Abstract
We present isogenic transgenic tobacco lines that carry at a given chromosomal position a β-glucuronidase (GUS) reporter gene either with or without the presence of the matrix-associated region known as the chicken lysozyme A element. Plants were generated with the Cre-lox site–specific recombination system using heterospecific lox sites. Analysis of GUS gene expression in plant populations demonstrates that the presence of the A element can shield against RNA silencing of the GUS gene. Protection was observed in two of three independent tobacco transformants. Plants carrying an A element 5′ of the GUS gene always had stable GUS activity, but upon removal of this A element, the GUS gene became silenced over time in two lines, notably when homozygous.
INTRODUCTION
RNA silencing occurs in a broad range of organisms and is well documented in transgenic plants (Matzke et al., 2000). Particularly in plants, it is a highly variable process (Meins, 2000) that can occur between two homologous transgenes, between a transgene and a homologous endogenous gene, or between a gene and an incoming virus (Fagard and Vaucheret, 2000). Its occurrence is influenced strongly by parameters such as complex integrations and (inverted) repeats of transgenes (Hobbs et al., 1993; Muskens et al., 2000), the integration site in the host genome (Meza et al., 2001, 2002), the stage of plant development (Kunz et al., 1996), and the environment (Meza et al., 2001; Szittya et al., 2003). Although silenced genes produce transcripts, these are degraded rapidly. The silencing is accompanied by the accumulation of 21- to 24-nucleotide small RNAs corresponding to both the sense and antisense strands of the target RNA (Hamilton and Baulcombe, 1999; Hutvagner et al., 2000; Zamore et al., 2000; Elbashir et al., 2001). These small interfering RNAs (siRNAs) are chopped from longer double-stranded RNA by an ATP-dependent RNase III–type enzyme known as Dicer (Bernstein et al., 2001; Nykanen et al., 2001), the plant equivalent of which is known as CAF (Jacobsen et al., 1999). The siRNAs then are incorporated into an endoribonuclease enzyme complex, in which they act as guides, restricting the ribonuclease to degrade only RNAs complementary to one of the two siRNA strands (Zamore, 2002). The small RNAs can have developmental functions (Hutvagner et al., 2001) but may not be the signaling molecules in the establishment of silencing (Mallory et al., 2001; Vance and Vaucheret, 2001). The silencing also is accompanied frequently by the DNA cytosine methylation of the silenced gene (Bender, 2001), is usually reset upon meiosis, but it may recur during development in every generation (Meins, 2000).
RNA silencing of single-copy transgenes is rarely documented and mainly concerns transgenes homologous with endogenous genes (Seymour et al., 1993; Jorgensen et al., 1996). Currently, the best documented example of RNA silencing in plants of a single-copy transgene that is not homologous with endogenous genes is probably the bacterial β-glucuronidase (GUS) gene driven by the 35S promoter of the doubled Cauliflower mosaic virus (dCaMV) (Elmayan and Vaucheret, 1996). GUS activity in single-copy tobacco lines was highly reduced in homozygous plants and in haploids derived from silenced lines. The influence of zygosity on gene silencing also has been documented for multicopy transgenes that often are homologous with endogenous genes. The homozygous state of the transgene can correlate with silencing (de Carvalho et al., 1992; Dorlhac de Borne et al., 1994) or increase silencing (Dehio and Schell, 1994; Neuhuber et al., 1994; Kunz et al., 1996; Tenllado and Diaz-Ruiz, 1999) relative to the hemizygous state.
Both for fundamental research and for applications, criteria are sought for how to obtain or minimize or prevent silencing in plants (Waterhouse et al., 2001). DNA elements with a chromatin insulator or boundary function to insulate the transgene from the repressing influences of neighboring chromatin are proposed to prevent silencing (Meyer, 1998; Allen et al., 2000). Matrix-associated regions (MARs) are candidates for such an insulating activity. MARs are DNA sequences that are thought to mediate the binding of chromatin to the proteinaceous nuclear matrix, thereby creating chromatin domains as topologically isolated units of gene regulation (Bode et al., 1995, 1996). The presence of the chicken lysozyme MAR element known as the A element around transgenes in tobacco results in the position-independent expression of the transgenes (Mlynarova et al., 1994, 1995; Jansen et al., 2002). This fact establishes the A element as a functional chromatin boundary, as defined by Udvardy (1999), in plants as it is in other systems (Bode et al., 1995; Strätling and Yu, 1999). Current assays for boundary element efficacy and the potential protection against gene silencing require the comparison of different constructs at different integration sites in large populations of plants or cells. Therefore, boundary action may be confounded with parameters beyond experimental control, such as different spectra of integration sites, complex integrations, boundary–transgene interactions, and boundary–plant species interactions (Brouwer et al., 2002).
A better assessment of boundaries could be by direct comparison of otherwise isogenic plants with and without boundary elements flanking a (trans)gene at the same chromosomal position. Here, we have generated isogenic transgenic tobacco lines with (+A) and without (−A) the 5′ A element with the help of heterospecific lox sites and the Cre-lox site–specific recombination system. The comparison of tobacco plants that carry at the same chromosomal position a transgene either with or without the flanking A element demonstrates that the presence of the A element can protect against RNA silencing of the GUS gene. Protection depends on the particular locus of integration. This finding indicates that the physical presence of a chromatin boundary element protects against RNA silencing. The RNA-silencing process is thought to occur primarily in the cytoplasm (Chicas and Macino, 2001), although there is recent evidence that nuclear processes are involved in at least some cases of RNA silencing, notably in yeast (Volpe et al., 2002). Therefore, the results presented here strengthen or establish the link between physical chromatin structure and the occurrence of RNA silencing.
RESULTS
Generation and Analyses of Tobacco Lines
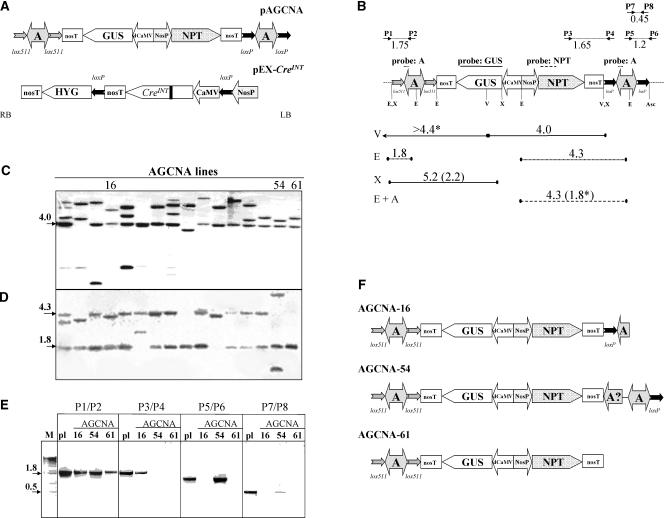
The T-DNA of the plant transformation vector pAGCNA consists of the GUS reporter gene driven by the CaMV d35S promoter and the nopaline synthase (NOS) promoter driven by the neomycin phosphotransferase (NPTII) selectable marker gene (Figure 1A). The A element is present at each T-DNA border. To be able to excise each A element independently with Cre while maintaining the GUS and NPTII genes, wild-type loxP sites around the left-border A element were combined with heterospecific lox511 sites around the right-border A element. A scheme of the T-DNA of the pEX-CreINT used for site-specific recombination (Mlynarova and Nap, 2003) is shown in Figure 1A.
Figure 1.
Structures of the T-DNA Regions of Vectors and Plant Lines.
(A) The right and left borders of the T-DNA are indicated with RB and LB, respectively. Promoters are indicated by large arrows that indicate the direction of transcription; coding sequences are indicated by pointed boxes. All genes contain the nos polyadenylation region (nosT). The chromatin boundary A elements are indicated by two-headed arrows. The neomycin phosphotransferase selectable marker gene (NPT) is driven by the nos promoter (NosP). The β-glucuronidase gene (GUS) is driven by the d35S promoter of CaMV (dCaMV). The chromatin boundary element is the chicken lysozyme MAR, known as the A element (A). The pairs of lox sites around the A elements are indicated by small arrows that indicate the orientation of the lox sites. The creINT gene is under the control of the single (i.e., not enhanced) 35S promoter of CaMV. The black bar in the Cre coding sequence indicates the introduced plant intron. After self-excision of 35S-creINT, the hygromycin phosphotransferase selectable marker gene (HYG) will be driven by the nos promoter (NosP).
(B) Structure of the AGCNA T-DNA (as in [A]) with the positions of the restriction enzymes EcoRI (E), VspI (V), XhoI (X), and AscI (Asc) indicated. The fragments used as probes are indicated with lines above the T-DNA. The expected hybridizing fragments are given as lines below the T-DNA, with full lines for the GUS probe, dashed lines for the NPTII probe, and dotted lines for the A element probe. The sizes of the expected fragments are given in kb. The lengths of the expected fragments after the removal of the A elements are shown in parentheses (when different from those of the parent line). The border fragments consisting of T-DNA and flanking plant DNA are marked with asterisks, and the known size of the T-DNA fragment is given.
(C) Phosphorimage of the hybridization of VspI-digested DNA from the primary AGCNA transgenic plant population probed with GUS. The three lines chosen for further analysis are indicated.
