Abstract
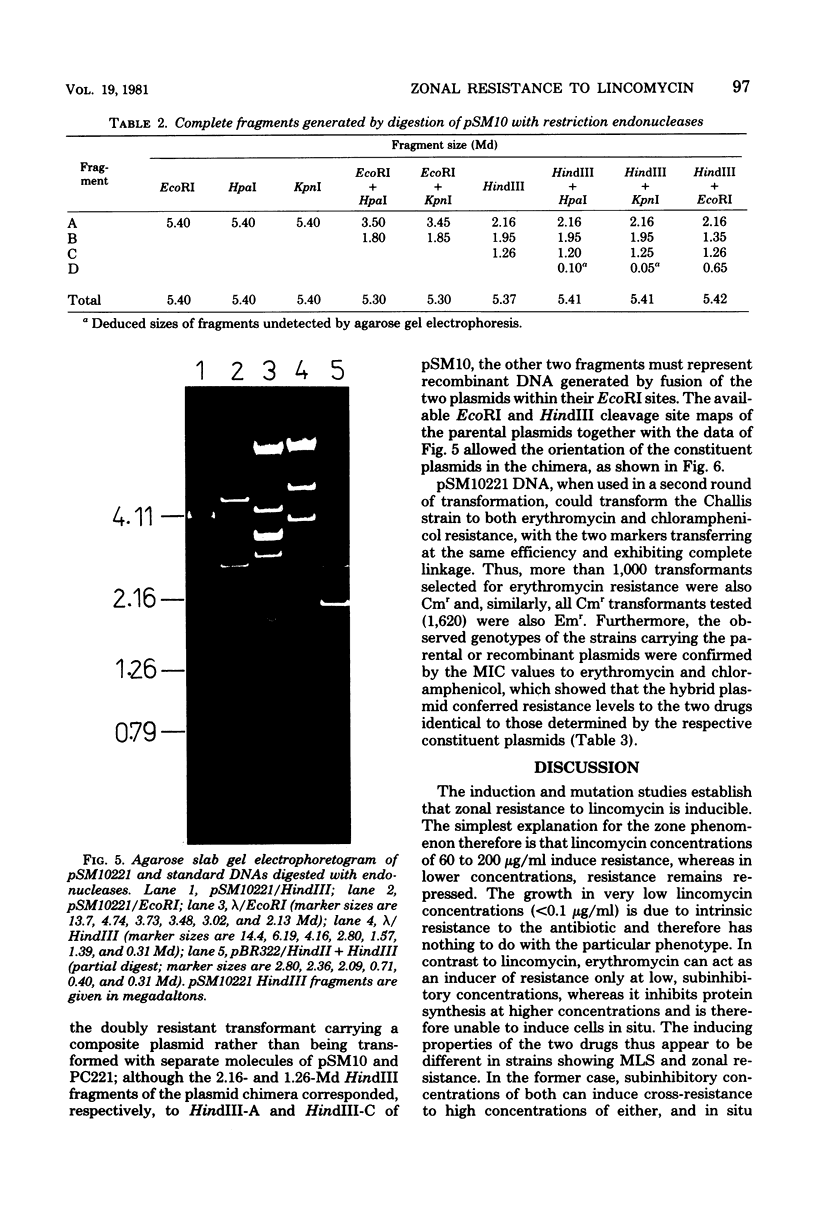
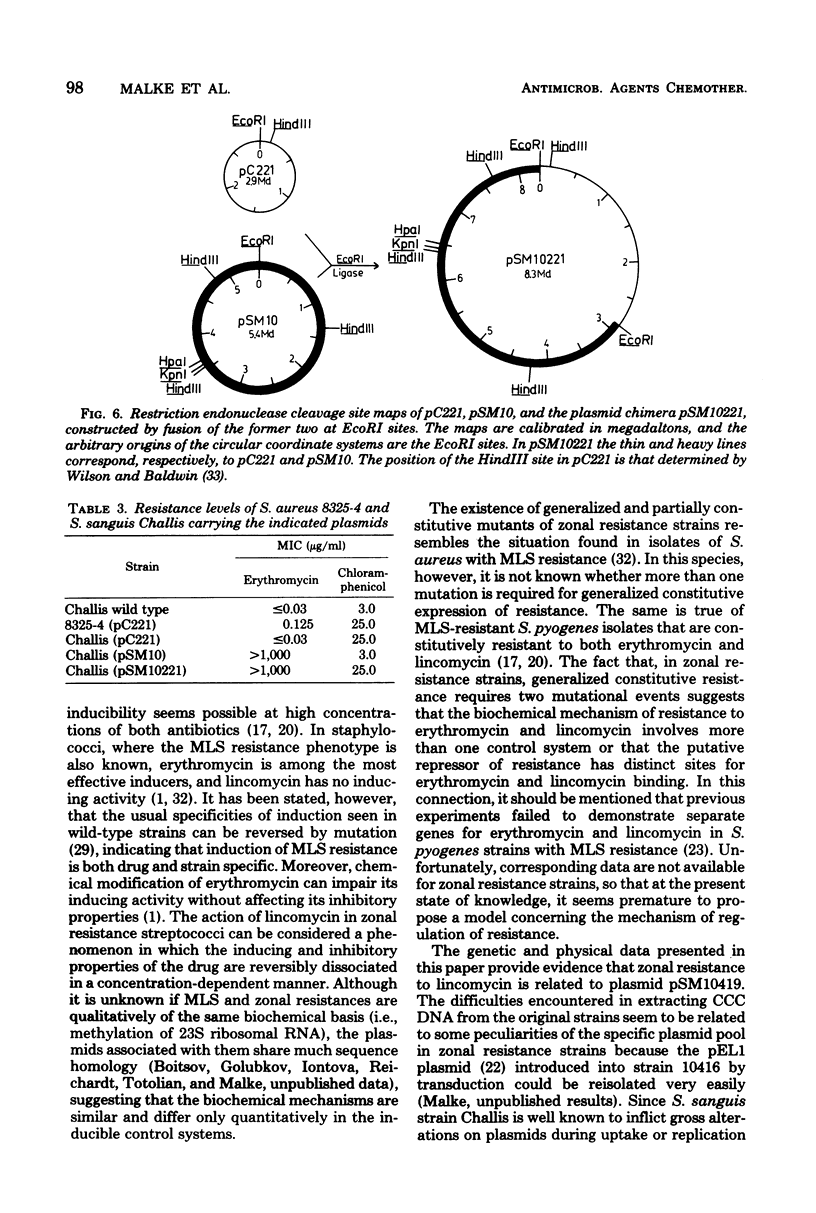
The phenomenon of zonal resistance to lincomycin, which is characteristic of most clinical isolates with lincomycin resistance in Streptococcus pyogenes, has been studied. These strains grow within a defined concentration range of lincomycin (approximately 60 to 200 microgram/ml), or at lincomycin concentrations below the minimal inhibitory concentration for susceptible strains. It is shown that the zonal growth phenomenon is a stable phenotype and results from induction of resistance only within the zonal concentration range of lincomycin. These strains also possess inducible resistance to erythromycin which is nonzonal in character. One-step mutations to constitutive resistance have been isolated which are of two types: constitutive for lincomycin or for erythromycin, but not for both. Those strains with constitutive erythromycin resistance retain their zonal resistance for lincomycin. Mutants doubly constitutive for both lincomycin and erythromycin can be obtained by a second mutational step from either of the singly constitutive mutants. Satellite deoxyribonucleic acid has been shown to be present in the zonal resistant strains. A plasmid, pSM10419, of 14.9 megadaltons, has been isolated from one of the doubly constitutive mutants and used to jointly transform Streptococcus sanguis strain Challis to constitutive resistance to both lincomycin and erythromycin. From this, a multicopy plasmid of reduced size, pSM10 (5.4 megadaltons), which retains its resistance phenotype, has been isolated and mapped with restriction endonucleases HindIII (three sites), EcoRI (one site), KpnI (one site), and HpaI (one site). The staphylococcal plasmid pC221 (2.9 megadaltons; chloramphenicol resistant) has been fused to pSM10 at the EcoRI site resulting in a chimeric plasmid, pSM10221 (8.3 megadaltons), which retains resistance to chloramphenicol, erythromycin, and lincomycin. pSM10 is therefore suggestive as an effective cloning vehicle for the genus Streptococcus.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen N. E. Macrolide resistance in Staphylococcus aureus: inducers of macrolide resistance. Antimicrob Agents Chemother. 1977 Apr;11(4):669–674. doi: 10.1128/aac.11.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak A. L., Christiansen C., Stenderup A. Bacterial genome sizes determined by DNA renaturation studies. J Gen Microbiol. 1970 Dec;64(3):377–380. doi: 10.1099/00221287-64-3-377. [DOI] [PubMed] [Google Scholar]
- Bazaral M., Helinski D. R. Characterization of multiple circular DNA forms of colicinogenic factor E-1 from Proteus mirabilis. Biochemistry. 1968 Oct;7(10):3513–3520. doi: 10.1021/bi00850a028. [DOI] [PubMed] [Google Scholar]
