations of the YFP signal in small (<1 μm diameter) circular or rod-shaped bodies (Figures 2A and 2B) that colocalized with Mitotracker Red (Figures 2D and 2E). Thus, aldolase- and enolase-YFP fusion proteins were located in the cytosol and associated with mitochondria. By contrast, YFP alone was located only in the cytosol and showed no association with mitochondria (Figures 2C and 2F).
Figure 2.
Localization of Aldolase and Enolase in Arabidopsis Protoplasts.
Arabidopsis protoplasts were transformed with constructs for the expression of an aldolase-YFP fusion, an enolase-YFP fusion, and YFP alone (for more details, see Methods). YFP fluorescence was visualized using a confocal microscope and compared with the distribution of the mitochondrion-specific probe Mitotracker Red (Molecular Probes).
(A) to (C) YFP fluorescence.
(D) to (F) Mitotracker Red fluorescence.
(A) and (D) show a protoplast expressing aldolase::YFP, (B) and (E) show a protoplast expressing enolase::YFP, and (C) and (F) show a protoplast expressing YFP alone as a control.
Purified Mitochondrial Preparations Contain Significant Activities of All Enzymes of Glycolysis
The proteomic analysis identified 7 of the 10 glycolytic enzymes. To confirm these results and to investigate whether the remaining three enzymes (phosphoglucose isomerase, phosphofructokinase, and phosphoglycerate kinase) also are present, we used standard spectrophotometric assays to measure the activity of the glycolytic enzymes in preparations of isolated mitochondria (Table 3). We found detectable activity of each of the 10 enzymes that constitute the glycolytic pathway. By comparing the activity of each enzyme recovered in the mitochondrial fraction with that in the crude cell homogenate, we were able to quantitate the proportion of each glycolytic enzyme that is associated with the mitochondrion. Activities of each enzyme measured in the mitochondrial fraction were expressed as a percentage of the activity in the crude cell homogenate and then recalculated to account for mitochondrial yield (Table 3).
Table 3.
Activities of Glycolytic Enzymes in Preparations of Arabidopsis Mitochondria
| Enzyme | Activity in the Mitochondrial Fraction (nmol·min−1·mg−1) |
Percentage of Total Activity Recovered in the Mitochondrial Fraction (% of total homogenate) |
Percentage of Total Activity Associated with Mitochondria |
Activity in the Mitochondrial Fraction after Treatment with Proteinase K (nmol·min−1·mg−1) |
|---|---|---|---|---|
| Alcohol dehydrogenase | 13 ± 4 | 0.01 ± 0.00 | 0 | − |
| UDP-glucose pyrophosphorylase | N.D. | N.D. | 0 | − |
| Hexokinase | 59 ± 5 | 0.6 ± 0.1 | 12 | N.D. |
| Phosphoglucose isomerase | 69 ± 3 | 0.2 ± 0.0 | 5 | N.D. |
| Phosphofructokinase | 12 ± 2 | 0.2 ± 0.0 | 3 | N.D. |
| Aldolase | 23 ± 2 | 0.2 ± 0.0 | 3 | N.D. |
| Triosephosphate isomerase | 266 ± 19 | 0.1 ± 0.0 | 3 | N.D. |
| Glyceraldehyde-3 P dehydrogenase | 48 ± 8 | 0.6 ± 0.2 | 10 | N.D. |
| Phosphoglycerate kinase | 19 ± 1 | 0.2 ± 0.0 | 4 | N.D. |
| Phosphoglyceromutase | 98 ± 8 | 0.2 ± 0.1 | 3 | N.D. |
| Enolase | 28 ± 2 | 0.2 ± 0.0 | 3 | N.D. |
| Pyruvate kinase | 17 ± 2 | 0.2 ± 0.0 | 5 | N.D. |
Glycolytic enzyme activities were assayed in a crude cell homogenate (total homogenate) or in mitochondria isolated from that cell homogenate. Alcohol dehydrogenase and UDP-glucose pyrophosphorylase also were assayed to give an indication of the cytosolic contamination of the mitochondrial preparation. Values are means ± se from three measurements. The percentage of each enzyme activity associated with mitochondria is calculated on the basis of the amount of total enzyme activity recovered in the mitochondrial fraction and by considering mitochondrial yield (for more details, see Methods). The accessibility of these enzymes to protease digestion was determined by the treatment of intact mitochondria with proteinase K and subsequent reassay of each enzyme. N.D., not detected.
The percentage of each enzyme associated with the mitochondrion varied between 3 and 12%. For two of the enzymes, hexokinase and glyceraldehyde-3-P dehydrogenase, there was a markedly higher proportion associated with the mitochondrion (12 and 10%, respectively), with between 3 and 5% of each of the other enzymes being mitochondrially associated. By contrast, the cytosolic marker enzyme UDP-glucose pyrophosphorylase was not detected in the mitochondrial fraction, indicating that the presence of glycolytic enzymes does not merely reflect cytosolic contamination. A small but detectable activity of another cytosolic marker enzyme, alcohol dehydrogenase, was observed, but the percentage of this activity associated with the mitochondrion was 1 order of magnitude lower than that of the glycolytic enzymes (Table 3). It is not clear why a small activity of alcohol dehydrogenase was detected but there was no detectable activity of UDP-glucose pyrophosphorylase. One possibility is that a small proportion of alcohol dehydrogenase also is associated specifically with the mitochondrion.
Glycolytic Enzymes Are Located on the Outside of the Mitochondrion
Subfractionation of mitochondria indicated that at least four of the glycolytic enzymes are located either in the mitochondrial IMS or associated with the OMM, but it was not possible to discriminate between the two locations. To resolve this issue of localization, the sensitivity of the glycolytic enzymes to protease treatment of intact mitochondria was investigated. If the enzymes are localized in the IMS, then they will be protected from protease digestion by the presence of the OMM. If, however, the enzymes are bound to the cytosolic face of the outer membrane, then they will be sensitive to protease digestion. We assayed for glycolytic enzyme activity in a mitochondrial sample that had been treated with proteinase K (Table 3). We detected no glycolytic enzymes after this treatment, indicating that the enzymes are located on the outside of the mitochondrion. As a control, we assayed the activity of cytochrome c oxidase (which is located in the IMS) and found that this activity was unaffected by the protease treatment (data not shown).