(D) Phosphorimage of the hybridization of EcoRI-digested DNA probed with the A element.
(E) PCR analysis of AGCNA-16, -54, and -61 lines using four primer pairs to verify the intactness of the T-DNA integration. The lanes labeled pl contain a PCR fragment obtained on pAGCNA plasmid DNA as a positive control.
(F) In planta configuration of the T-DNA in lines AGCNA-16, -54, and -61 based on DNA gel blot analysis and PCR.
Primary transformants carrying a single T-DNA locus were identified based on the 3:1 segregation of seeds on kanamycin and DNA gel blot analysis with a GUS probe. Sixteen single-locus lines showing highly stable GUS expression were analyzed in more detail. These were checked by DNA gel blot analysis and PCR for complete and intact T-DNA. None had a perfect left-border integration. Three lines with intact integrations at the right border (AGCNA-16, -54, and -61) and stable GUS expression in time and in the next generations were chosen for analyses. Their precise T-DNA configurations were determined by additional DNA gel blot and PCR analyses (Figures 1B to 1E). In Figure 1B, the positions of the restriction enzymes, probes, primers used, and expected resulting fragments are indicated. In all three lines, hybridization (Figures 1C and 1D) as well as PCR (Figure 1E) and sequencing of the right-border flanking DNA (see below for results) confirmed the presence of a single, complete GUS gene and A element at the right border of the T-DNA.
Unfortunately, for all three lines, incomplete integrations were shown at the left T-DNA border (Figures 1C to 1E). Line AGCNA-16 yielded a fragment smaller than the expected 4.3 kb for a left-border A element (Figure 1D). PCR analyses with the P5/P6 and P7/P8 primer pairs indicated that the 3′ part of the A element and the 3′ loxP site were not integrated (Figure 1E). Sequencing of the flanking DNA confirmed integration of only the 1.7-kb 5′ part of the left-border A element. In line AGCNA-54, hybridization with the A element showed the presence of one fragment smaller and one fragment larger than the predicted 4.3 kb (Figure 1D). PCR analysis using P3/P4 and P5/P6 primers confirmed the absence of the 5′ loxP site of the A element at the left border, although the 3′ loxP site was present (Figure 1E). In line AGCNA-54, apparently a complex and rearranged T-DNA structure was present at the left border. In the case of AGCNA-61, hybridization (Figure 1D), PCR (Figure 1E), and sequencing of flanking DNA indicated the absence of the entire A element at the left border.
The resulting detailed in planta T-DNA configurations of three transgenic lines are shown in Figure 1F. The particular configuration at the left T-DNA border will not allow Cre to remove the remaining part of the left-border A element by site-specific recombination from either AGCNA-16 or AGCNA-54. Because this DNA gel blot analysis is incomplete, we cannot completely exclude the possibility of right-border-centered inverted duplications of the T-DNA.
All three lines were subjected to in planta site-specific recombination to evaluate the influence of the removal of the right-border A element. For each line, homozygous offspring were identified and used to generate batches of hemizygous seeds by backcrossing to the wild type. Hemizygous plants were retransformed with Agrobacterium tumefaciens LBA4404 carrying pEX-creINT, a binary vector with a self-excising, intron-containing variant of the cre gene (Figure 1A). This variant of cre allows the highly efficient removal of its own loxP-flanked coding sequence as well as any other DNA flanked by lox sites. With the use of combined kanamycin/hygromycin selection, excision in tobacco was highly efficient (Mlynarova and Nap, 2003). For each tobacco line, a population of kanamycin- and hygromycin-resistant retransformants was generated. They were indicated with an R plus a number added to the line number, for example, AGCNA-16(R2).
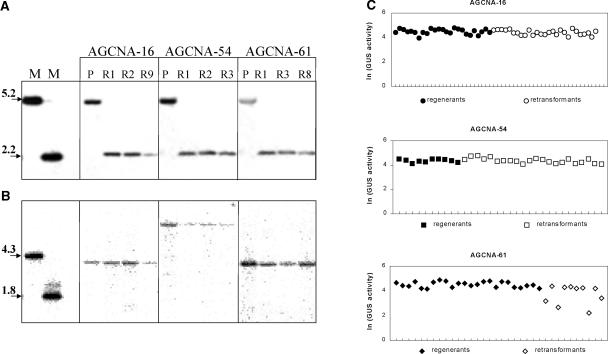
To evaluate the intrinsic expression stability of the individual T-DNA loci in tissue culture, populations of plants were generated by in vitro regeneration. For line AGCNA-16, 27 retransformed and 25 regenerated lines were obtained; for line AGCNA-54, 22 retransformants and 10 regenerants were obtained; and for line AGCNA-61, 10 retransformants and 24 regenerants were obtained. In none of the regenerated lines had the T-DNA configuration changed with respect to that of the parental line (data not shown). The excision of the right-border A element that flanked the GUS gene was analyzed in XhoI-digested genomic DNA probed with GUS (for the positions of restriction enzymes and expected fragment lengths, see Figure 1B). In all three parental lines, the 5.2-kb DNA fragment containing the A element was present, whereas all retransformants analyzed carried the 2.2-kb fragment predicted after A element excision (Figure 2A). This finding demonstrates the complete removal of the right-border A element from plant DNA in all retransformants. Hybridization studies confirmed that as a consequence of the incomplete integrations, the T-DNA configuration had not changed at the left border in any of the retransformed lines (Figure 2B). Retransformant and regenerant populations establish material that allows us to analyze the effect of the removal of the A element on GUS gene expression.
Figure 2.
Characteristics of Retransformed Plants.
(A) Phosphorimage of the hybridization of XhoI-digested genomic DNA probed with GUS. The positions of the restriction sites and expected fragments are indicated as in Figure 1B. The lanes labeled M contain restriction fragments from pAGCNA as relevant markers. The lanes labeled P contain DNA from parent lines before in planta recombination.
(B) Hybridization of EcoRI-AscI–digested DNA probed with NPTII.
(C) Natural logarithm (ln) of the GUS activity in leaves of individual plants obtained after in vitro regeneration of AGCNA-16 (closed circles) and after retransformation with pEX-creINT (open circles). For line AGCNA-54, GUS activity in regenerated (closed squares) and retransformed (open squares) plants is given. For AGCNA-61, GUS activity in regenerated (closed diamonds) and retransformed (open diamonds) plants is given.
In Planta Removal of the A Element Can Result in Severe GUS Gene Silencing in Offspring Plants
GUS activity was analyzed in all retransformed (−A) and regenerated (+A) plant populations (Figure 2C). For both AGCNA-16 and AGCNA-54, the variation in GUS activity in the leaves of the primary, hemizygous retransformants was as low as that in the corresponding regenerants. This finding indicates that for these two lines, tissue culture and subsequent removal of the A element had no detectable influence on GUS activity. The situation was different for line AGCNA-61. For this line, some but not all retransformed plants had a considerably reduced GUS activity. For this particular line, removal of the A element seems to create enhanced instability of expression of the GUS gene. This was not the result of enhanced general somaclonal variation in tissue culture, because the population of regenerants showed low variability. The instability could be caused by sequence homology between the AGCNA locus and the EX-creINT locus and/or by the process of in planta recombination. After self-excision of the cre gene, DNA regions of pEX-creINT homologous with the GUS-containing T-DNA locus remained in the plant. These regions could affect the stability of GUS gene expression. To further evaluate the influence of the presence of the A element, GUS activity was analyzed in offspring lacking the EX-creINT locus.
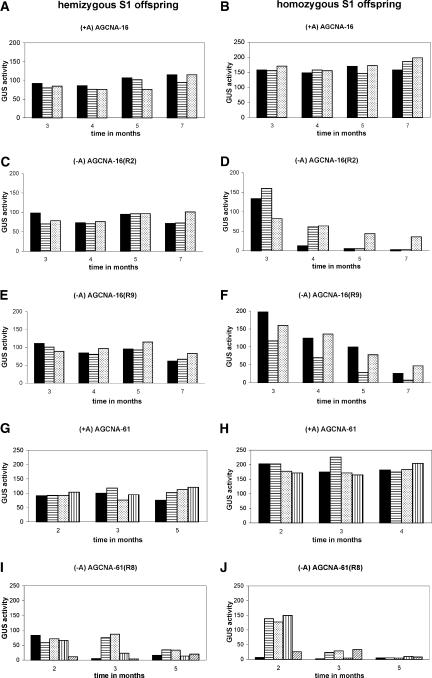
For each line, two primary retransformants and one regenerant were selfed. Offspring were selected for progeny analysis carrying an AGCNA locus from which the EX-creINT locus has segregated away. By means of PCR and germination assays on hygromycin-containing medium, the absence of the EX-creINT locus was determined (data not shown). GUS activity was followed over a period of 7 months in three to four hemizygous and homozygous individual S1 offspring plants. Results for line AGCNA-16 are shown in Figures 3A to 3F, and results for line AGCNA-61 are shown in Figures 3G to 3J. In all cases, the regenerated plants were stably active over the entire period and homozygous plants were approximately twice as active as the corresponding hemizygous plants (Figures 3A, 3B, 3G, and 3H). This finding establishes the fact that the three A element–containing loci are stable and show fully additive, Mendelian GUS gene expression. By contrast, the three lines differ in characteristics when the A element is removed from the T-DNA locus.
