Abstract
We report the characterization and partial purification of potato mitochondrial RNase Z, an endonuclease that generates mature tRNA 3′ ends. The enzyme consists of one (or more) protein(s) without RNA subunits. Products of the processing reaction are tRNA molecules with 3′ terminal hydroxyl groups and 3′ trailers with 5′ terminal phosphates. The main processing sites are located immediately 3′ to the discriminator and one nucleotide further downstream. This endonucleolytic processing at and close to the tRNA 3′ end in potato mitochondria suggests a higher similarity to the eukaryotic than to the prokaryotic tRNA 3′ processing pathway. Partial purification and separation of RNase Z from the 5′ processing activity RNase P allowed us to determine biochemical characteristics of the enzyme. The activity is stable over broad pH and temperature ranges, with peak activity at pH 8 and 30°C. Optimal concentrations for MgCl2 and KCl are 5 mM and 30 mM, respectively. The potato mitochondrial RNase Z accepts only tRNA precursors with mature 5′ ends. The precursor for tRNAPhe requires RNA editing for efficient processing by RNase Z.
In all genetic systems, mature functional tRNAs are generated by several processing reactions that include the removal of cotranscribed 5′ and 3′ extensions. Processing at the tRNA 5′ end is catalyzed by the ubiquitous endoribonuclease RNase P (1, 2) well characterized in prokaryotes (3). Although the reaction at the tRNA 5′ end is alike in bacteria, archaeas, and eukaryotes (nuclei and organelles), tRNA 3′ end maturation seems to be more variable.
In Escherichia coli an endonucleolytic cleavage several nucleotides downstream of the tRNA is followed by exonucleolytic removal of some of the residual nucleotides. Only after RNase P has then matured the 5′ terminus of the tRNA are the remaining extra 3′ residues removed. This step yields the mature tRNA molecule because the terminal CCAOH sequence is genomically encoded in E. coli (for review, see ref. 4).
The majority of nuclear and chloroplast 3′ processing enzymes have been found to be endonucleases cleaving at the tRNA 3′ end (5–10), although prokaryotic-like 3′ maturation with exonucleases being involved has also been observed (11–14). At present the general consensus appears to attribute a multistep processing pathway to prokaryotes, whereas in eukaryotes single-step reactions predominate.
Mitochondria may have retained a prokaryotic-like processing system because they are thought to be of prokaryotic origin according to the endosymbiont theory (15). On the other hand, they may have adapted the host nuclear enzymes for processing, precedence having been found with tRNA modifying enzymes in yeast mitochondria (16) and plant mitochondria (17). To clarify the nature and origin of the plant mitochondrial tRNA processing machinery, the enzymes involved have to be investigated and characterized in detail.
A special feature of mitochondrial tRNA maturation in plant cells is the involvement of RNA editing (18–20). Herein we demonstrate that RNA editing and 5′ end maturation have to precede tRNA 3′ processing in potato mitochondria. Furthermore, we show that the 3′ extension is removed from the precursor tRNA by an endonuclease. tRNA 3′ processing in potato mitochondria thus seems to be more related to the single-step eukaryotic than to the multistep prokaryotic pathway.
MATERIALS AND METHODS
Materials.
Enzymes were purchased from Boehringer Mannheim, chemicals were from Fluka or Merck, and radioactivity was from Amersham.
Substrate Preparation.
Templates for in vitro transcription were obtained by PCR amplification of plasmids containing the mitochondrial genes or cDNA sequences for tRNATyr and for tRNAPhe (21), respectively, from Oenothera berteriana. The template for in vitro transcription of tRNACys from Nicotiana rustica (GenBank accession no. X85221) was obtained by PCR amplification of total cellular DNA from N. rustica. Template pCys was amplified by using PCR primers CyCys (5′-TAATACGACTCACTATAGGGTCCATAGCTCAGTGGTAG-3′) and Cys-3 (5′-CTCGTATAATAGGAGACTGC-3′). The resulting transcript contains tRNACys (72 nt) and a 3′ trailer (60 nt). Template pTyrI was amplified between the PCR primers Y1 (5′-TAATACGACTCACTATAGGGAGAGTGGCTGAGTGGTCAAAAGCG-3′) and T3 (5′-ATTAACCCTCACTAAAG-3′). Transcription of pTyrI yields a 720-nt RNA containing tRNATyr (83 nt) and a 3′ trailer (637 nt). For template pTyrII, PCR primers Y1 (see above) and NAD2/A (5′-GATGGGGAATCTATAGATCG-3′) were used, the corresponding transcripts contains a 155-nt 3′ trailer and tRNATyr. For template pPheIa (5′ extended edited form), plasmid pPheI was linearized with XhoI. Transcription yields a precursor tRNA with a 117-nt 5′ leader, a 73-nt tRNAPhe, and a 66-nt 3′ trailer. Template pPheIb (5′ matured edited form) was amplified from plasmid pPheI with primers P9 (5′-TAATACGACTCACTATAGTTTAGGTAGCTCAGCTGG-3′) and T3 (see above); the resulting PCR product was digested with XhoI. Transcription yields a 139-nt precursor with a 73-nt tRNA and a 66-nt 3′ trailer. Template pPheIIa (5′ matured unedited form) was synthesized by using PCR primers P8 (5′-TAATACGACTCACTATAGTTCAGGTAGCTCAGCTGG-3′ and T3 (see above) from the genomic sequence of tRNAPhe. Template pPheIIb (edited form) was amplified with primers P9 (see above) and T3 (see above) from the cDNA clone containing the edited version of tRNAPhe. Both templates contain the tRNAPhe (73 nt) and a 114-nt 3′ trailer. Primers were designed such that the first 17 nt of primers Y1, CyCys, P8, and P9 contain the promoter sequence for the T7 RNA polymerase, initiating transcription at nucleotide +1 of tRNATyr, tRNACys, and tRNAPhe, respectively. Plasmid and genomic DNA (100 ng) were amplified by PCRs with 500 ng of each primer in 100 μl of a mixture containing 10 mM Tris⋅HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, all four dNTPs (each at 0.15 mM), and 2 units of Taq DNA polymerase, for 35 cycles of 30 sec at 95°C, 1 min at 52°C, and 30 sec 72°C for pCys; of 1 min at 95°C, 1 min at 39°C, and 1 min at 72°C for pTyrI; of 1 min at 95°C, 1 min at 42°C, and 30 sec at 72°C for pTyrII; and of 1 min at 95°C, 1 min at 35°C, and 30 sec at 72°C for pPheIb, pPheIIa, and pPheIIb. In vitro transcription and purification of the tRNA precursor substrates were carried out as described (22).