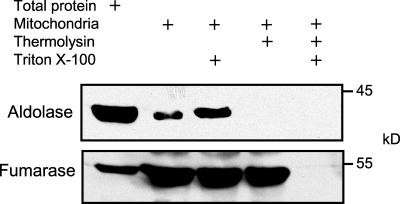
A complementary experiment was conducted in which the protease sensitivity of aldolase was assessed using an antiserum that specifically cross-reacts with cytosolic castor bean aldolase (Hodgson and Plaxton, 1998) (Figure 3). It was possible to detect a single protein with a molecular mass of 40,000 kD, which corresponds to the molecular mass of the aldolase that was observed in Arabidopsis mitochondria samples (Tables 1 and 2). This protein band was detected both in a total protein sample and also in a purified mitochondria sample (Figure 3). However, this band was not detected when mitochondria were incubated with thermolysin in the absence of detergent, indicating that aldolase is present on the outside of mitochondria (Figure 3). By contrast, the mitochondrial matrix enzyme fumarase was digested by thermolysin only when detergent was added, confirming that proteins that are contained within the mitochondrion are protected from protease digestion.
Figure 3.
Submitochondrial Localization of Aldolase Is Revealed by a Protease Protection Assay.
The protease thermolysin was added to isolated mitochondria in the presence or absence of Triton X-100 to rupture the mitochondrial membranes. The reaction was terminated by the addition of EGTA, and 50 μg of the protein sample was fractionated by SDS-PAGE before being transferred to a nitrocellulose membrane. The membrane was probed with antisera against aldolase and fumarase.
Metabolism of 1-13C–Fructose-1,6-Bisphosphate or U-13C–Glucose by Isolated Mitochondria Suggests the Presence of a Complete and Functional Glycolytic Pathway
The association of the enzymes of glycolysis with mitochondria does not necessarily mean that there is a functional glycolytic pathway, because many of these enzymes have alternative functions (Jeffery, 1999). To investigate the functionality of the glycolytic pathway that is associated with isolated Arabidopsis mitochondria, we conducted 13C-labeling experiments, supplying 13C-labeled glycolytic intermediates to isolated mitochondria. By analyzing the fate of the label using gas chromatography (GC)–MS, it was possible to investigate the metabolic activity of the glycolytic enzymes in these preparations. Although GC-MS does not allow the direct detection of glycolytic intermediates other than hexose phosphates and 3-phosphoglyceric acid, it can detect trichloroacetic acid (TCA)–cycle acids that result from the metabolism of glycolytically produced pyruvate. Thus, the appearance of 13C-labeled TCA-cycle acids when isolated mitochondria are supplied with 13C-labeled glycolytic precursors or intermediates would provide evidence for the operation of glycolysis.
We initially supplied 1-13C–fructose-1,6-bisphosphate to isolated mitochondria. Mitochondria were resuspended at a high concentration (∼80 μg mitochondrial protein/μL) in an osmotically balanced medium containing 1-13C–fructose-1,6-bisphosphate and the essential glycolytic cofactors ATP, ADP, and NAD+. These cofactors were supplied at concentrations equivalent to those found in vivo in heterotrophic plant cells (Farre et al., 2001). We also supplied thiamine pyrophosphate, which is an essential cofactor of the mitochondrial pyruvate dehydrogenase complex, inorganic phosphate to support mitochondrial ATP synthesis, and citrate (unlabeled) to initiate the TCA cycle. Incubation was for 16 h to ensure isotopic equilibration. At the end of the incubation, metabolites were extracted using methanol, and the presence of 13C-isotopomers was detected by GC-MS (Table 4).
Table 4.
Metabolism of 1-13C–Fructose-1,6-Bisphosphate and U-13C–Glucose by Isolated Arabidopsis Mitochondria
| Molar Percentage Enrichment of 13C
|
||
|---|---|---|
| Metabolite | From 1-13C–Fructose-1 6-Bisphosphate |
From U-13C Glucose |
| Fructose-6-P | 1.2 | 3.6 |
| Glucose-6-P | 1.4 | 7.6 |
| 3-Phosphoglyceric acid | 2.1 | N.D. |
| Citric acid | 2.2 | 12.3 |
| Succinic acid | 1.1 | 1.1 |
| Fumaric acid | 1.2 | 1.3 |
| Malic acid | 1.2 | 1.6 |
| Fatty acid 18:0 | 1.1 | 1.2 |
| Gluconic acid | 2.5 | 21.4 |
| Ala | 2.6 | 1.8 |
Isolated mitochondria were incubated with either 50 mM 1-13C–fructose-1,6-bisphosphate or 50 mM U-13C–glucose in 0.3 M mannitol, 10 mM Tes-NaOH, pH 7.5, 3 mM MgSO4, 10 mM NaCl, and 5 mM KH2PO4 containing the following cofactors: 0.6 mM ATP, 0.2 mM ADP, 0.3 mM NAD, 0.1 mM thiamine pyrophosphate, and 10 mM citrate. The final reaction mixture contained 5 mg of mitochondrial protein in a final volume of 60 μL. The reaction was incubated at 25°C for 16 h, and 13C-labeled metabolites were detected by GC-MS. N.D, not detected.