Figure 3.
Removal of the A Element Triggers Severe GUS Gene Silencing in S1 Offspring Plants.
The panels at the left give data for the hemizygous offspring, and the panels at the right give data for the corresponding homozygous offspring. Each bar gives the GUS activity of an individual plant. All retransformed offspring were selected for the physical absence of the EX-creINT locus.
(A) and (B) GUS activity in AGCNA-16 (+A).
(C) and (D) GUS activity in retransformed AGCNA-16(R2) (−A).
(E) and (F) GUS activity in retransformed AGCNA-16(R9) (−A).
(G) and (H) GUS activity in AGCNA-61 (+A).
(I) and (J) GUS activity in AGCNA-61(R8).
Retransformants of line AGCNA-54 showed stable GUS activity over the entire period in both the hemizygous and homozygous configurations (data not shown). In this locus, the presence or absence of the A element apparently had no impact. For both AGCNA-16(R2) and AGCNA-16(R9), GUS activity in hemizygous plants was as stable as that in the regenerated AGCNA-16 plants (Figures 3C and 3E). Homozygous plants were twice as active as the corresponding hemizygous plants at the beginning of the measurement period, but after ∼3 months, they gradually lost activity (Figures 3D and 3F). Seven-month-old AGCNA-16(R2) and AGCNA-16(R9) homozygous plants had on average a GUS activity of <5% of that of the corresponding +A homozygous plants. The presence of the A element in locus AGCNA-16 apparently protects against silencing in the homozygous state.
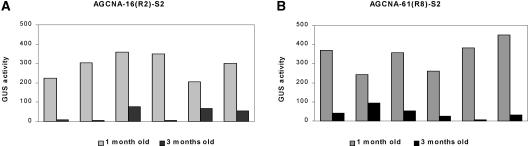
GUS activities of homozygous and hemizygous plants of retransformant AGCNA-61(R8) are shown in Figures 3I and 3J. Already 2-month-old plants showed quite variable activities that were reduced compared with those of the corresponding +A regenerants. Three-month-old homozygous plants were all silenced, as were most hemizygous plants. Offspring of retransformant AGCNA-61(R3) showed the same trend (data not shown). The presence of the A element in locus AGCNA-61 apparently protects against silencing in the hemizygous state, although silencing in the homozygous stage occurs earlier in plant development and is more severe. To investigate whether the silencing observed is influenced or caused by some epigenetic “memory” associated with the presence of the EX-creINT locus, homozygous plants already lacking this locus were selfed once more. For all six homozygous plants analyzed, GUS activity was reset in the S2 seedlings, followed by the same gradual GUS gene silencing observed in the preceding S1 generation. In Figure 4, the data obtained for S2 seedlings of AGCNA-16(R2) (Figure 4A) and AGCNA-61(R8) (Figure 4B) plants are shown.
Figure 4.
GUS Activity Is Reset in S2 Offspring.
(A) GUS activity in selfed S2 progeny of a homozygous AGCNA-16(R2) plant.
(B) GUS activity in selfed S2 progeny of an AGCNA-61(R8) plant.
Characterization of Flanking Plant DNA Shows the Presence of Repetitive Sequences in Cases of Silencing
The different reactions of the three GUS lines with respect to the removal of the A element could be interpreted as a manifestation of the “classic” position effect, supposedly as a result of the influence of the surrounding chromatin (Mlynarova et al., 1994). To determine if the characteristics of the flanking DNA could further suggest the mechanism(s) of gene silencing, plant DNA flanking the right border of the T-DNA was isolated by walk PCR and sequenced. From line AGCNA-16, a fragment of 400 bp was isolated with an AT% of 68%; from line AGCNA-61, a fragment of 850 bp (58% AT) was isolated; and from line AGCNA-54, a fragment of 1200 bp (53% AT) was isolated.
BLAST (Basic Local Alignment Search Tool) analysis gave a significant hit for only the AGCNA-54 flanking sequence. It consists of 400 bp with 89% amino acid identity to a tomato ovary cDNA clone (cLED35G16; EST268134). The right border of the AGCNA-54 T-DNA is integrated upstream of the 5′ region of this gene. Therefore, the reason that the AGCNA-54 locus is not susceptible to gene silencing, irrespective of the presence or absence of the A element, may be integration very close to an endogenous tobacco gene that supposedly is expressed in the appropriate cells. The flanking DNA of either AGCNA-61 or AGCNA-16 gave no significant hits with standard BLAST, but more detailed analysis showed that both flanking DNAs are repetitive in nature. Analysis of the AGCNA-16 flanking DNA with the Repbase database for repetitive elements (Jurka et al., 1996) revealed a fragment of 93 bp containing 62 matches (2 gaps and 31 mismatches) with the terminal inverted repeat of the Sol3 transposable element (Oosumi and Belknap, 1997). AGCNA-61 flanking DNA contains six direct copies of a 23-bp motif that was demonstrated previously in a DNA marker related to the sensitivity of tobacco to pathogens (E16011; Noguchi et al., 1999).
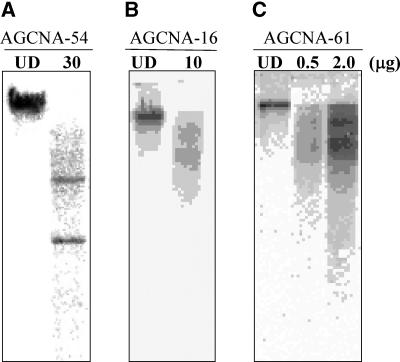
To experimentally verify the repetitive nature of the flanking sequences, they were used as probes in hybridization with tobacco genomic DNA (Figure 5). The AGCNA-54 flanking sequence showed two distinct hybridizing bands (Figure 5A), indicative of a low-copy area of the tobacco genome. Both AGCNA-16 and AGCNA-61 sequences produced the patterns characteristic of dispersed repetitive sequences (Figures 5B and 5C). The flanking DNA from AGCNA-61 gave an especially strong signal on genomic DNA when loaded in amounts as low as 0.5 μg (Figure 5C), indicating the highly repetitive nature of the DNA flanking the T-DNA locus in this line. When hybridized against Arabidopsis (10 μg) or potato (100 μg) genomic DNA, the same fragment showed no hybridization signal (data not shown), indicating that it is a repeated sequence highly specific for tobacco.
Figure 5.
Characterization of Flanking DNA from Three Tobacco Transformants.
Genomic DNA from wild-type tobacco was digested with EcoRI and probed with the flanking sequence isolated from the plant line as indicated at top. The lanes labeled UD contain undigested DNA. The amount of genomic DNA loaded (in μg) is indicated at the top.
(A) Line AGCNA-54.
(B) Line AGCNA-16.
(C) Line AGCNA-61.
The Silencing Phenomenon in −A Plants Is an Example of Post-Transcriptional RNA Silencing
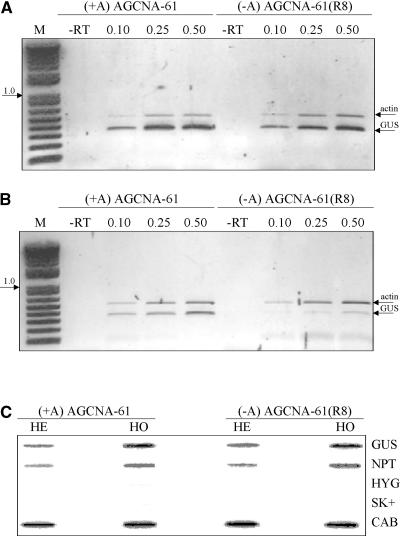
To investigate the molecular nature of the silencing involved, the transcriptional activity of the GUS gene was determined by semiquantitative reverse transcriptase–mediated (RT) PCR on nuclear RNA from active and silenced plants. First-strand cDNA generated with oligo(dT) was amplified by PCR using combined GUS and actin gene primer pairs, and care was taken that the PCR covered the linear range of amplification by varying the amount of input cDNA (see Methods). In nuclei of both lines, the amount of GUS gene transcript relative to the amount of actin gene transcripts was of the same order of magnitude whether from active or silenced plants (Figure 6A; results shown for AGCNA-61 and AGCNA-61[R8]), whereas in total RNA, GUS transcript relative to actin transcript was underrepresented in silenced plants but not in active plants (Figure 6B). The latter finding is in agreement with the results of steady state RNA gel blot analysis (see below).
Figure 6.
GUS Gene Silencing Is Post-Transcriptional.
(A) and (B) Photographs of ethidium bromide–stained agarose gels with the products obtained from semiquantitative RT-PCR analysis of nuclear (A) and total (B) RNA from homozygous active (+A) and silenced (−A) AGCNA-61 plants using primers specific for GUS and actin genes. The lanes labeled M contain the 1-kb DNA ladder (Life Technologies) as a size marker. The lane labeled −RT is the negative control in which reverse transcriptase was omitted from the reaction mixture; the lanes labeled 0.1, 0.25, and 0.5 indicate the different amounts of cDNA (in μL) used to cover the appropriate range for the PCR. The photographs had different exposure times and allow comparison within a panel only.