Purification of 3′ Endonuclease RNase Z.
Mitochondrial fractionation. Mitochondria were isolated from potato tubers (Solanum tuberosum L., cv. Agria); mitochondrial matrix and intermembrane space fractions were prepared as described (23). A mitochondrial protein fraction virtually devoid of cytoplasmic or amyloplastid contaminations was obtained by purifying mitochondria through Percoll gradients (24). Mitochondria were disrupted by sonification, and a high-speed supernatant (S100) was prepared by ultracentrifugation; 75 kg of potato tubers yielded 25 g of mitochondria and finally 600 mg of S100.
DEAE-Trisacryl column.
Ninety milliliters of DEAE-Trisacryl material (Sigma) was packed according to the manufacturer’s protocol. Matrix and intermembrane space fractions were loaded onto the column in buffer A (40 mM Tris⋅HCl, pH 7.6/10 mM MgCl2/2 mM DTT). The column was washed with buffer B (40 mM Tris⋅HCl, pH 7.6/10 mM MgCl2/0.05% Nonidet P-40/5% glycerol/2 mM DTT/0.5 phenylmethylsulfonyl fluoride) until no proteins were detectable in the flow-through. Retained proteins were eluted with a KCl step gradient (0.3 and 1 M KCl in buffer B) and collected in 10-ml fractions. Fractions of each salt step were pooled and concentrated with Centricon filtration units (Amicon). Protein concentrations were determined with a modified Bradford assay (Bio-Rad, Roth), and fractions were tested for processing activities.
Bio-Rex 70 column.
A 40-ml Bio-Rex 70 column (Bio-Rad) was packed according to the manufacturer’s protocol. The 0.3 M KCl fraction from the DEAE-Trisacryl column was diluted to 0.1 M KCl with buffer B and loaded onto the cation-exchange column. Fractions (10 ml) of each KCl gradient step were pooled and concentrated, and protein concentrations and activities were determined.
Heat treatment.
The RNase Z-active flow-through fraction (containing 0.1 M KCl) from the Bio-Rex column was heated to 60°C in a 90°C water bath and subsequently placed in a 60°C water bath for 15 min. After rapid cooling on ice for 30 min, centrifugation at 30,000 × g for 30 min pelleted the denatured molecules. The resulting supernatant was concentrated and dialyzed against buffer B.
Heparin column.
A 40-ml heparin column (Pharmacia) was packed according to the manufacturer’s instructions, equilibrated with buffer B, and loaded with the heat-treated fraction. A KCl step gradient was applied to elute proteins and 10-ml fractions were collected. RNase Z activity was eluted with 0.3 M KCl, and this fraction was concentrated and dialyzed against buffer B.
Hi-Trap blue column.
A prepacked 1-ml Hi-Trap blue column (Pharmacia) was equilibrated with buffer B. The RNase Z-active fraction obtained from the heparin column was applied to the Hi-Trap blue column. Proteins were eluted with a KCl step gradient and fractions of 1 ml were collected. Pools of each KCl step were concentrated and dialyzed, and protein concentrations and activities were determined. RNase Z activity was eluted between 0.6 and 0.8 M KCl. Aliquots from the RNase Z-active heat-denatured, Heparin, and Blue fractions were analyzed on an SDS/PAGE gel, as described (23) with the only modification that a 10% resolving gel system instead of a 15% gel was used.
Optimization of Processing Assays.
The initial processing assay was performed as described (22). Processing conditions and salt optima were determined with the flow-through fraction from the Bio-Rex column. Aliquots containing 50 μg of protein were incubated in a reaction volume of 100 μl with precursor tRNATyrI in different buffers, depending on the parameters examined. All experiments were repeated three times and the resulting data were averaged. For pH determination, the following buffers were used: Mes for pH 5.5 to pH 6.5, Pipes for pH 7, and Tris for pH 7.5 to pH 8.9. Processing reactions were terminated by phenol and chloroform extractions. Nucleic acids were precipitated, and reaction products were analyzed on 8% polyacrylamide (PAA) gels (25). Products were quantitated by measuring signal intensities of an exposed x-ray film with a Molecular Dynamics densitometer. After determination of optimal reaction conditions, the resulting buffer used for subsequent studies contained 40 mM Tris⋅HCl (pH 8.0), 5 mM MgCl2, 30 mM KCl, and 2 mM DTT. Reactions were incubated for 15 min at 30°C.
Analysis of RNase Z Composition.
Investigation for protein subunits. The in vitro processing assay was preincubated with 5 μl of proteinase K (100 μg) for 20 min at 4°C and then extracted with phenol and chloroform. The resulting aqueous phase was examined in a standard in vitro processing assay.
Investigation for RNA subunits.
The reaction mixture was preincubated with 1 mg of RNase A (immobilized on agarose beads, Sigma) for 15 min at 4°C. RNase beads were removed by repeated centrifugation. One microliter of RNasin (1 unit) was added to the supernatant and activity was tested in a standard in vitro processing assay. Controls for both reactions were treated identically except that neither proteinase K nor RNase A was added. Reaction products were analyzed on 8% PAA gels.