We detected a number of 13C-isotopomers, indicating that metabolism of the supplied label had occurred. The presence of label in the glycolytic intermediate 3-phosphoglyceric acid indicates that glycolytic metabolism downstream of aldolase had occurred. Furthermore, the presence of the labeled TCA-cycle acids citric, succinic, fumaric, and malic acid suggests that the supplied label had been metabolized right through to pyruvate, which then was taken up by the mitochondria. The presence of labeled glucose- and fructose-6-P suggests that gluconeogenic metabolism of fructose-1,6-bisphosphate also had occurred and indicates the presence of the enzyme pyrophosphate-fructose 6-phosphate phosphotransferase or possibly fructose-1,6-bisphosphatase. We also found 13C-isotopomers of a fatty acid (18:0), gluconic acid, and the amino acid Ala. It is not clear how label was incorporated into fatty acid, because this requires the uptake of malonate into the mitochondrion (Gueguen et al., 2000); however, it is possible that sufficient endogenous malonate is present in the isolated mitochondria to support this flux. The significant labeling of gluconic acid most likely is the result of the presence of labeled 6-phosphogluconate and indicates the presence of the enzyme 6-phosphogluconate dehydrogenase.
To determine whether the complete glycolytic pathway was present in the isolated mitochondrial fraction, we also supplied U-13C–glucose under the same conditions. Similar results were obtained as with 1-13C–fructose-1,6-bisphosphate, with extensive labeling of TCA-cycle acids, indicating the presence of a complete glycolytic pathway (Table 4). As a control, labeled glucose also was supplied in the absence of the glycolytic cofactors NAD+ and ATP. In the absence of these cofactors, we detected no labeling of TCA-cycle acids (data not shown). This finding demonstrates that glycolytic flux is responsible for the provision of labeled substrate to the mitochondrion. In addition, this control experiment allowed us to discount the possibility that bacteria might be responsible for the observed metabolism of glucose (because bacteria would not require the addition of exogenous glycolytic cofactors to metabolize glucose to TCA-cycle acids). Based on the labeling of fumarate, we estimate that the flux through the mitochondrial glycolytic pathway was 3 nmol·min−1·mg−1 mitochondrial protein. This is considerably less than the maximal rate of pyruvate oxidation by isolated mitochondria (which in our hands was ∼30 nmol·min−1·mg−1 mitochondrial protein). However, the flux estimate is likely to be an underestimate, because fumarate is not an end product. Moreover, the glycolytic pathway may not operate as efficiently in the in vitro conditions of our experiments as it would in vivo.
DISCUSSION
As part of a more extensive investigation of the plant mitochondrial proteome, we have been using mass spectrometry to identify proteins in a mitochondrial fraction of Arabidopsis cells (Millar et al., 2001). Here, we present the unexpected finding that seven glycolytic enzymes are present in this fraction, raising the possibility that the glycolytic pathway is associated with mitochondria, possibly to ensure direct delivery of pyruvate to the site of its consumption. As with any such analysis of proteins associated with purified organelles, the possibility of contamination by other subcellular compartments remains. However, our method of purification of mitochondria from Arabidopsis cells provides a much higher level of purity than is generally achieved: two sequential density gradients each followed by a series of sedimentary wash centrifugation steps were used to isolate mitochondria that are entirely free of cytosol and peroxisomes and that contain <0.2% plastid contamination (Millar et al., 2001).
Although slight, this plastid contamination could be significant, because the enzymes of glycolysis are present either as cytosolic or plastidic isozymes (Dennis et al., 1997); therefore, it is possible that the glycolytic enzymes we observed are simply contaminating enzymes from the plastid. There are two lines of evidence that lead us to believe that this is not the case. First, of the seven glycolytic enzymes, we identified isoforms of each that match to entries in the TIGR Arabidopsis protein database (http://www.tigr.org) that are annotated as cytosolic. These annotations are based on the lack of N-terminal organellar targeting sequences in these proteins and allow a clear distinction to be made between cytosolic and plastidic glycolytic enzymes (the latter all contain N-terminal targeting sequences). We did find one isoform of pyruvate kinase that contained a plastidic targeting sequence, but we emphasize that we also found a second cytosolic isoform. It is not clear whether the plastidic isoform of pyruvate kinase is the result of slight plastidic contamination or whether this protein is targeted to both plastid and mitochondrion (Small et al., 1998). The second line of evidence that the glycolytic enzymes are not plastidic in origin comes from the expression of YFP fusion proteins in protoplasts. YFP fusions of enolase and aldolase clearly were associated with mitochondria as well as distributed throughout the cytosol (Figure 2). We found no evidence that these proteins were plastidic in location.
Because the majority of the glycolytic proteins in the Arabidopsis mitochondrial fraction do not have organellar targeting sequences, the question arises of how the association of these proteins with the mitochondrion came about and which compartment of the mitochondrion they reside in. We identified four of the glycolytic proteins (aldolase, glyceraldehyde-3-P dehydrogenase, phosphoglycerate mutase, and enolase) in a subfraction of mitochondria that consists of IMS and OMM proteins (Table 2). Because our analysis of the IMS/OMM proteome was not exhaustive, it is possible that the other glycolytic proteins also are present in this compartment. Given that the uptake of nucleus-encoded proteins into the mitochondrial IMS requires targeting sequences (Diekert et al., 1999) that are not present in these proteins, we favored the interpretation that these glycolytic proteins are attached to the outside of the mitochondrion. The fact that the incubation of isolated mitochondria with a nonspecific protease (in the absence of detergent) led to the loss of detectable activity of each of the 10 glycolytic enzymes confirmed this hypothesis (Table 3). Quantitative assessments of the activity of glycolytic enzymes in the mitochondrial fraction demonstrated that every enzyme in the glycolytic pathway is present and that between 3 and 12% of the total cellular activity of these enzymes is associated with the mitochondrion. This is a significant proportion of the total glycolytic capacity, and the proportion of this pathway associated with the mitochondrion in vivo may be even higher, because it is likely that substantial amounts of the glycolytic enzymes are washed off or dislodged during the isolation of the mitochondria from the other cellular constituents.
The mechanism by which glycolytic enzymes might be attached to the OMM is not known, although several of the enzymes have predicted hydrophobic regions that could form membrane-spanning domains (data not shown). Alternatively, other protein–lipid interactions may occur. It is interesting that the hexo- kinase (At1g50460) has a predicted membrane-lipoprotein lipid attachment site (Prosite; http://ca.expasy.org/prosite). Indeed, the mitochondrial association of hexokinase with the OMM has long been recognized in animal systems, in which it is thought that hexokinase interacts with the outer membrane porin (Wilson, 1980). It is possible that plant hexokinase is anchored similarly to the OMM (Galina et al., 1995) and that the other glycolytic enzymes are tethered by protein–protein interactions with hexokinase and/or each other. It also has been suggested that cytosolic proteins with a basic pI may interact electrostatically with the phospholipids of the OMM (Hartmann et al., 1993).