(C) Phosphorimage of slot-blot hybridization results with nascent RNA synthesized by run-on transcription in nuclei isolated from leaves of active hemizygous (HE) and homozygous (HO) AGCNA-61 plants as well as silenced hemizygous (HE) and homozygous (HO) AGCNA-61(R8) plants. The slot-blot filters contain linearized SK+ plasmids with cloned sequences as indicated: GUS, NPTII, HYG (hygromycin), and CAB (chlorophyl a/b binding protein). The empty plasmid pSK+, indicated by SK+, was added as a negative control as well.
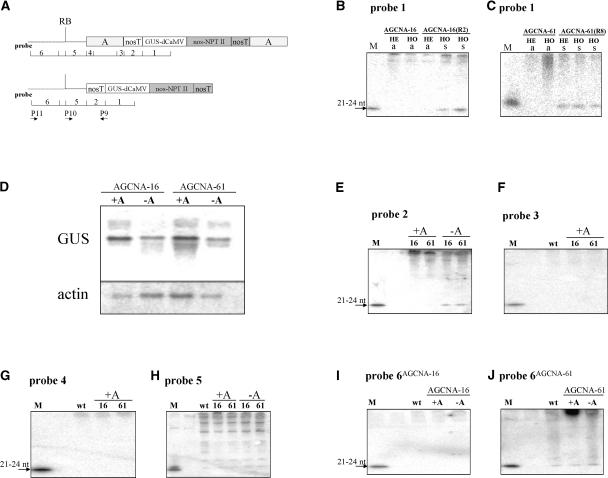
In addition, transcription in hemizygous and homozygous AGCNA-61 (+A, active) and AGCNA-61(R8) (−A, silenced) plants was examined by nuclear run-on analysis. The results (Figure 6C) show that GUS transcription in −A silenced lines was similar to GUS transcription in +A active lines. This finding confirmed the PCR results and established the fact that the gene silencing observed after the removal of the chromatin boundary was post-transcriptional RNA silencing. Moreover, the RNA fractions of plant AGCNA-16(R2) and AGCNA-61(R8) were analyzed for the presence of the 21- to 24-nucleotide siRNAs that are considered to be associated with post-transcriptional silencing. The various probes used in the analysis of RNA from +A and −A plants are shown in Figure 7A. In GUS-silenced material, either AGCNA-16(R2) homozygous plants (Figure 7B) or AGCNA-61(R8) hemizygous or homozygous plants (Figure 7C) siRNA species homologous with GUS mRNA were detected, whereas such siRNA species were not observed in the corresponding active plants.
Figure 7.
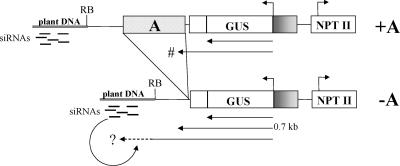
GUS Gene Silencing Involves Plant Flanking DNA.
(A) Schemes of the T-DNA in +A and −A transgenic plants with the positions of probes 1 to 6 used for the detection of siRNAs. The primers used for cDNA synthesis and RT-PCR are indicated. RB, right border.
(B) Phosphorimage of the small RNA fraction from active (a) hemizygous (HE) and homozygous (HO) AGCNA-16 plants and silenced (s) homozygous (HO) AGCNA-16(R2) plants after hybridization with probe 1. Small RNA only is seen in the silenced plants. nt, nucleotides.
(C) The same as in (B) in a silenced HE and HO AGCNA-61(R8) plant.
(D) RNA gel blot analysis of total RNA using GUS and actin gene probes. For analysis, a 10-fold higher amount of RNA from silenced plants was used to compensate for the reduced steady state level of RNA.
(E) to (J) Detection of siRNAs with the probes indicated at top. Line AGCNA-16 is indicated as 16, and line AGCNA-61 is indicated as 61. Flanking DNA of AGCNA-16 is indicated as probe 6AGCNA-16, and flanking DNA of AGCNA-61 is indicated as probe 6AGCNA-61. In all cases, lanes labeled M contain an oligomer from a particular probe (ranging in size from 21 to 24 nucleotides) that functions as a size marker and a positive control for hybridization. The lanes labeled wt contain RNA from wild-type tobacco.
The precise mechanism by which the generation of siRNAs is triggered is a matter of debate, but it may involve aberrant transcription. At present, it is unknown how or when an RNA molecule classifies as “aberrant” within the general characteristics of the transcription machinery. The silencing is not associated with extensive methylation of the coding sequence, and no methylation of the d35S promoter was observed (data not shown). To compare the quality of transcripts in the silenced lines with the RNA in active lines, RNA gel blot analysis was performed using the full GUS gene as a probe (Figure 7D). From silenced plants, a 10-fold higher amount of total RNA was used to compensate for the greatly reduced steady state levels of RNA compared with those levels in active plants (see Methods). The preparations showed no obvious major difference in the quality of GUS transcripts between active and silenced lines by visual inspection of the RNA gel blots, RT-PCR (Figure 6), or yield in radioactive cDNA synthesis (data not shown).
The analysis of the occurrence of siRNAs beyond the GUS coding region showed that the sequence of the nos polyadenylation region is detectably present in the siRNA fraction of −A plants, although in markedly lower amounts than the GUS-coding siRNAs (Figure 7E). In active +A plants, neither the nos polyadenylation region, nor the A element next to the nos polyadenylation region (Figure 7F), nor the most 5′ sequence of the A element (Figure 7G) was detected in the siRNA fraction. In all plants analyzed, including wild-type tobacco, probing with the T-DNA part to the right border (Figure 7A, probe 5) resulted unexpectedly in a whole ladder of small RNAs, including putative siRNAs (Figure 7H). Because this result also was observed in RNA from wild-type tobacco, it is not possible to conclude that these small RNAs originate from the T-DNA. The flanking DNA of AGCNA-16 was not detectable in the siRNA fraction (Figure 7I); the flanking DNA of AGCNA-61 was present in the siRNA from +A, −A, and wild-type plants (Figure 7J).
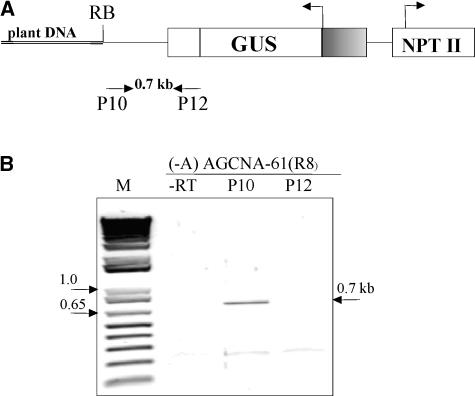
RNA gel blot analysis of wild-type tobacco RNA using flanking DNA (from either AGCNA-16 or AGCNA-61) as a probe showed no hybridization, indicating that the flanking DNA is not expressed regularly in tobacco (data not shown). RT-PCR (Figure 8B, lane P12) and experiments with radioactive cDNA synthesis (data not shown) to demonstrate read-through transcription from flanking DNA (read-in) did not result in any signal above background, suggesting that the silencing is not characterized by extensive read-in transcription from plant flanking DNA. Read-out transcription, however, was observed by analysis of nuclear RNA from silenced homozygous AGCNA-61(R8) plants. RT-PCR analysis did reveal the presence of read-out transcripts reaching at least the right border of the T-DNA (Figure 8B, lane P10). Attempts to directly demonstrate read-out transcription by PCR farther into the flanking DNA using three different primers designed on the flanking DNA failed, possibly because of the repetitive nature of the flanking DNA. Nevertheless, these results indicate that read-out transcripts in combination with flanking sequences are involved in triggering the RNA-silencing pathway.
Figure 8.
Read-Out Transcription from the GUS Transgene in −A Silenced Plants.
(A) The positions of primers used to detect a read-out transcription from the GUS transgene (P10 primer used for reverse transcription) or a read-in transcription from plant DNA (P12 primer used for reverse transcription).
(B) Nuclear RNA from a silenced AGCNA-61(R8) homozygous plant was used for reverse transcription with P10 (lane P10) or P12 (lane P12) primers followed by PCR using P10 and P12 primers. The fragment corresponding to a read-out transcript is indicated (0.7 kb). The lane labeled M contains the 1-kb DNA ladder; the lane labeled −RT contains the negative control in which the reverse transcriptase was omitted.
DISCUSSION
The influence of the chicken lysozyme MAR known as the A element on GUS gene expression was analyzed in three independently transformed single-copy tobacco lines. With the help of site-specific recombination involving pairs of heterospecific lox sites, well-defined plant material was generated (Figures 1 and 2). This material allowed the comparison of GUS gene expression with and without an upstream A element at a given locus of integration.
In the presence of the 5′ A element, GUS activity was stable over time in both the hemizygous and homozygous plants in all three lines. Upon removal of the 5′ A element, the stability of GUS gene expression differed markedly among the three lines. In −A retransformants of line AGCNA-54, removal of the 5′ A element did not affect GUS activity at all. In −A derivatives of line AGCNA-16, GUS activity was stable in hemizygous plants, but in homozygous plants it gradually silenced to a very low level. In line AGCNA-61, GUS activity destabilized in a supposedly stochastic manner in hemizygous plants and became severely silenced in homozygous plants. In both cases, the active +A and silenced −A plants differed in the presence of the 5′ A element.