Characterization of Processing Products.
tRNA nucleotidyltransferase reactions. For examination of tRNA nucleotidyltransferase activity, ATP (25 mM), CTP (25 mM), and [α-32P]CTP (40 μCi; 1 Ci = 37 GBq) were added to the standard in vitro processing assay with unlabeled pre-tRNATyrII. The S100 fraction (50 μg of protein), which contains tRNA nucleotidyltransferase activity (23), was used as control.
Analysis of tRNA 3′ end.
In vitro processing assays were performed with unlabeled tRNATyrII precursor. The tRNATyrII precursor was omitted from the control reaction, which was otherwise treated like the precursor-containing reaction mixture. Processing products were separated on 8% PAA gels and mature tRNA molecules were excised according to the migration of processing products of a parallel assay with radiolabeled substrates, eluted in buffer C (0.5 M NH4OAc, pH 5.0/0.1 mM EDTA/0.1% SDS), precipitated, and dissolved in 29 μl of H2O. Eluted unlabeled tRNA products were incubated with [32P]pCp (10 μCi) and T4 RNA ligase (20 units) in a final volume of 40 μl with pCp buffer [50 mM Tris⋅HCl, pH 7.5/10 mM DTT/BSA (0.5 mg/ml)], 4 μl of dimethyl sulfoxide, and 1 unit of RNasin at 4°C for 16 h. Products were analyzed on 8% PAA gels.
Investigation of the 5′ end of the 3′ trailer.
The tRNATyrII precursor was transcribed from template pTyrII in the presence of [α-32P]CTP and gel-purified (22). After processing, reaction products were separated on an 8% PAA gel, and the trailer was eluted, precipitated, washed, and dissolved in 6 μl of H2O. To this solution, 2 μl of NH4OAc (50 mM, pH 4.5), 1 μl of RNase T2 (50 milliunits), and 1 μl of RNase A (100 ng) were added, and the reaction mixture was incubated for 4 h at 37°C to generate 3′-monophosphate nucleosides (Np). Two microliters was withdrawn, dried, and redissolved in 1 μl of a mixture of unlabeled 5′-monophosphate nucleosides (pA, pC, pG, and pU, each at 5 mg/ml), as markers. The solution was applied to a cellulose thin layer plate. Solvent for the first dimension was the mixture isobutyric acid/concentrated ammonia/water (57.7:3.8:38.5, vol/vol) at pH 4.3, and solvent for the second dimension was the mixture isopropanol/concentrated HCl/water (70:15:15, vol/vol). Thin layer plates were air-dried and exposed to x-ray films.
Determination of Cleavage Site.
After processing the 3′ trailer was excised from the PAA gel. A control containing only protein and no precursor was treated the same way to identify signals resulting from endogenous RNA molecules. Primer extension was carried out as described (26) from primer ND2–8: 5′ GGA TAC AAT TTC ACG GCA TGG 3′.
RESULTS
An Endonuclease Is Responsible for tRNA 3′ Processing in Plant Mitochondria.
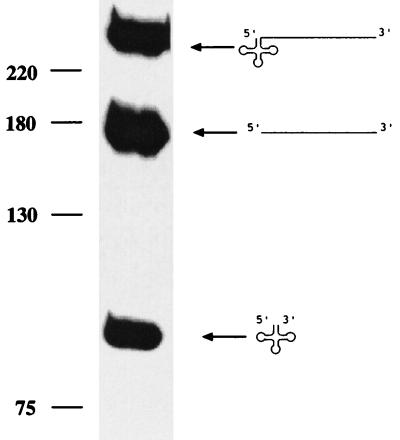
To circumvent exonucleolytic degradation of the 3′ trailer by unspecific exonucleases, pre-tRNATyrII (transcribed from template pTyrII) contains a rather long 3′ trailer of 155 nt. Because some of the described 3′ tRNA processing enzymes accept only 5′ matured tRNA precursors as substrates, the pre-tRNA is synthesized without 5′ leader sequences. Incubation of this substrate with a potato mitochondrial protein fraction (Bio-Rex 70 flow-through fraction) yielded two processing products (Fig. 1), an RNA of about 83 nt, corresponding in length to the tRNATyr, and a second molecule of about 155 nt, corresponding to the 3′ trailer. Therefore, tRNA 3′ end maturation is accomplished by an endonuclease cleaving at or very close to the mature tRNA 3′ end. We termed this tRNA-specific endonuclease RNase Z.
Figure 1.
In vitro processing of pre-tRNATyrII with an RNase Z-active fraction. The RNase Z-containing flow-through fraction from the Bio-Rex 70 column (a cation exchanger) was incubated with the pre-tRNATyrII substrate. A size marker (DNA) is indicated at left, sizes in nucleotides. Precursor (238 nt) and products (tRNA, 83 nt; 3′ trailer, 155 nt) are shown schematically at the right.
Partial Purification of RNase Z.