The idea that glycolytic enzymes might bind to organelles or might be concentrated in regions of high demand for ATP or pyruvate is not without precedent. As well as the association of hexokinase with mitochondria, glyceraldehyde-3-P dehydrogenase and phosphofructokinase also have been shown to be bound to mitochondria in Tetrahymena pyriformis (Srere, 1987). In addition, aldolase has been observed to associate with the cytoskeleton in muscle tissue (Pagliaro and Taylor, 1988), possibly as a rapid means of regulating glycolysis according to substrate supply (Ali et al., 1998). Aldolase also has been reported to interact with V-type ATPases, leading to the suggestion that glycolysis may be localized to regions of high ATP demand (Lu et al., 2001). In plants, phosphoglycerate kinase, glyceraldehyde-3-P dehydrogenase, and aldolase have been shown to be present in the nuclei of pea leaf cells (Anderson et al., 1995), and glyceraldehyde-3-P dehydrogenase and aldolase also were shown to associate with the cytoskeleton in developing maize endosperm (Azama et al., 2003). Very recently, glycolytic enzymes were identified in a proteomic survey of human mitochondria (Taylor et al., 2003), although the functional significance of this observation was not investigated further. We demonstrate here that the entire glycolytic pathway is associated with plant mitochondria. For two enzymes, enolase and aldolase, this association has been confirmed in vivo. Furthermore, we demonstrate that the association of glycolytic enzymes with mitochondria may be functionally significant.
The logical explanation for the association of glycolytic enzymes with the mitochondrion is that the complete glycolytic pathway is associated with the OMM or IMS to ensure the provision of pyruvate directly to its site of consumption. By forming an association between glycolysis and the mitochondrion, the enzymes of glycolysis could provide pyruvate at a high concentration directly to the mitochondrion, where it will be taken up as a substrate for respiration. In support of this idea, we provide evidence that highly purified mitochondria are capable of the metabolism of glucose to form TCA-cycle intermediates (Table 4). We supplied 13C-labeled fructose-1,6-bisphosphate or glucose plus appropriate cofactors to isolated mitochondria and after a 16-h incubation determined the distribution of the label using GC-MS. Mitochondria were isolated from sterile cell suspension cultures, and sterile solutions were used throughout to minimize contamination of the mitochondrial fraction with bacteria. The fact that we detected no metabolites, labeled or unlabeled, that are uniquely produced by bacteria suggests that bacterial metabolism does not contribute significantly to the observed distribution of 13C. In addition, the fact that we detected no label in TCA-cycle acids when exogenous glycolytic cofactors were omitted from the incubation further suggests that it is unlikely that bacteria are responsible for the observed metabolism.
In the presence of glycolytic cofactors, the appearance of label in TCA-cycle intermediates is strongly suggestive of the metabolism of glucose to pyruvate, which is then taken up by the mitochondria to enter the TCA cycle. With the exception of hexose phosphates and 3-phosphoglyceric acid, we detected no labeled glycolytic intermediates. This is most likely caused by a quantitative loss of these metabolites during the derivatization process required for GC-MS analysis. Although the mitochondrial fraction we used was highly purified, it was slightly contaminated by plastids; therefore, it is possible that the observed metabolism of glucose could occur in part in the plastid. Although plastids cannot metabolize glucose directly, they can take up and metabolize the glucose-6-P that would be produced by a mitochondrial hexokinase. Plastidic isoforms of glycolytic enzymes then could metabolize glucose-6-P to form pyruvate, which could leave the plastid on a phosphoenolpyruvate/pyruvate transporter and then enter the mitochondrion. However, because of kinetic considerations regarding plastid transporter proteins, we consider this metabolic route to be extremely unlikely, because the high concentration of inorganic phosphate (5 mM) in the incubation medium would effectively inhibit glucose-6-P uptake into the plastid by competing for the binding site of the plastidic hexose phosphate transporter (Fischer and Weber, 2002).
In summary, we have provided evidence that the enzymes of glycolysis are present in a highly purified mitochondrial fraction of Arabidopsis cells. Protease protection assays reveal that these enzymes are associated with the outside of the mitochondrion. The association of aldolase and enolase with mitochondria was confirmed in vivo by the expression of YFP fusion proteins in protoplasts. On the basis of these data, we propose that the glycolytic pathway can associate with the outside of the mitochondrion in plant cells. 13C-labeling experiments are consistent with the idea that these enzymes form a complete and functional glycolytic sequence. It is conceivable that these enzymes interact directly with one another through protein–protein binding to form a glycolytic metabolon (Srere, 1987; Moorhead and Plaxton, 1992; Moorhead et al., 1994), although we provide no direct evidence for this. Alternatively, given that glycolytic enzymes have many well-characterized secondary functions, it is possible that the association of these enzymes with the mitochondrion is entirely unconnected with glycolysis. For example, glyceraldehyde-3-P dehydrogenase also functions as a protein phosphotransferase/kinase and plays roles in DNA repair and membrane fusion (Sirover, 1999). Similarly, phosphoglucose isomerase also has a number of secondary functions that fulfill roles in cellular differentiation and tumor cell motility (Jeffery, 1999). In general, these secondary functions of glycolytic enzymes are regulatory in nature (Jeffery, 1999); therefore, it is conceivable that glycolytic proteins may be important in regulating mitochondrial functions that require ancillary outer membrane proteins (such as tRNA uptake and mitochondrial division).
METHODS
Chemicals
Unless stated otherwise, all chemicals were from Sigma-Aldrich (Poole, UK). 13C isotopes were from Campro Scientific (Berlin, Germany). Mitotracker Red was from Molecular Probes Europe (Leiden, The Netherlands). Coupling enzymes were from Roche Diagnostics (Lewes, UK).