- Behnke D., Malke H., Hartmann M., Walter F. Post-transformational rearrangement of an in vitro reconstructed group-A streptococcal erythromycin resistance plasmid. Plasmid. 1979 Oct;2(4):605–616. doi: 10.1016/0147-619x(79)90058-1. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Clewell D. B., Franke A. E. Characterization of a plasmid determining resistance to erythromycin, lincomycin, and vernamycin Balpha in a strain Streptococcus pyogenes. Antimicrob Agents Chemother. 1974 May;5(5):534–537. doi: 10.1128/aac.5.5.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Yagi Y., Dunny G. M., Schultz S. K. Characterization of three plasmid deoxyribonucleic acid molecules in a strain of Streptococcus faecalis: identification of a plasmid determining erythromycin resistance. J Bacteriol. 1974 Jan;117(1):283–289. doi: 10.1128/jb.117.1.283-289.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courvalin P. M., Carlier C., Croissant O., Blangy D. Identification of two plasmids determining resistance to tetracycline and to erythromycin in group D streptococcus. Mol Gen Genet. 1974;132(3):181–192. doi: 10.1007/BF00269391. [DOI] [PubMed] [Google Scholar]
- Currier T. C., Nester E. W. Isolation of covalently closed circular DNA of high molecular weight from bacteria. Anal Biochem. 1976 Dec;76(2):431–441. doi: 10.1016/0003-2697(76)90338-9. [DOI] [PubMed] [Google Scholar]
- Dixon J. M., Lipinski A. E. Infections with beta-Hemolytic Streptococcus resistant to lincomycin and erythromycin and observations on zonal-pattern resistance to lincomycin. J Infect Dis. 1974 Oct;130(4):351–356. doi: 10.1093/infdis/130.4.351. [DOI] [PubMed] [Google Scholar]
- Dixon J. M., Lipinski A. E. Resistance of group A beta-hemolytic streptococci to lincomycin and erythromycin. Antimicrob Agents Chemother. 1972 Apr;1(4):333–339. doi: 10.1128/aac.1.4.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene P. J., Heyneker H. L., Bolivar F., Rodriguez R. L., Betlach M. C., Covarrubias A. A., Backman K., Russel D. J., Tait R., Boyer H. W. A general method for the purification of restriction enzymes. Nucleic Acids Res. 1978 Jul;5(7):2373–2380. doi: 10.1093/nar/5.7.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershfield V. Plasmids mediating multiple drug resistance in group B streptococcus: transferability and molecular properties. Plasmid. 1979 Jan;2(1):137–149. doi: 10.1016/0147-619x(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Hyder S. L., Streitfeld M. M. Inducible and constitutive resistance to macrolide antibiotics and lincomycin in clinically isolated strains of Streptococcus pyogenes. Antimicrob Agents Chemother. 1973 Sep;4(3):327–331. doi: 10.1128/aac.4.3.