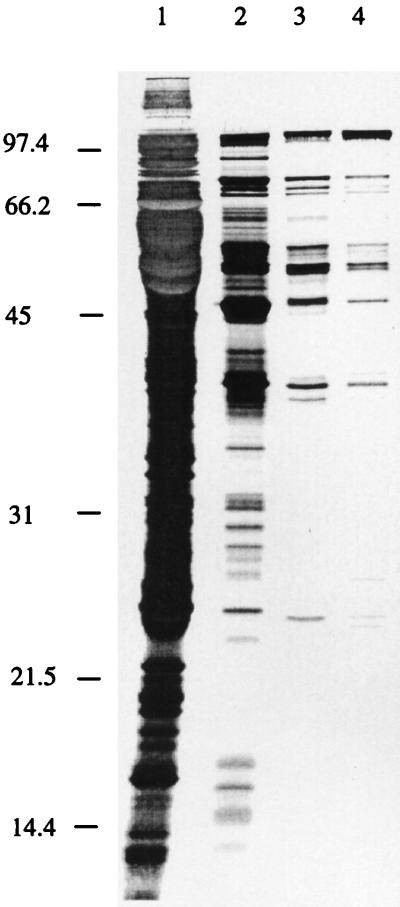
Crude mitochondrial lysates from potato tuber contain 5′ and 3′ tRNA-processing activities, as well as other tRNA modifying enzymes, all together able to process mitochondrial tRNA precursors to mature tRNAs (23). For a detailed analysis of the 3′ tRNA processing activity, we initiated purification of this enzyme RNase Z. The S100 fraction containing matrix and intermembrane space from Percoll gradient-purified mitochondria was applied to an anion-exchange DEAE-Trisacryl column. RNase Z activity was found to coelute with RNase P at 0.3 M KCl. This fraction was further purified on a cation-exchange column (Bio-Rex 70), which separates RNase Z (being in the flow-through containing 0.1 M KCl) from RNase P, the latter eluting between 0.2 and 0.4 M KCl. After heat treatment, the flow-through fraction was loaded onto a heparin column, from which RNase Z was eluted with 0.3 M KCl. From the subsequent HiTrap blue column, RNase Z was eluted between 0.6 and 0.8 M KCl, whereas the bulk of the residual protein was not retained by the column. SDS/PAGE analysis of the last three purification steps shows the degree of purification with comparatively few proteins remaining in the Hitrap Blue fractions (Fig. 2). We expect that only few additional purification steps will be required to yield an RNase Z protein preparation amenable to N-terminal sequence analysis to identify the respective gene. The RNase Z-active Bio-Rex 70 fraction was used to characterize the biochemical properties and requirements of this activity in detail.
Figure 2.
SDS/PAGE of RNase Z-active fractions. To investigate the degree of purification, aliquots of RNase Z-active fractions were loaded onto an SDS/PAGE gel. Molecules were visualized by silver staining. Lanes: 1, 30 μg of the heat-treated fraction; 2, 5.5 μg of the heparin fraction; 3, 500 ng of the 0.6 M KCl Blue fraction; 4, 500 ng of the 0.8 M KCl Blue fraction. Lane 1, showing the protein pattern in the heat-treated fraction, was exposed for a shorter time than the other lanes to allow identification of individual bands. A protein size marker is indicated at the left.
Salt, pH, and Temperature Requirements of the 3′ Processing Enzyme.
To determine optimal reaction conditions for RNase Z, the influence of varying pH, temperature, and salt concentrations was investigated. RNase Z is found active over rather broad temperature and pH ranges, peaking at 30°C and pH 8.0. Although divalent cations in low concentrations (optimum at 5 mM Mg2+) are essential for activity, Mg2+ and Ca2+ concentrations greater than 50 mM inhibit the reaction. Monovalent cations are not essential but enhance processing at low concentrations. Processing is inhibited by KCl and NH4Cl concentrations greater than 200 mM.
Enzyme Composition.
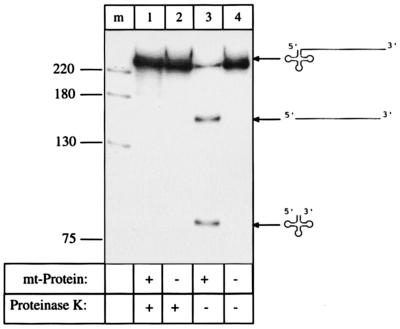
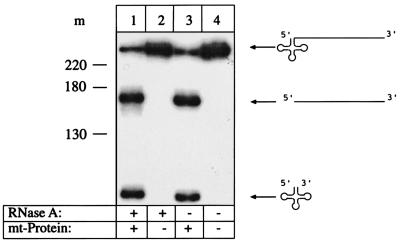
To analyze whether RNase Z is an all-protein enzyme or a ribonucleoprotein containing also RNA subunit(s) like RNase P, the RNase Z-active fraction was incubated with either proteinase K or RNase A prior to the in vitro processing assay. Preincubation with proteinase K eliminates processing activity (Fig. 3), confirming that RNase Z needs one or more protein(s) for activity. RNase A treatment on the other hand showed no effect (Fig. 4), suggesting that RNase Z does not require RNA subunit(s) for activity.
Figure 3.
Analysis of enzyme composition. Preincubation of the mitochondrial fraction with proteinase K abolishes processing activity (lane 1). The control processing assay without proteinase K preincubation shows normal processing activity (lane 3). Control reactions without mitochondrial protein but with tRNA precursor preincubated with proteinase K (lane 2) and with tRNA precursor without addition of mitochondrial protein and without proteinase K (lane 4) show no processing. Lane m is a DNA size marker, sizes in nucleotides. Precursors and products are shown schematically at the right.
Figure 4.
Analysis of enzyme composition. RNase A preincubation of the mitochondrial fraction does not inhibit the processing activity. The reaction with RNase A preincubation (lane 1) shows the same processing activity as a reaction without (lane 3). Controls are a reaction with addition of RNase A but without mitochondrial protein (lane 2) and a reaction with tRNA precursor only and without mitochondrial protein and RNase A (lane 4). A DNA size marker is shown at the left (m), sizes in nucleotides. Precursors and products are shown schematically at the right.
Characterization of 3′ Processing Products.
Maturation of tRNAs includes addition of the CCAOH sequence to the tRNA, which requires a 3′ hydroxyl group at the tRNA terminus. tRNAs released from the precursor by RNase Z can thus only be immediate substrates for the CCA adding nucleotidyltransferase if they contain a 3′ hydroxyl group. To determine whether the potato mitochondrial RNase Z does generate a 3′ hydroxyl terminus at the tRNA, 3′ and 5′ termini of the processing products were examined. Because a cofractionated tRNA nucleotidyltransferase activity would impede the direct analysis of the RNase Z-derived tRNA 3′ end, the respective Bio-Rex 70 fraction was analyzed for tRNA nucleotidyltransferase activity (data not shown). The mitochondrial S100 fraction shown previously to contain tRNA nucleotidyltransferase activity (23) was used as positive control. Incubation of unlabeled precursor tRNATyrII and [α-32P]CTP with the S100 fraction yields a labeled tRNATyr, but no labeled tRNA molecules can be detected upon incubation with the RNase Z-active Bio-Rex fraction, suggesting that there is no or only very little tRNA nucleotidyltransferase activity in the Bio-Rex fraction.