Plant Material
Heterotrophic Arabidopsis thaliana cells were cultured in 250-mL conical flasks under aseptic conditions in Murashige and Skoog (1962) basal medium supplemented with 3% (w/v) sucrose, 0.5 mg/L naphthaleneacetic acid, and 0.05 mg/L kinetin (May and Leaver, 1993). The cell cultures were maintained in the dark at 22°C with shaking at 150 rpm. At 6 to 7 days, each flask (120 mL) contained 8 to 10 g fresh weight of cells, and growth was approximately in the middle of the log phase. Subculture of 20 mL of culture to 100 mL of fresh medium began the cycle again.
Isolation and Subfractionation of Mitochondria
Highly purified mitochondria were isolated using two sequential density gradients according to Millar et al. (2001). Subfractionation of mitochondria to obtain an intermembrane space/outer mitochondrial membrane fraction was exactly as described by Sweetlove et al. (2001).
Two-Dimensional Gel Electrophoresis and Identification of Proteins by Mass Spectrometry
Proteins were fractionated using isoelectric focusing as a first dimension and SDS-PAGE as a second dimension, as described by Millar et al. (2001). Proteins were excised from the gel, digested with trypsin, and identified using tandem mass spectrometry (MS/MS) according to Sweetlove et al. (2002). Alternatively, aliquots of 50 μg of mitochondrial protein were precipitated in acetone, and the protein pellets were air-dried. A digestion solution consisting of 100 mM Tris-HCl, pH 8.6, and 50 μg/mL trypsin was added to a final volume of 45 μL and incubated for 16 h at 37°C. Peptide extracts were bound onto a microbore HPLC C18 column (Agilent, Palo Alto, CA) and eluted over 6 h with a linear acetonitrile gradient from 2 to 80% (v/v) in water. Mass spectra and collision MS/MS data from elution runs were analyzed with Analyst QS and BioAnalyst software (Applied Biosystems, Sydney, Australia). An in-house database comprising the TIGR and NCBI Arabidopsis protein sets was searched with resulting MS/MS data at error tolerances of ±0.15 for MS and ±0.05 for MS/MS. Where MS/MS peptide matches were <98%, manual interpretations of the spectra were performed to confirm the protein match.
Expression of Yellow Fluorescent Protein Fusions of Enolase and Aldolase in Arabidopsis Protoplasts
Aldolase and enolase full-length sequences were amplified from Arabidopsis total cDNA with the specific primers ALN5 (5′-TCACCCGGGACACAATGTCTGCCTTCACAAGC-3′) and ALN3 (5′-CGTCTAGAGTACTTGTAATCCTTCAC-3′) for aldolase and ENN5 (5′-ATCCCC-GGGACACAATGGCTACTATCACCGTTG-3′) and ENN3 (5′-CTTCTAGAGTAGGGTTCCACAGGTTTG-3′) for enolase and cloned into SmaI and XbaI sites in frame with the N terminus of yellow fluorescent protein (YFP) in a pGHiB-derived binary vector (obtained from H. Batoko, Department of Plant Sciences, University of Oxford, UK). To prepare protoplasts, 20 mL of a 6-day-old Arabidopsis cell suspension culture was centrifuged at 42g for 10 min at room temperature, and the cells were resuspended carefully in plasmolysis solution (0.4 M mannitol, 3% [w/v] sucrose, and 8 mM CaCl2, pH 5.6) and incubated for 30 min at room temperature. Cells then were pelleted by centrifugation at 42g for 10 min at room temperature, resuspended in enzyme solution (1% [w/v] cellulase and 0.25% [w/v] macerozyme diluted in plasmolysis solution), and incubated in the dark at room temperature for 1 h and for another 1 to 2 h on an orbital shaker at 20 rpm.
The resulting protoplasts were filtered through a nylon mesh (100 μm), washed by the addition of 30 mL of 0.4 M mannitol/W5 solution (5 mM glucose, 154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 1.5 mM Mes, pH 5.6), and centrifuged at 42g for 10 min at room temperature. Protoplasts then were washed twice in 10 mL of mannitol/Mg solution (0.4 M mannitol, 0.1% [w/v] Mes, and 15 mM MgCl2, pH 5.7) and finally resuspended in 10 mL of mannitol/Mg solution. To transform protoplasts, 50 μg of plasmid DNA plus 200 μg of herring sperm carrier DNA, cleaned by three cycles of phenol/chloroform extraction and ethanol precipitation, were resuspended in 50 μL of water. This solution was deposited in droplets on a Petri dish next to 300 μL of protoplast solution (2 × 106 protoplasts) and 350 μL of polyethylene glycol (PEG) solution (0.4 M mannitol, 100 mM CaNO3, and 40% [w/v] PEG-4000). The protoplasts were gently brought together with the DNA and then the PEG-CMS solutions. Protoplasts then were diluted successively with 600 μL, 1 mL, 2 mL, and 4 mL of 0.4 M mannitol/W5 solution added in droplets every 3 min. The diluted protoplasts were harvested by centrifugation at 20g for 5 min at room temperature. Transformed protoplasts were resuspended in 2 mL of culture medium (0.4 M sucrose, 1× Murashige and Skoog [1962] basal medium, and 250 mg/L xylose) and cultured in the dark at 20°C for up to 48 h. Transformed protoplasts were visualized by confocal microscopy with a CLSM 410 confocal microscope (Zeiss, Wellwyn, UK) set to measure an emission band of 520 to 565 nm for YFP and an emission band of 599 nm for Mitotracker Red. The software LSM dummy (Zeiss) was used for postacquisition image processing.