Detailed DNA gel blot analysis of the three lines, however, showed that they differed in the T-DNA configuration at the left border as a result of incomplete T-DNA transfer (Figure 1F). The fully active AGCNA-54 carries a partly duplicated 3′ A element, the silenced AGCNA-61 carries no 3′ A element, and AGCNA-16, which is silenced only in the homozygous state, carries only a part of the 3′ A element. Therefore, the difference in stability between the lines could be attributable to the presence of the full 3′ A element. However, line AGCNA-61 showed full stability of the expression of the GUS gene in the presence of only a 5′ A element, suggesting no or only a minor role of the 3′ A element at this genomic position. Moreover, the part of the 3′ A element present in line AGCNA-16 was shown in other systems to behave as a genuine insulator (Phi-Van and Strätling, 1996), suggesting that the difference between AGCNA-16 and AGCNA-54 is not the result of the presence of the 3′ A element. More lines with and without a 3′ A element at different genomic positions will be necessary to resolve this point completely, but the data accumulated to date suggest that the role of the 3′ A element, if any, will be minor.
The silencing also is not caused by intrinsic instability in tissue culture, as shown by regeneration experiments (Figure 2C). An epigenetic influence of homologous sequences of the EX-creINT locus also can be excluded. Analysis of GUS activity was performed in S1 offspring physically lacking this locus, and the results showed a complete reset upon an additional meiosis in the S2 offspring. It also has been proposed that upon T-DNA integration, transgenes become epigenetically marked depending on their particular position in the genome and may become (transcriptionally?) silenced as a result of that marking (Day et al., 2000). If so, the absence of silencing in the presence of the A element would indicate that the presence of this element either prevents or shields against this type of epigenetic marking.
The difference between the three lines in the absence of the A element reflects the classic position effect: the GUS gene behaves differently depending on the particular site of integration in the genome. This can be the result of the influence of the surrounding DNA and/or chromatin configuration. Although it also could be hypothesized that the removal of the 5′ A element reveals different epigenetic states of the GUS gene in the three lines, the full resetting of the activity/silencing events in subsequent generations suggests that either such time-course resetting of the GUS gene is implied in such a hypothetical epigenetic mark or that there is no such epigenetic mark. Therefore, we conclude that it is the physical presence of the 5′ A element that is responsible for the absence of GUS gene silencing.
To evaluate the role of the surrounding DNA, for all three lines, the plant DNA flanking the T-DNA integration was isolated and sequenced. In the fully stable line AGCNA-54, the flanking DNA is a low-copy, supposedly expressed, tobacco gene. By contrast, in the lines that become silenced upon the removal of the 5′ A element, the flanking DNA is repetitive. Because the AT contents of the insertion sites of both AGCNA-54 (stable) and AGCNA-61 (silenced) are not very different (53 versus 58%, respectively), and the A element is more AT rich than both (62%), the data do not support the hypothesis of the existence of a scanning mechanism for invasive DNA based on AT (or GC) content differences (Kumpatla et al., 1998). The observation that both silenced lines have flanking DNA that is repetitive may be biologically relevant. A potential relationship between the instability of transgene expression and the presence of surrounding repetitive DNA also was suggested by Iglesias et al. (1997) on the basis of the analysis of four other tobacco lines. Recent results from Meza et al. (2002) also suggest that silencing in Arabidopsis single-copy lines is caused by specific features in the surrounding plant DNA, but data with respect to the repetitive nature of such flanking DNA have not been presented. It is tempting to speculate that repetitive flanking DNA may cause the instability of gene expression, but additional lines are required to establish a statistically significant correlation. The analysis of flanking DNA at integration sites in relation to the stability of gene expression warrants more attention.
The chicken lysozyme A element is by operational definition a MAR and is classified as a true chromatin boundary element (Udvardy, 1999). If protecting, it was expected to protect against transcriptional silencing (e.g., by preventing DNA heterochromatization). The GUS silencing in −A plants is the post-transcriptional type of silencing known as RNA silencing, which is characterized by approximately equal amounts of GUS gene transcript in silenced and active nuclei (Figures 6A and 6C). Also, 21- to 24-nucleotide small RNAs homologous with GUS are present. There is only minor methylation of the GUS coding region associated with silencing, no apparent methylation of the promoter sequence is seen, and the silencing is fully reset after meiosis. The putative relationship between DNA or chromatin structure and RNA silencing was suggested previously by a RecQ DNA helicase being necessary for quelling in Neurospora crassa (Cogoni and Macino, 1999) and by the Arabidopsis ddm1 mutant influencing post-transcriptional 35S GUS silencing (Morel et al., 2000). DDM1 is a member of the SNF2/SWI2 family of chromatin-remodeling proteins (Jeddeloh et al., 1999), suggesting that methylation and chromatin structure can act as regulators of RNA silencing. The involvement of a nuclear step was supported further by the finding that nuclear localization is required for the inhibition of RNA silencing by the viral protein Cmv2b (Lucy et al., 2000). In fission yeast, RNA silencing steps are shown to be involved in the formation and maintenance of heterochromatin (Volpe et al., 2002), further integrating chromatin structure and RNA silencing. The results shown here demonstrate the direct involvement of the physical presence of a MAR in the prevention of RNA silencing. This finding establishes a direct demonstration of the protective role of the presence of a true chromatin boundary element against RNA silencing in plants.
Of particular interest is the molecular mechanism by which the presence of the A element prevents the occurrence of RNA silencing in tobacco plants. For the analysis of the molecular mechanism, we concentrated on line AGCNA-61, which showed the most severe silencing. The gradual RNA silencing in −A plants suggests a GUS gene dose effect. This may indicate the existence of a threshold of a silencing trigger that is built up during growth and development (Zamore, 2002). This threshold may be related to the high transcription rate of the d35S promoter. Promoter strength was shown to correlate with the frequency and degree of silencing in petunia (Que et al., 1997). In homozygous GUS plants, the threshold possibly is reached more readily than in hemizygous plants and triggers an autoregulatory, self-amplifying RNA degradation pathway to target homologous RNAs (Meins, 2000). The presence of the A element may prevent the accumulation of sufficient levels of trigger molecules.
It is now well accepted that silencing mechanisms converge at a double-stranded RNA (dsRNA) stage. The siRNAs visualized are generated via dsRNA precursors that trigger the autocatalytic generation of siRNAs. The presence of the A element could prevent the generation of such dsRNA. Unfortunately, such dsRNA is or can be present in very low, hence undetectable, amounts, and indirect approaches are still required. A straightforward way to generate dsRNA would be read-in (also called read-through) transcription from plant sequences adjacent to the site of integration into the GUS sequence (Sijen and Kooter, 2000; Matzke et al., 2001). However, the flanking DNA in the silenced tobacco line AGCNA-61 is not present in the steady state RNA fraction of tobacco leaf cells, and with PCR we failed to detect any read-in transcription in silenced tobacco plants. If there is any read-in transcription, it should be a rare and short-lived event that cannot be detected with the RNA analysis techniques used.
By contrast, however, read-out transcription from the d35S-driven GUS transgene in AGCNA-61 was detected relatively easily. Termination in plants is considered to be regulated less strictly than in mammalian systems. Whereas animal genes have a single poly(A) site, in plants, the position of polyadenylation can be heterogenous within a single transcription unit, leading to the production of mRNA populations with a variety of end points (Rothnie, 1996). Primary transcripts of 300 bp beyond a given poly(A) site have been documented in tobacco (Depicker et al., 1996), and after such a poly(A) signal, transcription gradually declines to zero (Litiere et al., 1999). In −A silenced AGCNA-61 plants, the nos polyadenylation region sequence occurs in the siRNA fraction. This finding may indicate that this sequence can be subject to the RNA-silencing mechanism. Although we were unable to demonstrate read-out into the flanking DNA, because of technical complications with the high repetitive nature of the flanking DNA, there is no obvious reason why transcription would cease immediately after the end of the nos polyadenylation region. Previous results (Depicker et al., 1996; Litiere et al., 1999) provide experimental support for the assumption that in our case transcription can continue into flanking DNA as well. Moreover, the repetitive flanking DNA of line AGCNA-61 also occurs in the siRNA fraction, indicating that this sequence itself is likely to be subject to RNA silencing. The occurrence of read-out in the case of AGCNA-16 plants was not analyzed in detail. The results indicated no read-out above detection levels, which could be taken to agree with the less severe nature of the silencing observed.
From these results, the following tentative model emerges for the triggering of GUS gene silencing in AGCNA-61 plants without the 5′ A element and the protective role of that A element (Figure 9). On the basis of largely theoretical considerations, Zamore (2002) postulated a similar model. Our data can be seen as experimental support for such a model. In GUS-silenced plants, read-out transcription continues to include the nos polyadenylation region, additional vector sequences, and the repetitive flanking DNA. The read-out transcript becomes via RNA-dependent RNA polymerase converted to dsRNA, using flanking DNA-derived siRNA for attenuation. This drags all sequences contained in the transcript to the silencing machinery. This process resembles the quelling of the endogenous al-1 gene of N. crassa (Cogoni et al., 1996; Catalanotto et al., 2002). Chimeric transcripts with sequences of both the al-1 gene and vector generated siRNAs derived from the vector sequence.