The tRNA 3′ end generated by RNase Z in vitro was then analyzed by incubating an unlabeled pre-tRNATyrII with the Bio-Rex 70 fraction. The tRNA products generated by this reaction were gel-purified and incubated with [α-32P]pCp and T4 RNA ligase. A single product of tRNATyr size is detectable on an 8% PAA gel (data not shown). The successful pCp labeling of the 3′ end of tRNATyr with RNA ligase indicates an accessible 3′ terminal hydroxyl group essential for this ligation. Further indirect evidence for this 3′ hydroxyl group is the efficient addition of the CCAOH triplet to the tRNA processing product by tRNA nucleotidyltransferase activity in the S100 fraction.
For analysis of the 5′ terminal group of the 3′ trailer, the precursor tRNA was transcribed in the presence of [α-32P]CTP instead of [α-32P]UTP to label the 5′ terminal nucleotide (a cytidine) of the 3′ trailer. After in vitro processing, the 3′ trailer was eluted from a denaturing gel and digested with RNase T2 and RNase A to yield 3′-monophosphate nucleosides (Np). The nucleotide mixture was separated by two-dimensional thin layer chromatography (data not shown). In addition to the four expected 3′-monophosphate nucleosides, a fifth nucleotide is detected, whose location on the two-dimensional chromatography corresponds to pCp (27). RNase Z thus leaves a 5′ phosphoryl group on the trailer and a 3′ hydroxyl group on the tRNA product.
Determination of Cleavage Site.
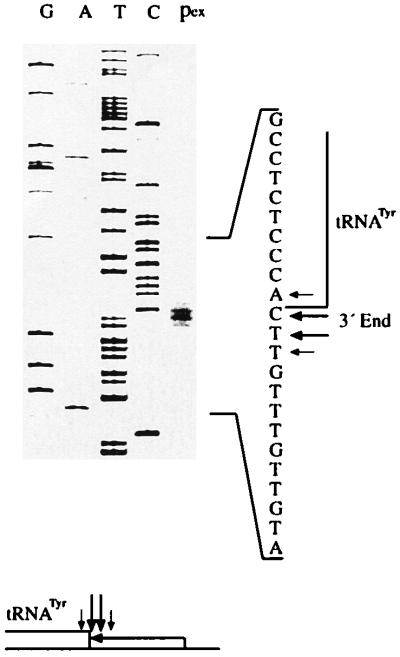
The exact site of the RNase Z cleavage was determined by primer-extension analysis of the purified tRNATyr 3′ trailer molecules, generated by incubation with RNase Z. Primer extension with this trailer generated two prominent and two less abundant cDNA products (Fig. 5). One of the two prominent cDNA species represents molecules cut exactly 3′ to the discriminator nucleotide, and the other reflects 3′ trailers processed 1 nt further downstream. The two less-abundant cDNA molecule populations represent RNAs cleaved 2 nt downstream and 1 nt upstream, respectively, of the discriminator. The 3′ endonuclease thus processes the tRNA precursor at the tRNA 3′ end with some scattering.
Figure 5.
Cleavage site determination by primer extension. Sequencing reactions (lanes G, A, T, and C) and primer extension (lane pex) were started from primer ND2–8. To the right is the coding strand sequence of the pTyrII template. Prominent cleavage sites are indicated with longer arrows and minor sites are indicated with shorter arrows. The main processing sites lie 3′ to the discriminator and 1 nt further downstream. Minor cleavage sites are located 1 nt further upstream of the discriminator and 2 nt further downstream.
Processing of a Nuclear Precursor.
Because plant mitochondrial genomes do not encode the whole set of tRNAs required for function, some of them have to be imported from the nucleus (28). So far, it is not known whether these tRNAs are imported as precursor molecules or as mature tRNAs. To determine whether the mitochondrial RNase Z is able to process nuclear pre-tRNAs, a 5′ matured pre-tRNACys from N. rustica was incubated with the potato mitochondrial extract. The RNase Z-active fraction generates two products, the mature tRNACys and the intact 3′ trailer (data not shown). Thus the mitochondrial RNase Z also accepts nuclear pre-tRNAs as substrates.
5′ Extended Pre-tRNAs Are Not Processed by RNase Z.
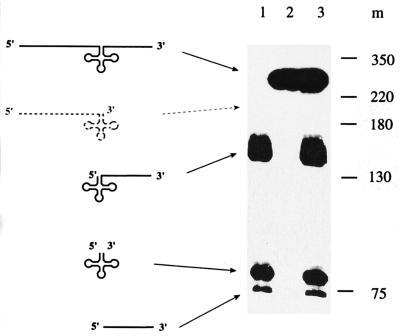
To investigate the influence of the 5′ leader in tRNA precursors on processing by the potato mitochondrial RNase Z, cleavage of 5′ extended (pre-tRNAPheIa) and 5′ matured (pre-tRNAPheIb) pre-tRNAPhe by RNase Z-active fractions devoid of 5′ processing activity was analyzed. Although the 5′ matured precursor is cleaved efficiently and accurately (Fig. 6), no processing is detected with the 5′ extended precursor. This indicates that the potato mitochondrial RNase Z requires a 5′ matured precursor for efficient processing and is inhibited by 5′ leader sequences.
Figure 6.