Enzyme Assays
Glycolytic enzymes were assayed spectrophotometrically according to Burrell et al. (1994). Hexokinase was assayed according to Dai et al. (1999). UDP-glucose and alcohol dehydrogenase were assayed according to Sweetlove et al. (1996). Enzyme activities were measured in a crude cell homogenate (“total homogenate”) and in purified mitochondria. Total enzyme activities in the mitochondrial fraction were expressed as a percentage of that in the total homogenate and corrected for mitochondrial yield (based on the recovery of cytochrome c oxidase activity). The distribution of glycolytic activity between plastid and cytosol in the total homogenate also was considered, such that the activity of each enzyme recovered in the mitochondrial fraction was expressed as a percentage of extraplastidic glycolytic activity. Values for the proportion of glycolytic activity that is plastidial were taken from published information on heterotrophic soybean cell suspensions (Macdonald and ap Rees, 1983), pea roots (Borchert et al., 1993), and pea cotyledons (Denyer and Smith, 1988).
Protease Protection Assays
Intact mitochondria (suspended in 0.3 M mannitol, 10 mM Tes-KOH, pH 7.5, and 0.1 mM CaCl2) were subjected to protease treatment by the addition of either proteinase K (10 units/mg mitochondrial protein) or thermolysin (10 μg/mg mitochondrial protein) for 1 h at 25°C. Proteases were removed by three rounds of washing of the mitochondria. Washed mitochondria were resuspended in 0.3 M mannitol and 10 mM Tes-KOH, pH 7.5, containing protease inhibitors (Roche Diagnostics).
After protease treatment, the activities of glycolytic enzymes were determined as detailed above (see Enzyme Assays). Alternatively, proteins were fractionated by SDS-PAGE and transferred to a nitrocellulose membrane. Protein gel blot analysis was performed with an affinity-purified polyclonal antibody raised in rabbit against castor seed cytosolic aldolase (obtained from W.C. Plaxton, Queen's University, Kingston, Canada) at a dilution of 1:40 and with polyclonal antibodies raised in rabbit against potato fumarase (Nast and Muller-Rober, 1996) at a dilution of 1:1000. Sheep anti-rabbit antibodies conjugated with horseradish peroxidase (Amersham Biosciences, Little Chalfont, UK) were used as secondary antibodies and revealed with enhanced chemiluminescence reagent (Pierce Biotechnology, Tattenhall, UK).
13C-Labeling Experiments
Isolated mitochondria were incubated with either 50 mM 1-13C–fructose-1,6-bisphosphate (Omicron Biochemicals) or 50 mM U-13C–glucose (Campro Scientific) in 0.3 M mannitol, 10 mM Tes-NaOH, pH 7.5, 3 mM MgSO4, 10 mM NaCl, and 5 mM KH2PO4 containing the following cofactors: 0.6 mM ATP, 0.2 mM ADP, 0.3 mM NAD, 0.1 mM thiamine pyrophosphate, and 10 mM citrate. The final reaction mixture contained 5 mg of mitochondrial protein in a final volume of 60 μL. The reaction was incubated at 25°C for 16 h. The reaction was terminated, and metabolites were extracted in 100% methanol at 70°C for 15 min (Roessner et al., 2000). After centrifugation, the resulting supernatant was dried under vacuum, and the resulting residue was derivatized for 120 min at 37°C (in 50 μL of 20 mg/mL methoxyamine hydrochloride in pyridine) followed by a 30-min treatment at 37°C with 50 μL of N-methyl-N-(trimethylsilyl)trifluoroacetamide. Gas chromatography–MS analysis of the derivatized samples was performed exactly as described by Roessner et al. (2001). The molar percentage enrichments were evaluated by determination of the intensities of the 12C spectral fragments, and the isotopic spectral fragments of nonlabeled standards were compared with the fragmentation patterns of metabolites detected in the chromatograms of the 13C-fed mitochondria after minor modifications to the procedure developed for the evaluation of NMR spectra (Des Rosiers et al., 1991).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Lee J. Sweetlove, lee.sweetlove@plants.ox.ac.uk.
Acknowledgments
We thank H. Batoko for assistance with confocal microscopy and Ruth Holtzapffel for assistance in maintaining cell cultures. The financial support of the European Community (P.G.), the Australian Research Council (A.H.M.), the Max Planck Gesellschaft (A.R.F.), and the Biotechnology and Biological Science Research Council and the European Community (QLG1-CT-2001-00966) (L.J.S. and C.J.L.) is gratefully acknowledged.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.012500.
References
- Ali, M., Rellos, P., and Cox, T.M. (1998). Hereditary fructose intolerance. J. Med. Genet. 35, 353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, L.E., Wang, X., and Gibbons, J.T. (1995). Three enzymes of carbon metabolism or their antigenic analogs in pea leaf nuclei. Plant Physiol. 108, 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Azama, K., Abe, S., Sugimoto, H., and Davies, E. (2003). Lysine-containing proteins in maize endosperm: A major contribution from cytoskeleton-associated carbohydrate-metabolizing enzymes. Planta, in press. [DOI] [PubMed]
- Balk, J., and Leaver, C.J. (2001). The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 13, 1803–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk, J., Leaver, C.J., and McCabe, P.F. (1999). Translocation of cytochrome c from mitochondria to cytosol occurs during heat-induced programmed cell death in cucumber plants. FEBS Lett. 463, 151–154. [DOI] [PubMed] [Google Scholar]
- Bardel, J., Louwagie, M., Jaquinod, M., Jourdain, A., Luche, S., Rabilloud, T., Macherel, D., Garin, J., and Bourguignon, J. (2002). A survey of the plant mitochondrial proteome in relation to development. Proteomics 2, 880–898. [DOI] [PubMed] [Google Scholar]
- Bartoli, C.G., Pastori, G.M., and Foyer, C.H. (2000). Ascorbate biosynthesis in mitochondria is linked to the electron transport chain between complexes III and IV. Plant Physiol. 123, 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert, S., Harborth, J., Schunemann, D., Hoferichter, P., and Heldt, H.W. (1993). Studies of the enzymatic capacities and transport properties of pea root plastids. Plant Physiol. 101, 303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell, M.M., Mooney, P.J., Blundy, M., Carter, D., Wilson, F., Green, J., Blundy, K.S., and ap Rees, T. (1994). Genetic manipulation of 6-phosphofructokinase in potato tubers. Planta 194, 95–101. [Google Scholar]
- Dai, N., Schaffer, A., Petreikov, M., Shahak, Y., Giller, Y., Ratner, K., Levine, A., and Granot, D. (1999). Overexpression of Arabidopsis hexokinase in tomato plants inhibits growth, reduces photosynthesis, and induces rapid senescence. Plant Cell 11, 1253–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis, D., Huang, H., and Negm, F. (1997). Glyolysis, the pentose phosphate pathway and anaerobic respiration. In Plant Metabolism, D. Dennis, D. Turpin, D. Lefebvre, and D. Layzell, eds (Harlow, UK: Longman), pp. 108–123.