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc D. J., Hassell F. P. Transformation of Streptococcus sanguis Challis by plasmid deoxyribonucleic acid from Streptococcus faecalis. J Bacteriol. 1976 Oct;128(1):347–355. doi: 10.1128/jb.128.1.347-355.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malke H. Genetics of resistance to macrolide antibiotics and lincomycin in natural isolates of Streptococcus pyogenes. Mol Gen Genet. 1974;135(4):349–367. doi: 10.1007/BF00271149. [DOI] [PubMed] [Google Scholar]
- Malke H., Jacob H. E., Störl K. Characterization of the antibiotic resistance plasmid ERL1 from Streptococcus pyogenes. Mol Gen Genet. 1976 Mar 30;144(3):333–338. doi: 10.1007/BF00341732. [DOI] [PubMed] [Google Scholar]
- Malke H. Phage A25-mediated transfer induction of a prophage in Streptococcus pyogenes. Mol Gen Genet. 1973 Sep 12;125(3):251–264. doi: 10.1007/BF00270747. [DOI] [PubMed] [Google Scholar]
- Murray K., Murray N. E. Phage lambda receptor chromosomes for DNA fragments made with restriction endonuclease III of Haemophilus influenzae and restriction endonuclease I of Escherichia coli. J Mol Biol. 1975 Nov 5;98(3):551–564. doi: 10.1016/s0022-2836(75)80086-6. [DOI] [PubMed] [Google Scholar]
- Panet A., van de Sande J. H., Loewen P. C., Khorana H. G., Raae A. J., Lillehaug J. R., Kleppe K. Physical characterization and simultaneous purification of bacteriophage T4 induced polynucleotide kinase, polynucleotide ligase, and deoxyribonucleic acid polymerase. Biochemistry. 1973 Dec 4;12(25):5045–5050. doi: 10.1021/bi00749a003. [DOI] [PubMed] [Google Scholar]
- Stiffler P. W., Sweeney H. M., Cohen S. Co-transduction of plasmids mediating resistance to tetracycline and chloramphenicol in Staphylococcus aureus. J Bacteriol. 1974 Nov;120(2):934–934. doi: 10.1128/jb.120.2.934-944.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden B., De Troy B., Roberts R. J., Sambrook J. Agarose slab-gel electrophoresis equipment. Anal Biochem. 1975 Sep;68(1):36–46. doi: 10.1016/0003-2697(75)90676-4. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Weisblum B. Mutant of Staphylococcus aureus with lincomycin- and carbomycin-inducible resistance to erythromycin. Antimicrob Agents Chemother. 1974 May;5(5):538–540. doi: 10.1128/aac.5.5.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum B., Siddhikol C., Lai C. J., Demohn V. Erythromycin-inducible resistance in Staphylococcus aureus: requirements for induction. J Bacteriol. 1971 Jun;106(3):835–847. doi: 10.1128/jb.106.3.835-847.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C. R., Baldwin N. J. Characterization and construction of molecular cloning vehicles within Staphylococcus aureus. J Bacteriol. 1978 Oct;136(1):402–413. doi: 10.1128/jb.136.1.402-413.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]