Figure 9.
Model for the Protective Role of the A Element in RNA Silencing.
In GUS-silenced (−A) plants, read-out transcription from the GUS transgene continues to include the nos polyadenylation region, vector sequences, and plant flanking DNA. These chimeric transcripts could be converted to dsRNA via RNA-dependent RNA polymerase using flanking DNA–derived siRNA for attenuation and drag all sequences contained in the transcript into the silencing machinery. The protective role of the A element is in preventing read-out transcription from reaching the flanking DNA.
In Caenorhabditis elegans, siRNAs were observed that correspond to target sequences not present in the triggering dsRNA (Ketting et al., 2001). Spreading of RNA targeting into adjacent regions also occurs in plants (Vaistij et al., 2002). Unexpectedly, the vector part of the T-DNA adjacent to the nos polyadenylation region gives several small RNA species in transgenic plants as well as the wild type. The latter finding indicates that sufficiently homologous sequences are present in the tobacco genome. Although this particular vector sequence may be involved in silencing, the fully stable GUS gene expression in tobacco line AGCNA-54 shows that it is not only the vector sequence involved. Therefore, flanking DNA must be involved in triggering silencing. In −A plants of AGCNA-61, the silencing can spread into the GUS gene using endogenous siRNAs from flanking DNA as primers. Such endogenous siRNAs were demonstrated to exist in the case of AGCNA-61 (Figure 7), but we hypothesize that they exist below detection levels for AGCNA-16 as well. The fact that AGCNA-16 is not silenced in the hemizygous state may indicate that the amount of read-out and/or the amount of endogenous siRNA is not sufficient to trigger the silencing machinery. More lines need to be generated and analyzed to establish a solid relationship between the nature of the flanking DNA, the presence of endogenous siRNA fractions, and the termination of transcription.
If GUS gene silencing in −A plants is caused by read-out transcription to the flanking DNA, the protective role of the A element is to prevent read-out transcription from reaching the flanking DNA. The A element sequence does not occur in the endogenous siRNA fraction and will not drag the GUS-containing transcript in the silencing machinery. Similar experiments with a 1.7-kb scs element did not result in protection against the gradual RNA silencing of the GUS gene in tobacco (L. Mlynarova, unpublished data). Apart from discussions about the precise role or action of the scs element, which was proposed previously to function as a boundary element in Drosophila (Kellum and Schedl, 1992) and was shown to contain a gene locus (Avramova and Tikhonov, 1999), these results indicate that not all DNA sequences can protect against the GUS gene silencing documented here.
The protective action of the A element in preventing read-out transcription is not merely a matter of additional physical distance but is attributable to the particular sequence characteristics of the A element. The particular characteristics of the A element may target the GUS gene to more favorable positions in the interphase nucleus (Gerasimova et al., 2000) for RNA metabolism (Nayler et al., 1998), such as transcription termination (Proudfoot et al., 2002). The element may create a block that prevents transcriptional read-out into plant flanking DNA through the formation of a localized repressive chromatin configuration (Strätling and Yu, 1999). This may involve plant equivalents of the repressor methyl-CpG binding protein MeCP2. In a mammalian system, this protein was shown to bind to the A element and recruit corepressor complexes containing histone deacetylases (Strätling and Yu, 1999). As a result of the blocking action of the A element, T-DNA molecules integrated at inactive chromatin regions can survive a selection process and become available for analysis. Normally, only integration in active chromatin would overcome selection procedures based on gene expression. Less selection bias for T-DNA in active chromatin regions will help in elucidating the relationships between chromatin structure and gene function.
METHODS
Recombinant DNA
Standard procedures were used for DNA digestion, cloning, and analysis and the preparation of the plasmids pAGCNA and pEX-creINT (Sambrook et al., 1989). Binary vectors are named according to the T-DNA structures they contain. The structures of the T-DNAs of plant transformation vectors used are shown in Figure 1A. Binary vectors were conjugated into Agrobacterium tumefaciens LBA4404 as described previously (Mlynarova et al., 1994).
Plant Material
Tobacco (Nicotiana tabacum cv Petit Havana SR1) was transformed with A. tumefaciens LBA4404 carrying the plasmid of interest as described (Mlynarova et al., 1994). A concentration of 50 μg/mL kanamycin was applied during transformation with pAGCNA. Upon retransformation with pEX-creINT, 20 μg/mL hygromycin was applied in addition to kanamycin. Regenerants were obtained in parallel, without A. tumefaciens incubation. Retransformants and regenerants were selfed and germinated on kanamycin-containing MS medium (Murashige and Skoog, 1962). Plants without the NPTII gene were discarded. Hemizygous and homozygous offspring were grown under the same conditions in a fully climatized greenhouse. The zygotic state of individual plants was determined afterward by segregation of selfed seeds on kanamycin-containing medium.
DNA Analyses of Transgenic Plants
Genomic DNA was isolated from 5-month-old plants and analyzed by DNA gel blot analysis as described (Mlynarova et al., 1994). As probes, the full-length 1.8-kb GUS gene, a 0.6-kb fragment of the NPTII gene, and a 0.6-kb 5′ part of the A element were used. Integration around the right T-DNA borders was analyzed with PCR using the primers P1 (5′-GTAAAACGACGGCCAGT-3′) and P2 (5′-GAAATGCGCGATTTCATT- GGG-3′). For analysis of the left border configuration, three primer pairs were used: P3 (5′-TGCCCGACGGCGATGATC-3′)/P4 (5′-GTACCATAA- CTTCGTATAGCATACATTATACGAAGTTATC-3′; identical to the bottom strand of the loxP site); P5 (5′-CCCAATGAAATCGCGCATTTC-3′)/P6 (5′-GGAAACAGCTATGACCATG-3′); and P7 (5′-GACTGTTAAGCA- ATTTGCTG-3′)/P8 (5′-TCGACATAACTTCGTATAGCATACATTATACGA- AGTTATT-3′; identical to the bottom strand of the loxP site). See Figure 1B for the relative positions of these primers. The first PCR cycle of 3 min at 94°C was followed by 35 cycles of 45 s at 94°C, 45 s at 55°C, and 2 min at 72°C. The last cycle was performed for 7 min at 72°C.
Plant flanking DNA was isolated by walk PCR (Balzergue et al., 2001). Before ligation of an asymmetrical adaptor sequence, genomic DNA (0.5 μg) was restricted with EcoRV. Two successive PCRs nested for both adaptor and T-DNA primers were performed with primers AP1 (5′-GGATCCTAATACGACTCACTATAGGGC-3′) and AP2 (5′-CTATAGGGC- TCGAGCGGC-3′) for the adaptor sequence. Primers specific for the sequence of the right T-DNA were RB-SP1 (5′-CTGGCCGTCGTTTTACAACGTC-3′) and RB-SP2 (5′-CCCGTCAAGCTCTAAATCGGG-3′). The first PCR cycle of 2 min at 94°C was followed by 30 cycles of 30 s at 94°C, 45 s at 65°C, and 2 min at 72°C. The last cycle was performed for 7 min at 72°C. PCR products were cloned into pGEM T-Easy (Promega) and sequenced over both DNA strands using standard procedures on an ABI 3700 DNA analyzer with BigDye terminator cycle sequencing (Applied Biosystems, Foster City, CA). Walk PCR also was used to determine the exact T-DNA configuration at the left borders of lines AGCNA-16 and AGCNA-61 using the A element primers A-SP1 (5′-CACAGTTGCAGCATGCTAACG-3′) and A-SP2 (5′-CAGCTGTTTACGGCA- CTGCC-3′) for line AGCNA-16 and the NPTII primers N-SP1 (5′-CTGGCACGACAGGTTTCC-3′) and N-SP2 (5′-GAGTTAGCTCACTCATTA- GGC-3′) for line AGCNA-61.
RNA Analyses of Transgenic Plants
Total RNA was isolated with Trizol reagent (Life Technologies, Grand Island, NY). For nuclear RNA isolation, the nuclei were obtained according to van Blokland et al. (1994). RNA concentrations were determined fluorimetrically using PicoGreen reagent (Molecular Probes, Eugene, OR) in a microplate fluorescence reader (Fluoroskan 11, LabSystems, Franklin, MA) according to the manufacturer's recommendations. RNA gel blot analysis was performed as described (Mlynarova et al., 2002). From silenced −A plants, 50 μg of total RNA was used on gels. From active +A plants, 5 μg of total RNA was used together with 45 μg of total RNA from untransformed tobacco. Small RNA species were detected in 50 to 150 μg of total RNA on 8% PAGE gels by hybridization with PCR fragment probes as described previously (Hutvagner et al., 2000). Radioactively labeled cDNA was synthesized with 1 to 3 μg of total RNA using primer P9 (5′-GGACTGGCATGAACTTCGG-3′) and Carboxydothermus hydrogenoformans polymerase (Roche, Mannheim, Germany) at 54°C in the presence of α-32P-dATP.