Processing of 5′ extended precursor tRNAs. 5′ extended and 5′ matured precursor tRNAs were incubated with Bio-Rex 70 fractions to compare respective processing efficiencies. Lanes: 1, incubation of the 5′ matured pre-tRNAPheIb; 2, incubation of the 5′ extended pre-tRNAPheIa; 3, incubation of both pre-tRNAPheIa and pre-tRNAPheIb together; m, DNA marker sizes in nucleotides. Precursors and products are shown schematically at the left. Processing of the 5′ extended precursor would yield a 191-nt product (corresponding to the 5′ leader and the tRNA, shown schematically as dotted molecule) and a 66-nt product (the 3′ trailer).
RNA Editing Has to Precede Processing by RNase Z.
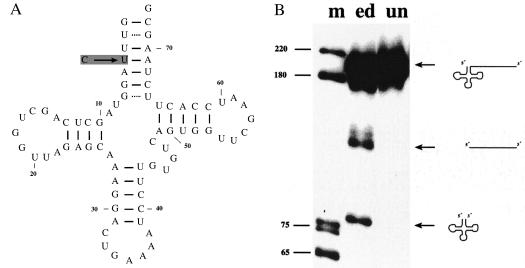
In plant mitochondria, several tRNAs have to be edited to become functional molecules (19, 20). RNA editing has been shown to be essential for 5′ tRNA processing (29, 30), the observation being that tRNAs are not excised from unedited precursors. These studies investigated processing of pre-tRNAs containing 5′ extensions. Because at least for the potato mitochondrial RNase Z, 5′ maturation is essential, the immediate influence of RNA editing on RNase Z has to be analyzed with 5′ matured precursors. Therefore, 5′ matured forms of edited (pre-tRNAPheIIb) and unedited (pre-tRNAPheIIa) pre-tRNAPhe (Fig. 7A) were incubated with potato mitochondrial RNase Z. Although the edited precursor is efficiently processed by RNase Z, only minor cleavage activity is observed with the unedited version (Fig. 7B), suggesting that this RNA editing event is required not only for 5′ processing but also for the 3′ endonuclease RNase Z.
Figure 7.
Processing of unedited and edited pre-tRNAPhe. (A) Cloverleaf structure of tRNAPhe. The editing event is indicated by an arrow. The genomically encoded nucleotide at position 4 of the tRNA is a cytidine, RNA editing converts this cytidine to a uridine in the tRNA. (B) Edited and unedited versions of pre-tRNAPheII were incubated with the Bio-Rex 70 fraction. Lanes: un, incubation of the unedited pre-tRNAPhe; ed, incubation with the edited pre-tRNAPhe; m, DNA marker, sizes in nucleotides. Precursors and products are shown schematically at the right.
DISCUSSION
In this paper, we report the characterization and partial purification of a mitochondrial endonuclease from plants that cleaves tRNA precursors at the tRNA 3′ end; we have designated this endonuclease RNase Z.
Biochemical Characterization.
Divalent cations are required for activity, optimal concentration for Mg2+ and Ca2+ ions being 5 mM. Monovalent cations are not essential for activity but do enhance processing efficiency. Similar reaction parameters have been described for the analogous activities from rat liver mitochondria (7) and Xenopus laevis nuclei (8). The RNase Z activity remains active over broad pH and temperature ranges, indicative of a considerably stable enzyme. The purification steps established for the potato mitochondrial enzyme separated RNase Z from other tRNA processing activities, such as RNase P and terminal tRNA nucleotidyltransferase, confirming that the 5′ and 3′ tRNA processing activities are different enzymes.
Enzyme Composition.
3′ processing is abolished by treatment with proteinase but is not affected by RNase digestion. Thus RNase Z seems to consist of one or more protein subunit(s) without RNA. Although it cannot be excluded that a possible RNA subunit is protected from RNase A degradation by the respective protein(s). An all-protein character has been observed for the nuclear 3′ endonuclease from Xenopus and pig (3, 8) and similarly the plant mitochondrial RNase Z is likely to be an all-protein enzyme.
Analysis of Processing Products.
RNase Z cleavage generates terminal 5′ phosphoryl and 3′ hydroxyl groups that are analogous to RNase P but different from endonucleases RNase A and RNase T1, which both leave 5′ hydroxyl and 3′ phosphoryl groups. The resulting 3′ hydroxyl tRNA terminus facilitates direct addition of the CCA triplet without further intermediate enzymatic steps.
Cleavage Site Selection of RNase Z.
Major in vitro cleavage sites are located 3′ to the discriminator and 1 nt further downstream, identical to the major in vivo cleavage sites (26). Thus the in vitro processing assay established herein faithfully mimics the in vivo situation and allows characterization of the 3′ processing activity. Cleavage 1 nt downstream of the discriminator leaves a cytidine residue at the tRNA 3′ end that could serve as the first base of the mature tRNA-CCAOH terminus. Studies with other 3′ tRNA endonucleases have shown significant sensitivity toward the identity of the first 5′ trailer nucleotides. With the rat liver mitochondrial 3′ tRNase, staggered cleavage of a pre-tRNA containing CCA at the 5′ end of the trailer was observed (7). Studies with mammalian nuclear 3′ tRNase showed that pre-tRNAs containing a cytidine as the 5′ trailer nucleotide are cleaved less efficiently (32). Extensive studies with pre-tRNAs containing different 3′ trailer sequences should reveal processing dependence by the potato mitochondrial RNase Z on the primary structure of the 3′ trailer.
tRNAs Could Be Imported as Precursors.
Plant mitochondria have to import tRNAs from the nucleus to complete their tRNA set (28). Import of tRNAs as precursors requires that the mitochondrial processing activities are able to cleave the nuclear precursors, and indeed the potato mitochondrial RNase Z does correctly process a plant nuclear pre-tRNACys. The similar competence of the plant mitochondrial RNase P toward nuclear pre-tRNAs (33, 34) suggests that tRNA import could occur in the precursor RNA form, the imported tRNA being completely excised from the precursor by the mitochondrial 5′ and 3′ processing activities.
RNase Z Cleaves Only 5′ Matured Precursors.