- Denyer, K., and Smith, A.M. (1988). The capacity of plastids from developing pea cotyledons to synthesize acetyl CoA. Planta 173, 172–182. [DOI] [PubMed] [Google Scholar]
- Des Rosiers, C., David, F., Carneau, M., and Bruengraber, H. (1991). Non-homogenous labeling of liver mitochondrial acetyl CoA. J. Biol. Chem. 266, 1574–1578. [PubMed] [Google Scholar]
- Diekert, K., Kispal, G., Guiard, B., and Lill, R. (1999). An internal targeting signal directing proteins into the mitochondrial intermembrane space. Proc. Natl. Acad. Sci. USA 96, 11752–11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douce, R. (1985). Mitochondria in Higher Plants: Structure, Function, and Biogenesis. (New York: Academic Press).
- Douce, R., and Neuberger, M. (1989). The uniqueness of plant mitochondria. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40, 371–414. [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Farre, E.M., Tiessen, A., Roessner, U., Geigenberger, P., Trethewey, R.N., and Willmitzer, L. (2001). Analysis of the compartmentation of glycolytic intermediates, nucleotides, sugars, organic acids, amino acids, and sugar alcohols in potato tubers using a nonaqueous fractionation method. Plant Physiol. 127, 685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, K., and Weber, A. (2002). Transport of carbon in non-green plastids. Trends Plant Sci. 7, 345–351. [DOI] [PubMed] [Google Scholar]
- Galina, A., Reis, M., Albuquerque, M.C., Puyou, A.G., Puyou, M.T., and de Meis, L. (1995). Different properties of the mitochondrial and cytosolic hexokinases in maize roots. Biochem. J. 309, 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, S.A., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100. [DOI] [PubMed] [Google Scholar]
- Green, D.R., and Reed, J.C. (1998). Mitochondria and apoptosis. Science 281, 1309–1312. [DOI] [PubMed] [Google Scholar]
- Gueguen, V., Macherel, D., Jaquinod, M., Douce, R., and Bourguignon, J. (2000). Fatty acid and lipoic acid biosynthesis in higher plant mitochondria. J. Biol. Chem. 275, 5016–5025. [DOI] [PubMed] [Google Scholar]
- Hanson, M.R. (1991). Plant mitochondrial mutations and male sterility. Annu. Rev. Genet. 25, 461–486. [DOI] [PubMed] [Google Scholar]
- Hartmann, C.M., Gehring, H., and Christen, P. (1993). The mature form of imported mitochondrial proteins undergoes conformational changes upon binding to isolated mitochondria. Eur. J. Biochem. 218, 905–910. [DOI] [PubMed] [Google Scholar]
- Heazlewood, J.L., Howell, K.A., Whelan, J., and Millar, A.H. (2003). Towards an analysis of the rice mitochondrial proteome. Plant Physiol. 132, 230–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson, R.J., and Plaxton, W.C. (1998). Purification and characterization of cytosolic fructose-1,6-bisphosphate aldolase from endosperm of germinated castor oil seeds. Arch. Biochem. Biophys. 355, 189–196. [DOI] [PubMed] [Google Scholar]
- Jang, J.C., Leon, P., Zhou, L., and Sheen, J. (1997). Hexokinase as a sugar sensor in higher plants. Plant Cell 9, 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery, C.J. (1999). Moonlighting proteins. Trends Biochem. Sci. 24, 8–11. [DOI] [PubMed] [Google Scholar]
- Koller, A., Washburn, M.P., Lange, B.M., Andon, N.L., Deciu, C., Haynes, P.A., Hays, L., Schieltz, D., Ulaszek, R., Wei, J., Wolters, D., and Yates, J.R., III (2002). Proteomic survey of metabolic pathways in rice. Proc. Natl. Acad. Sci. USA 99, 11969–11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruft, V., Eubel, H., Jansch, L., Werhahn, W., and Braun, H.P. (2001). Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol. 127, 1694–1710. [PMC free article] [PubMed] [Google Scholar]
- Kushnir, S., Babiychuk, E., Storozhenko, S., Davey, M.W., Papenbrock, J., De Rycke, R.R., Engler, G., Stephan, U.W., Lange, H., Kispal, G., Lill, R., and Van Montagu, M.M. (2001). A mutation of the mitochondrial ABC transporter Sta1 leads to dwarfism and chlorosis in the Arabidopsis mutant starik. Plant Cell 13, 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi, M. (1999). Plant mitochondrial carriers: An overview. Cell. Mol. Life Sci. 56, 918–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link, A.J., Eng, J., Schieltz, D.M., Carmack, E., Mize, G.J., Morris, D.R., Garvik, B.M., and Yates, J.R., III (1999). Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17, 676–682. [DOI] [PubMed] [Google Scholar]
- Lu, M., Holliday, L.S., Zhang, L., Dunn, W.A., Jr., and Gluck, S.L. (2001). Interaction between aldolase and vacuolar H+-ATPase: Evidence for direct coupling of glycolysis to the ATP-hydrolyzing proton pump. J. Biol. Chem. 276, 30407–30413. [DOI] [PubMed] [Google Scholar]
- Macdonald, F.D., and ap Rees, T. (1983). Enzyme properties of amyloplasts from suspension cultures of soybean. Biochim. Biophys. Acta 755, 81–89. [Google Scholar]
- Mackenzie, S., and McIntosh, L. (1999). Higher plant mitochondria. Plant Cell 11, 571–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, M., and Leaver, C. (1993). Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol. 103, 621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.H., Sweetlove, L.J., Giege, P., and Leaver, C.J. (2001). Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol. 127, 1711–1727. [PMC free article] [PubMed] [Google Scholar]