GUS expression was normalized by actin obtained in a cDNA synthesis reaction using primers from tobacco actin RNA: ACfor (5′-GGTGTCAGCCACACTGTCCC-3′) and ACrev (5′-CTTCATGCTGCAAGGAGCCAG-3′). For reverse transcriptase–mediated (RT) PCR analysis, RNA was treated with amplification-grade DNase I (Life Technologies) according to the manufacturer's instructions. For RT-PCR to investigate the type of silencing (Figure 6), first-strand cDNA was synthesized with SuperScript II reverse transcriptase (200 units; Life Technologies) for 1 h at 43°C using an oligo(dT) primer. This was followed by PCR (30 cycles) with SuperTaq polymerase using GUS-specific primers (GUSfor, 5′-GCAGGAGAAACTGCATCAG-3′; GUSrev, 5′-CGATAATTTATCCTAGTTTGC-3′) and actin primers (see above) in the same reaction. To cover the range of amplification in which we assume a linear relationship between the amount of template cDNA and the reaction product, the range of 0.1 to 1 μL of cDNA solution was used.
Run-on transcription assays were performed essentially as described by van Blokland et al. (1994). Nuclei were isolated from ∼25 g of leaves of 6-month-old greenhouse-grown tobacco plants. For each assay, ∼5 × 107 nuclei were used. The labeled RNA transcripts were isolated using Trizol reagent. Slot blots of linearized plasmids (5 μg/slot) were prepared as described by Sambrook et al. (1989). The linearized plasmids comprise the GUS gene (GUS) and the NPTII gene (NPT) both cloned in pSK+. As negative controls, the hygromycin gene in pSK+ (HYG) as well as plasmid pSK+ itself were included. An SK+ plasmid containing the cloned cDNA of a potato chlorophyl a/b binding protein Lhca3.St.1 gene (CAB) was included as an internal standard. Hybridization was performed overnight at 50°C in DIG Easy Hybridization Solution (Roche). Hybridizing signals were visualized and quantified using a Bas2000 phosphorimager (Fuji, Tokyo, Japan) with BasReader and TINA software.
For RT-PCR detection of read-out transcription, 1 μg of nuclear RNA was used, but first-strand cDNA was synthesized with primer P10 (5′-CACTGATAGTTTGTGAACCATC-3′). For RT-PCR detection of read-in transcription, 1 μg of nuclear RNA was used. First-strand cDNA was synthesized as described above with primer P12 (5′-CACCATCGTCGGCTACAGC-3′). In both cases, first-strand cDNA synthesis was followed by PCR (31 cycles) using primers P10 and P12. In all analyses, parallel reactions without reverse transcriptase served as negative controls. RT-PCR products were visualized on 1% agarose gels with ethidium bromide.
Protein Analysis of Transgenic Plants
GUS activity was determined in leaves of tobacco plants as described previously (Mlynarova et al., 1994).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Jan-Peter Nap, j.p.h.nap@plant.wag-ur.nl.
Acknowledgments
We acknowledge the Plant Research International greenhouse team for plant care. We thank Erik Coppoolse (Plant Research International), Ton Bisseling (Molecular Biology, Wageningen University), Jan Kooter (Free University, Amsterdam), and anonymous reviewers for critical and constructive comments on earlier versions of the manuscript. This research was supported by The Netherlands Organization for Scientific Research in the framework of various program subsidies (L.M.) as well as by a personal fellowship (A.H.) and Program Subsidy 347 (J.-P.N. and A.L.), both from the Dutch Ministry for Agriculture, Nature Management, and Fisheries.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.012070.
References
- Allen, G.C., Spiker, S., and Thompson, W.F. (2000). Use of matrix attachment regions (MARs) to minimize transgene silencing. Plant Mol. Biol. 43, 361–376. [DOI] [PubMed] [Google Scholar]
- Avramova, Z., and Tikhonov, A. (1999). Are scs and scs′ ‘neutral’ chromatin domain boundaries of the 87A7 locus in vivo? Trends Genet. 15, 138–139. [DOI] [PubMed] [Google Scholar]
- Balzergue, S., et al. (2001). Improved PCR-walking for large-scale isolation of plant T-DNA borders. Biotechniques 30, 496–503. [DOI] [PubMed] [Google Scholar]
- Bender, J. (2001). A vicious cycle: RNA silencing and DNA methylation in plants. Cell 106, 129–132. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. [DOI] [PubMed] [Google Scholar]
- Bode, J., Schlake, T., Rios-Ramirez, M., Mielke, C., Stengert, M., Kay, V., and Klehr-Wirth, D. (1995). Scaffold/matrix-attached regions: Structural properties creating transcriptionally active loci. Int. Rev. Cytol. 162A, 389–454. [DOI] [PubMed] [Google Scholar]
- Bode, J., Stengert-Iber, M., Kay, V., Schlake, T., and Dietz-Pfeilstetter, A. (1996). Scaffold/matrix-attached regions: Topological switches with multiple regulatory functions. Crit. Rev. Eukaryot. Gene Expr. 6, 115–138. [DOI] [PubMed] [Google Scholar]
- Brouwer, C., Bruce, W., Maddock, S., Avramova, Z., and Bowen, B. (2002). Suppression of transgene silencing by matrix attachment regions in maize: A dual role for the maize 5′ ADH1 matrix attachment region. Plant Cell 14, 2251–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto, C., Azzalin, G., Macino, G., and Cogoni, C. (2002). Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev. 16, 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas, A., and Macino, G. (2001). Characteristics of post-transcriptional gene silencing. EMBO Rep. 2, 992–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni, C., Irelan, J.T., Schumacher, M., Schmidhauser, T.J., Selker, E.U., and Macino, G. (1996). Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. EMBO J. 15, 3153–3163. [PMC free article] [PubMed] [Google Scholar]
- Cogoni, C., and Macino, G. (1999). Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science 286, 2342–2344. [DOI] [PubMed] [Google Scholar]
- Day, C.D., Lee, E., Kobayashi, J., Holappa, L.D., Albert, H., and Ow, D.W. (2000). Transgene integration into the same chromosome location can produce alleles that express at a predictable level, or alleles that are differentially silenced. Genes Dev. 14, 2869–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho, F., Gheysen, G., Kushnir, S., Van Montagu, M., Inze, D., and Castresana, C. (1992). Suppression of β-1,3-glucanase transgene expression in homozygous plants. EMBO J. 11, 2595–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehio, C., and Schell, J. (1994). Identification of plant genetic loci involved in a posttranscriptional mechanism for meiotically reversible transgene silencing. Proc. Natl. Acad. Sci. USA 91, 5538–5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depicker, A., Ingelbrecht, I., van Houdt, H., De Loose, M., and Van Montagu, M. (1996). Post-transcriptional reporter transgene silencing in transgenic tobacco. In Mechanisms and Applications of Gene Silencing, D. Grierson, G. W. Lycett, and G. A. Tucker, eds (Nottingham, UK: Nottingham University Press), pp. 71–84.