The potato mitochondrial RNase Z does not cleave a 5′ extended tRNA precursor in vitro. 5′ processing catalyzed by RNase P thus has to precede 3′ processing by RNase Z in a potato mitochondrial extract. In this respect the potato mitochondrial RNase Z resembles most of the tRNA 3′ endonucleases characterized in other organisms (5, 7, 8, 11, 35–38), including carrot nuclei (9). The opposite processing order (3′ processing preceding 5′ processing) has been reported for Bombyx mori (10) and wheat germ nuclear/cytoplasmic extracts (39), whereas in pea and spinach chloroplast (40), as well as in wheat mitochondria (4), 5′ and 3′ processing activities have been found to be independent of each other. The differences in processing orders observed between the wheat (4) and the herein reported potato mitochondrial 3′ endonucleases and between the two reported plant nuclear processing systems (carrot and wheat germ) could suggest species-specific differences (between monocots and dicots) or might be attributed to the different substrates used.
RNA Editing Has to Precede tRNA 3′ Processing.
RNase Z clearly prefers an edited tRNA precursor for 3′ processing of tRNAPhe. The C⋅A mismatch in the acceptor stem of the unedited pre-tRNAPhe seems to interfere with substrate recognition by RNase Z, similar to the highly inefficient processing of this precursor by RNase P (29). Both activities, RNase P and RNase Z, therefore, appear to depend on a correctly folded acceptor stem. The tRNA processing pathway in potato mitochondria thus contains two checkpoints for edited and correctly folded tRNAs. Unedited molecules that slip through the first checkpoint and are processed by RNase P will get stuck at the second checkpoint, which is RNase Z.
Processing Pathway for Mitochondrial Precursor tRNAs.
In summarizing these results, we propose the following maturation pathway for pre-tRNAPhe in potato mitochondria. The processing pathway is initiated by editing of the precursor molecule, which is subsequently cleaved by RNase P to yield the mature 5′ end. RNase Z releases the tRNA from the precursor and finally the CCA triplett is added by the terminal tRNA nucleotidyltransferase. Modification of tRNA nucleotides is not necessary for cleavage by RNase Z and RNase P in vitro. So far, it is not known whether tRNA nucleotide modification precedes the other processing steps or takes place anywhere during the pathway. Precursors and tRNAs have been shown to be substrates for modification enzymes (42), so there may be no general rule for when the different modifications are introduced.
The Potato Mitochondrial RNase Z Has Eukaryotic Properties.
The results reported herein suggest that in plant mitochondria tRNA 3′ ends are generated in a single enzymatic step. The potato mitochondrial 3′ processing thus resembles the single-step maturation observed in nuclear/cytoplasmic extracts and differs from the multistep cleavage event in prokaryotes. Because in mammalian and yeast mitochondria, 3′ processing is also performed in a single-step reaction (5, 7, 43), these organelles most likely lost their prokaryotic 3′ processing activity and adapted the nuclear host-derived enzyme. A similar process has probably occurred in chloroplast evolution, because these plant organelles also use a eukaryotic-like 3′ processing pathway (36, 40, 44). Plant cells appear to contain similar RNase Z activities in each compartment: nucleus (refs. 9, 39, 44, and 45 and M. Mayer and A.M., unpublished results), mitochondrion (ref. 4 and this work), and chloroplast (36, 40, 44), all three enzymes possibly encoded by very similar or even a single nuclear gene(s) and routed to the different organelles after translation in the cytoplasm. It will thus be very interesting to identify the respective nuclear gene(s) in plants and to analyze the sorting of each of the encoded proteins.
Acknowledgments
We thank all members of our lab for helpful discussions and critical reading of the manuscript. We appreciate the gift of pCys from Margit Mayer. Martina Falk was a great help in peeling kilograms of potato tubers and isolating the Percoll-purified mitochondria used in this study. We also thank Elli Bruckbauer for her expert technical assistance. Work described in this study was supported by the Deutsche Forschungsgemeinschaft, Anfangsförderung der Universität Ulm, and Landesschwerpunkt-programm Baden-Württemberg.
ABBREVIATION
- PAA
polyacrylamide
References
- 1.Darr S C, Brown J W, Pace N R. Trends Biochem Sci. 1992;17:178–182. doi: 10.1016/0968-0004(92)90262-8. [DOI] [PubMed] [Google Scholar]
- 2.Altman S, Kirsebom L, Talbot S. In: tRNA: Structure, Biosynthesis and Function. Söll D, RajBhandary U, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 67–78. [Google Scholar]
- 3.Brown J,W, Pace N R. Nucleic Acids Res. 1992;20:1451–1456. doi: 10.1093/nar/20.7.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deutscher M P. In: tRNA: Structure, Biosynthesis and Function. Söll D, RajBhandary U, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 51–65. [Google Scholar]
- 5.Chen J-Y, Martin N C. J Biol Chem. 1988;263:13677–13682. [PubMed] [Google Scholar]
- 6.Hagenbüchle O, Larson D, Hall G I, Sprague K U. Cell. 1979;18:1217–1229. doi: 10.1016/0092-8674(79)90234-4. [DOI] [PubMed] [Google Scholar]