- Moller, I. (1986). Membrane-bound NAD(P)H dehydrogenases in higher plant cells. Annu. Rev. Plant Physiol. 37, 309–334. [Google Scholar]
- Moller, I.M. (2001). Plant mitochondria and oxidative stress: Electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 561–591. [DOI] [PubMed] [Google Scholar]
- Moorhead, G.B., Hodgson, R.J., and Plaxton, W.C. (1994). Copurification of cytosolic fructose-1,6-bisphosphatase and cytosolic aldolase from endosperm of germinating castor oil seeds. Arch. Biochem. Biophys. 312, 326–335. [DOI] [PubMed] [Google Scholar]
- Moorhead, G.B., and Plaxton, W.C. (1992). Evidence for an interaction between cytosolic aldolase and the ATP- and pyrophosphate-dependent phosphofructokinases in carrot storage roots. FEBS Lett. 313, 277–280. [DOI] [PubMed] [Google Scholar]
- Mouillon, J.M., Ravanel, S., Douce, R., and Rebeille, F. (2002). Folate synthesis in higher plant mitochondria: Coupling between the dihydropterin pyrophosphokinase and the dihydropteroate synthase activities. Biochem. J. 363, 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Nast, G., and Muller-Rober, B. (1996). Molecular characterization of potato fumarate hydratase and functional expression in Escherichia coli. Plant Physiol. 112, 1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliaro, L., and Taylor, D.L. (1988). Aldolase exists in both the fluid and solid phases of cytoplasm. J. Cell Biol. 107, 981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson, C.A., and Vanlerberghe, G.C. (2002). Transgenic plant cells lacking mitochondrial alternative oxidase have increased susceptibility to mitochondria-dependent and -independent pathways of programmed cell death. Plant Physiol. 129, 1908–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner, U., Luedemann, A., Brust, D., Fiehn, O., Linke, T., Willmitzer, L., and Fernie, A.R. (2001). Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13, 11–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner, U., Wagner, C., Kopka, J., Trethewey, R.N., and Willmitzer, L. (2000). Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J. 23, 131–142. [DOI] [PubMed] [Google Scholar]
- Santoni, V., Rabilloud, T., Doumas, P., Rouquie, D., Mansion, M., Kieffer, S., Garin, J., and Rossignol, M. (1999). Towards the recovery of hydrophobic proteins on two-dimensional electrophoresis gels. Electrophoresis 20, 705–711. [DOI] [PubMed] [Google Scholar]
- Schnable, P.S., and Wise, R.P. (1998). The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3, 175–180. [Google Scholar]
- Sirover, M.A. (1999). New insights into an old protein: The functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim. Biophys. Acta 1432, 159–184. [DOI] [PubMed] [Google Scholar]
- Small, I., Wintz, H., Akashi, K., and Mireau, H. (1998). Two birds with one stone: Genes that encode products targeted to two or more compartments. Plant Mol. Biol. 38, 265–277. [PubMed] [Google Scholar]
- Srere, P.A. (1987). Complexes of sequential metabolic enzymes. Annu. Rev. Biochem. 56, 89–124. [DOI] [PubMed] [Google Scholar]
- Sweetlove, L.J., Burrell, M.M., and ap Rees, T. (1996). Characterization of transgenic potato (Solanum tuberosum) tubers with increased ADPglucose pyrophosphorylase. Biochem. J. 320, 487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove, L.J., Heazlewood, J.L., Herald, V., Holtzapffel, R., Day, D.A., Leaver, C.J., and Millar, A.H. (2002). The impact of oxidative stress on Arabidopsis mitochondria. Plant J. 32, 891–904. [DOI] [PubMed] [Google Scholar]
- Sweetlove, L.J., Mowday, B., Hebestreit, H.F., Leaver, C.J., and Millar, A.H. (2001). Nucleoside diphosphate kinase III is localized to the inter-membrane space in plant mitochondria. FEBS Lett. 508, 272–276. [DOI] [PubMed] [Google Scholar]
- Taylor, S.W., Fahy, E., Zhang, B., Glenn, G.M., Warnock, D.E., Wiley, S., Murphy, A.N., Gaucher, S.P., Capaldi, R.A., Gibson, B.W., and Ghosh, S.S. (2003). Characterization of the human heart mitochondrial proteome. Nat. Biotechnol. 21, 281–286. [DOI] [PubMed] [Google Scholar]
- Vander Heiden, M.G., Chandel, N.S., Li, X.X., Schumacker, P.T., Colombini, M., and Thompson, C.B. (2000). Outer mitochondrial membrane permeability can regulate coupled respiration and cell survival. Proc. Natl. Acad. Sci. USA 97, 4666–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe, G.C., and McIntosh, L. (1997). Alternative oxidase: From gene to function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 703–734. [DOI] [PubMed] [Google Scholar]
- Washburn, M.P., Wolters, D., and Yates, J.R., III (2001). Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247. [DOI] [PubMed] [Google Scholar]
- Werhahn, W., Niemeyer, A., Jansch, L., Kruft, V., Schmitz, U.K., and Braun, H.P. (2001). Purification and characterization of the preprotein translocase of the outer mitochondrial membrane from Arabidopsis: Identification of multiple forms of TOM20. Plant Physiol. 125, 943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J.E. (1980). Brain hexokinase, the prototype ambiquitous enzyme. Curr. Top. Cell. Regul. 16, 1–54. [DOI] [PubMed] [Google Scholar]
- Wolters, D.A., Washburn, M.P., and Yates, J.R., III (2001). An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 73, 5683–5690. [DOI] [PubMed] [Google Scholar]
- Yu, J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp indica). Science 296, 79–92. [DOI] [PubMed] [Google Scholar]