- Dorlhac de Borne, F., Vincentz, M., Chupeau, Y., and Vaucheret, H. (1994). Co-suppression of nitrate reductase host genes and transgenes in transgenic tobacco plants. Mol. Gen. Genet. 243, 613–621. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Elmayan, T., and Vaucheret, H. (1996). Expression of single copies of a strongly expressed 35S transgene can be silenced post-transcriptionally. Plant J. 9, 787–797. [Google Scholar]
- Fagard, M., and Vaucheret, H. (2000). Systemic silencing signal(s). Plant Mol. Biol. 43, 285–293. [DOI] [PubMed] [Google Scholar]
- Gerasimova, T.I., Byrd, K., and Corces, V.G. (2000). A chromatin insulator determines the nuclear localization of DNA. Mol. Cell 6, 1025–1035. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J., and Baulcombe, D.C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Hobbs, S.L.A., Warkentin, T.D., and DeLong, M.O. (1993). Transgene copy number can be positively or negatively associated with transgene expression. Plant Mol. Biol. 21, 17–26. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G., McLachlan, J., Pasquinelli, A.E., Balint, E., Tuschl, T., and Zamore, P.D. (2001). A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293, 834–838. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G., Mlynarova, L., and Nap, J.P. (2000). Detailed characterization of the posttranscriptional gene-silencing-related small RNA in a GUS gene-silenced tobacco. RNA 6, 1445–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias, V.A., Moscone, E.A., Papp, I., Neuhuber, F., Michalowski, S., Phelan, T., Spiker, S., Matzke, M., and Matzke, A.J.M. (1997). Molecular and cytogenetic analyses of stably and unstably expressed transgene loci in tobacco. Plant Cell 9, 1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, S.E., Running, M.P., and Meyerowitz, E.M. (1999). Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126, 5231–5243. [DOI] [PubMed] [Google Scholar]
- Jansen, R.C., Nap, J.P., and Mlynarova, L. (2002). Errors in genomics and proteomics. Nat. Biotechnol. 20, 19. [DOI] [PubMed] [Google Scholar]
- Jeddeloh, J.A., Stokes, T.L., and Richards, E.J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22, 94–97. [DOI] [PubMed] [Google Scholar]
- Jorgensen, R.A., Cluster, P.D., English, J., Que, Q., and Napoli, C.A. (1996). Chalcone synthase cosuppression phenotypes in petunia flowers: Comparison of sense vs. antisense constructs and single-copy vs. complex T-DNA. Plant Mol. Biol. 31, 957–973. [DOI] [PubMed] [Google Scholar]
- Jurka, J., Klonowski, P., Dagman, V., and Pelton, P. (1996). CENSOR: A program for identification and elimination of repetitive elements from DNA sequences. Comput. Chem. 20, 110–122. [DOI] [PubMed] [Google Scholar]
- Kellum, R., and Schedl, P. (1992). A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol. 12, 2424–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. (2001). Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumpatla, S.P., Chandrasekharan, M.B., Iyer, L.M., Li, G., and Hall, T.C. (1998). Genome intruder scanning and modulation systems and transgene silencing. Trends Plant Sci. 3, 97–104. [Google Scholar]
- Kunz, C., Schob, H., Stam, M., Kooter, J.M., and Meins, F. (1996). Developmentally regulated silencing and reactivation of tobacco chitinase transgene expression. Plant J. 10, 437–450. [Google Scholar]
- Litiere, K., van Eldik, G.J., Jacobs, J.J., Van Montagu, M., and Cornelissen, M. (1999). Posttranscriptional gene silencing of gn1 in tobacco triggers accumulation of truncated gn1-derived RNA species. RNA 5, 1364–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucy, A.P., Guo, H.S., Li, W.X., and Ding, S.W. (2000). Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J. 19, 1672–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C., Ely, L., Smith, T.H., Marathe, R., Anandalakshmi, R., Fagard, M., Vaucheret, H., Pruss, G., Bowman, L., and Vance, V.B. (2001). HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell 13, 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke, M.A., Matzke, A.J., Pruss, G.J., and Vance, V.B. (2001). RNA-based silencing strategies in plants. Curr. Opin. Genet. Dev. 11, 221–227. [DOI] [PubMed] [Google Scholar]
- Matzke, M.A., Mette, M.F., and Matzke, A.J. (2000). Transgene silencing by the host genome defense: Implications for the evolution of epigenetic control mechanisms in plants and vertebrates. Plant Mol. Biol. 43, 401–415. [DOI] [PubMed] [Google Scholar]
- Meins, F., Jr. (2000). RNA degradation and models for post-transcriptional gene-silencing. Plant Mol. Biol. 43, 261–273. [DOI] [PubMed] [Google Scholar]
- Meyer, P. (1998). Stabilities and instabilities in transgene expression. In Transgenic Plant Research, K. Lindsey, ed (Amsterdam: Harwood Academic Publishers), pp. 263–275.
- Meza, T.J., Kamfjord, D., Håkelien, A.M., Evans, I., Gadager, L.H., Mandal, A., Jakobsen, K.S., and Aalen, R.B. (2001). The frequency of silencing in Arabidopsis thaliana varies highly between progeny of siblings and can be influenced by environmental factors. Transgenic Res. 10, 53–67. [DOI] [PubMed] [Google Scholar]
- Meza, T.J., Stangeland, B., Mercy, I.S., Skårn, M., Nymoen, D.A., Berg, A., Butenko, M.A., Håkelien, A.M., Haslekås, C., Meza-Zepeda, L.A., and Aalen, R.B. (2002). Analyses of single-copy Arabidopsis T-DNA-transformed lines show that the presence of vector backbone sequences, short inverted repeats and DNA methylation is not sufficient or necessary for the transgene silencing. Nucleic Acids Res. 30, 4556–4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynarova, L., Jansen, R.C., Conner, A.J., Stiekema, W.J., and Nap, J.P. (1995). The MAR-mediated reduction in position effect can be uncoupled from copy number–dependent expression in transgenic plants. Plant Cell 7, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynarova, L., Loonen, A., Heldens, J., Jansen, R.C., Keizer, P., Stiekema, W.J., and Nap, J.P. (1994). Reduced position effect in mature transgenic plants conferred by the chicken lysozyme matrix-associated region. Plant Cell 6, 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynarova, L., Loonen, A., Mietkiewska, E., Jansen, R.C., and Nap, J.P. (2002). Assembly of two transgenes in an artificial chromatin domain gives highly coordinated expression in tobacco. Genetics 160, 727–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynarova, L., and Nap, J.P. (2003). A self-excising Cre recombinase allows efficient recombination of multiple ectopic heterospecific lox sites in transgenic tobacco. Transgenic Res. 12, 45–57. [DOI] [PubMed] [Google Scholar]
- Morel, J.-B., Mourrain, P., Beclin, C., and Vaucheret, H. (2000). DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr. Biol. 10, 1591–1594. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Muskens, M.W., Vissers, A.P., Mol, J.N., and Kooter, J.M. (2000). Role of inverted DNA repeats in transcriptional and post-transcriptional gene silencing. Plant Mol. Biol. 43, 243–260. [DOI] [PubMed] [Google Scholar]
- Nayler, O., Strätling, W., Bourquin, J.P., Stagljar, I., Lindemann, L., Jasper, H., Hartmann, A.M., Fackelmayer, F.O., Ullrich, A., and Stamm, S. (1998). SAF-B protein couples transcription and pre-mRNA splicing to SAR/MAR elements. Nucleic Acids Res. 26, 3542–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhuber, F., Park, Y.D., Matzke, A.J., and Matzke, M.A. (1994). Susceptibility of transgene loci to homology-dependent gene silencing. Mol. Gen. Genet. 244, 230–241. [DOI] [PubMed] [Google Scholar]
- Noguchi, S., Tajima, T., Yamamoto, Y., Ohno, T., and Kubo, T. (1999). Deletion of a large genomic segment in tobacco varieties that are resistant to potato virus Y (PVY). Mol. Gen. Genet. 262, 822–829. [DOI] [PubMed] [Google Scholar]
- Nykanen, A., Haley, B., and Zamore, P.D. (2001). ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107, 309–321. [DOI] [PubMed] [Google Scholar]
- Oosumi, T., and Belknap, W.R. (1997). Characterization of the Sol3 family of nonautonomous transposable elements in tomato and potato. J. Mol. Evol. 45, 137–144. [DOI] [PubMed] [Google Scholar]
- Phi-Van, L., and Strätling, W.H. (1996). Dissection of the ability of the chicken lysozyme gene 5′ matrix attachment region to stimulate transgene expression and to dampen position effect. Biochemistry 35, 10735–10742. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N.J., Furger, A., and Dye, M.J. (2002). Integrating mRNA processing with transcription. Cell 108, 501–512. [DOI] [PubMed] [Google Scholar]
- Que, Q.D., Wang, H.Y., English, J.J., and Jorgensen, R.A. (1997). The frequency and degree of cosuppression by sense chalcone synthase transgenes are dependent on transgene promoter strength and are reduced by premature nonsense codons in the transgene coding sequence. Plant Cell 9, 1357–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothnie, H.M. (1996). Plant mRNA 3′-end formation. Plant Mol. Biol. 32, 43–61. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Seymour, G.B., Fray, R.G., Hill, P., and Tucker, G.A. (1993). Down-regulation of two non-homologous endogenous tomato genes with a single chimaeric sense gene construct. Plant Mol. Biol. 23, 1–9. [DOI] [PubMed] [Google Scholar]
- Sijen, T., and Kooter, J.M. (2000). Post-transcriptional gene-silencing: RNAs on the attack or on the defense? Bioessays 22, 520–531. [DOI] [PubMed] [Google Scholar]
- Strätling, W.H., and Yu, F. (1999). Origin and roles of nuclear matrix proteins: Specific functions of the MAR-binding protein MeCP2/ARBP. Crit. Rev. Eukaryot. Gene Expr. 9, 311–318. [DOI] [PubMed] [Google Scholar]
- Szittya, G., Silhavy, D., Molnar, A., Havelda, Z., Lovas, A., Lakatos, L., Banfalvi, Z., and Burgyan, J. (2003). Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J. 22, 633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenllado, F., and Diaz-Ruiz, J.R. (1999). Complete resistance to pepper mild mottle tobamovirus mediated by viral replicase sequences partially depends on transgene homozygosity and is based on a gene silencing mechanism. Transgenic Res. 8, 83–93. [Google Scholar]
- Udvardy, A. (1999). Dividing the empire: Boundary chromatin elements delimit the territory of enhancers. EMBO J. 18, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij, F.E., Jones, L., and Baulcombe, D.C. (2002). Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14, 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blokland, R., van der Geest, N., Mol, J.N.M., and Kooter, J.M. (1994). Transgene-mediated suppression of chalcone synthase expression in Petunia hybrida results from an increase in RNA turnover. Plant J. 6, 861–877. [Google Scholar]
- Vance, V., and Vaucheret, H. (2001). RNA silencing in plants: Defense and counterdefense. Science 292, 2277–2280. [DOI] [PubMed] [Google Scholar]
- Volpe, T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I.S., and Martienssen, R.A. (2002). Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M., Wang, M.B., and Lough, T. (2001). Gene silencing as an adaptive defence against viruses. Nature 411, 834–842. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D. (2002). Ancient pathways programmed by small RNAs. Science 296, 1265–1269. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. (2000). RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101, 25–33. [DOI] [PubMed] [Google Scholar]