- 7.Manam S, Van Tuyle G C. J Biol Chem. 1987;262:10272–10279. [PubMed] [Google Scholar]
- 8.Castaño J G, Tobian J A, Zasloff M. J Biol Chem. 1985;260:9002–9008. [PubMed] [Google Scholar]
- 9.Franklin S E, Zwick M G, Johnson J D. Plant J. 1995;7:553–563. doi: 10.1046/j.1365-313x.1995.7040553.x. [DOI] [PubMed] [Google Scholar]
- 10.Garber R L, Gage L P. Cell. 1979;18:817–828. doi: 10.1016/0092-8674(79)90134-x. [DOI] [PubMed] [Google Scholar]
- 11.Garber R L, Altman S. Cell. 1979;17:389–397. doi: 10.1016/0092-8674(79)90165-x. [DOI] [PubMed] [Google Scholar]
- 12.Engelke D R, Gegenheimer P, Abelson J. J Biol Chem. 1985;260:1271–1279. [PubMed] [Google Scholar]
- 13.Papadimitriou A, Gross H J. Eur J Biochem. 1996;242:747–759. doi: 10.1111/j.1432-1033.1996.0747r.x. [DOI] [PubMed] [Google Scholar]
- 14.Solari A, Deutscher M P. Mol Cell Biol. 1983;3:1711–1717. doi: 10.1128/mcb.3.10.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray M W. Trends Genet. 1989;5:294–299. doi: 10.1016/0168-9525(89)90111-x. [DOI] [PubMed] [Google Scholar]
- 16.Martin N C, Hopper A K. Biochimie. 1994;76:1161–1167. doi: 10.1016/0300-9084(94)90045-0. [DOI] [PubMed] [Google Scholar]
- 17.Mireau H, Lancelin D, Small I D. Plant Cell. 1996;8:1027–1039. doi: 10.1105/tpc.8.6.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price D H, Gray M W. In: RNA Modification and RNA Editing. Benne R, Grosjean H, editors. Washington, DC: Am. Soc. Microbiol.; 1998. , in press. [Google Scholar]
- 19.Maréchal-Drouard L, Ramamonjisoa D, Cosset A, Weil J H, Dietrich A. Nucleic Acids Res. 1993;21:4909–4919. doi: 10.1093/nar/21.21.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Binder S, Marchfelder A, Brennicke A. Mol Gen Genet. 1994;244:67–74. doi: 10.1007/BF00280188. [DOI] [PubMed] [Google Scholar]
- 21.Binder S, Schuster W, Grienenberger J-M, Weil J-H, Brennicke A. Curr Genet. 1990;17:353–358. doi: 10.1007/BF00314884. [DOI] [PubMed] [Google Scholar]
- 22.Marchfelder A, Schuster W, Brennicke A. Nucleic Acids Res. 1990;18:1401–1406. doi: 10.1093/nar/18.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchfelder A, Brennicke A. Plant Physiol. 1994;105:1247–1254. doi: 10.1104/pp.105.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neuburger M, Journet E P, Bligny R, Carde J P, Douce R. Arch Biochem Biophys. 1982;217:312–323. doi: 10.1016/0003-9861(82)90507-0. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 26.Binder S, Marchfelder A, Brennicke A, Wissinger B. J Biol Chem. 1992;267:7615–7623. [PubMed] [Google Scholar]
- 27.Nishimura S. In: tRNA: Structure, Properties, and Recognition. Schimmel P R, Söll D, Abelson J N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1979. pp. 551–552. [Google Scholar]
- 28.Maréchal-Drouard L, Small I, Desprez T, Masson J, Ramamonjisoa D, Souciet G, Cosset A:, Pelletier G, Weil J H, Dietrich A. In: Plant Mitochondria. Brennicke A, Kück U, editors. New York: VCH; 1993. pp. 131–136. [Google Scholar]
- 29.Marchfelder A, Brennicke A, Binder S. J Biol Chem. 1996;271:1898–1903. doi: 10.1074/jbc.271.4.1898. [DOI] [PubMed] [Google Scholar]
- 30.Maréchal-Drouard L, Cosset A, Remacle C, Ramamonjisoa D, Dietrich A. Mol Cell Biol. 1996;16:3504–3510. doi: 10.1128/mcb.16.7.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nashimoto M. Nucleic Acids Res. 1995;23:3642–3647. doi: 10.1093/nar/23.18.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nashimoto M. Nucleic Acids Res. 1997;25:1148–1154. doi: 10.1093/nar/25.6.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marchfelder A. Mol Biol Rep. 1996;22:151–156. doi: 10.1007/BF00988721. [DOI] [PubMed] [Google Scholar]
- 34.Marchfelder A, Brennicke A. Biochem Mol Biol Int. 1993;29:621–633. [PubMed] [Google Scholar]
- 35.Frendewey D, Dingermann T, Cooley L, Söll D. J Biol Chem. 1985;260:449–454. [PubMed] [Google Scholar]
- 36.Yamaguchi-Shinozaki K, Shinozaki K, Sugiura M. FEBS Lett. 1987;215:132–136. [Google Scholar]
- 37.Wang M J, Davis N W, Gegenheimer P. EMBO J. 1988;7:1567–1574. doi: 10.1002/j.1460-2075.1988.tb02981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Connor J P, Peebles C L. Mol Cell Biol. 1991;11:425–439. doi: 10.1128/mcb.11.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arends S, Schön A. Eur J Biochem. 1997;244:635–645. doi: 10.1111/j.1432-1033.1997.t01-1-00635.x. [DOI] [PubMed] [Google Scholar]
- 40.Marion-Poll A, Hibbert C S, Radebaugh C A, Hallick R B. Plant Mol Biol. 1988;11:45–56. doi: 10.1007/BF00016013. [DOI] [PubMed] [Google Scholar]
- 41.Hanic-Joyce P J, Gray M W. J Biol Chem. 1990;265:13782–13791. [PubMed] [Google Scholar]
- 42.Björk G R. In: tRNA: Structure, Biosynthesis and Function. Söll D, RajBhandary U, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. pp.165–205. [Google Scholar]
- 43.Rossmanith W, Tullo A, Potuschak T, Karwan R, Sbisa E. J Biol Chem. 1995;270:12885–12891. doi: 10.1074/jbc.270.21.12885. [DOI] [PubMed] [Google Scholar]
- 44.Oommen A, Li X, Gegenheimer P. Mol Cell Biol. 1992;12:865–875. doi: 10.1128/mcb.12.2.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stange N, Beier H. EMBO J. 1987;6:2811–2818. doi: 10.1002/j.1460-2075.1987.tb02577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]