Abstract
Caulobacter crescentus divides asymmetrically generating two distinct cell types at each cell division: a stalked cell competent for DNA replication, and a swarmer cell that is unable to initiate DNA replication until it differentiates into a stalked cell later in the cell cycle. The CtrA protein, a member of the response regulator family of the two-component signal transduction system, controls multiple cell cycle processes in Caulobacter and is present in swarmer cells but absent from stalked cells. We report that CtrA binds five sites within the chromosome replication origin in vitro. These sites overlap an essential DnaA box and a promoter in the origin that is essential for replication initiation. Analysis of mutant alleles of ctrA and point mutations in one of the CtrA binding sites in the origin demonstrate that CtrA represses replication in vivo. CtrA-mediated repression at the origin thus restricts replication to the stalked cell type. Thus, the direct coupling of chromosome replication with the cell cycle is mediated by the ubiquitous two-component signaling proteins.
Keywords: CtrA response regulator, Caulobacter crescentus, cell cycle control
The coordination of cell cycle progression and cell differentiation with the initiation of DNA replication is a fundamental, yet poorly understood problem in bacteria. The bacterium Caulobacter crescentus provides an amenable system for studying this problem because cell division in this organism is obligatorily asymmetric, and cellular differentiation is an integral part of the cell cycle (1–3). The predivisional cell yields two distinct progenies upon division; the progeny stalked cell immediately initiates DNA replication but the progeny swarmer cell does not replicate its chromosome until shedding its flagellum and differentiating into a stalked cell later in the cell cycle (refs. 4 and 5; Fig. 1). Thus, the chromosomes in the two poles of the predivisional cell exhibit differential control of replication potential.
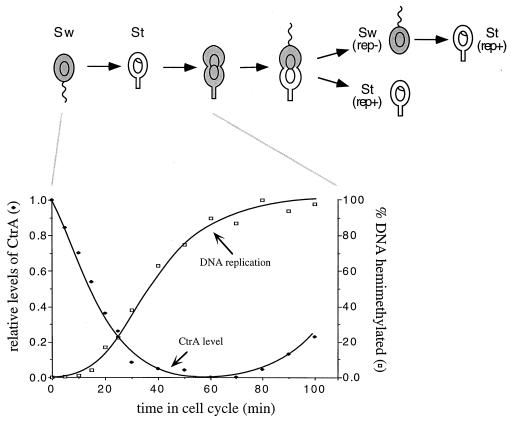
Figure 1.
Initiation of chromosome replication during the Caulobacter cell cycle negatively correlates with CtrA protein abundance. The flagellated swarmer cell (Sw) differentiates into a stalked cell (St) that initiates chromosome replication and asymmetric cell division yielding replicating (rep+) and nonreplicating (rep−) progeny (4, 5). The shading shows that CtrA is restricted to the swarmer cells and to the swarmer compartment of the predivisional cell (17). The graphs present the analysis of synchronized cells assayed simultaneously for CtrA protein abundance and the percentage of the chromosome replicated as the swarmer cells differentiate into stalked cells. The relative abundance of CtrA was determined by Western blot analysis of samples taken at the indicated times in a synchronous cell cycle, and replication state was determined by an assay of the relative amount of fully methylated and hemimethylated DNA as described in the text.
To define the molecular basis of the spatial control of replication initiation in the predivisional cell and the temporal control of replication initiation during the swarmer-to-stalked cell transition, the chromosome replication origin, termed Cori, was cloned and shown to support autonomous plasmid replication (6). Cori plasmid replication, like chromosome replication, initiates specifically in nascent stalked cells, indicating that cell type-specific factors act through regulatory sequences in Cori (6).
The minimal origin region contains many essential regulatory elements, including a strong transcriptional promoter (ref. 7; Fig. 2A), potential binding sites for the DnaA replication initiator (6, 8), and five repeats of a novel GTTAA-N7-TTAA, 9-mer motif (Fig. 2A, sites a–e). Transcription from the strong promoter occurs in stalked cells but not swarmer cells, and two of the five 9-mer sequences overlap the origin promoter (6). In vivo assays of origin promoter activity in synchronized cell populations suggested that expression of the origin promoter is repressed in the swarmer portion of the predivisional cell (6). Thus, the 9-mer sequences are likely binding sites for a repressor of the origin promoter and, consequently, replication initiation.
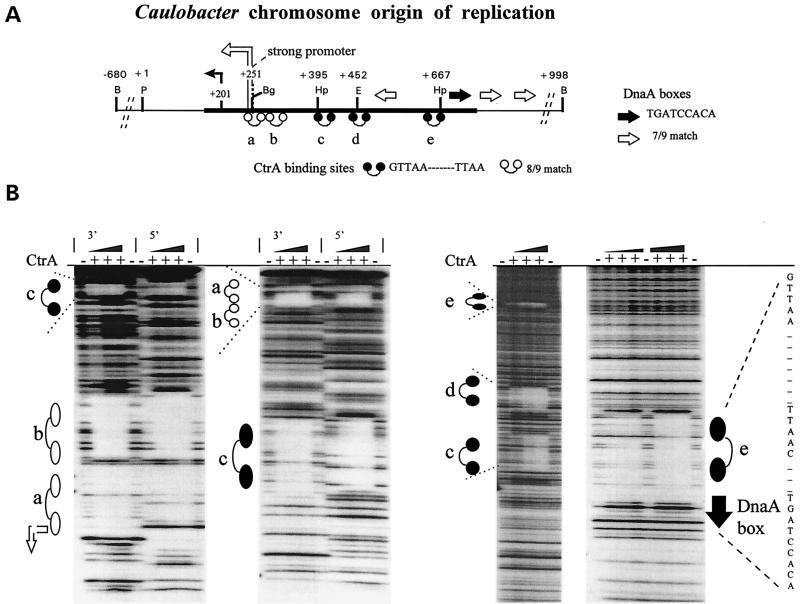
Figure 2.
CtrA protein binds five sites within the Caulobacter chromosome replication origin (Cori). (A) Schematic of Cori. The thick horizontal line shows the minimal DNA sequences that support autonomous Cori plasmid replication (7). The dumbbell-shaped symbols denote the 9-mer motif GTTAA-N7-TTAA sequences (a–e), and short horizontal arrows mark DnaA boxes (6, 8). Bent arrows mark the RNA start sites for the “weak” (+201) and “strong” (+251) promoters (7). Restriction endonuclease sites: B, BamHI; Bg, BglII; E, EcoRI; Hp, HpaI; P, PstI. (B) DNase I protection footprinting experiments. + or − indicates that His6–CtrA protein (50 μg/ml maximum) was added or omitted from the footprinting reactions.
We recently demonstrated that several cell cycle-regulated genes have promoters with a similar 9-mer sequence motif that is recognized by the essential cell cycle regulatory protein CtrA (cell cycle transcription regulator) (9). CtrA is a global regulator that controls the transcription of genes encoding flagellar proteins (9), a DNA methyltransferase (9), and cell division proteins (9, 10). Transcription from the origin promoter increased when CtrA levels were reduced in vivo (9), suggesting that CtrA might regulate this essential replication element (7). CtrA is a member of the “response regulator” superfamily of transcription factors (9) activated by aspartyl phosphorylation in the two-component signal transduction pathways (11, 12). Such proteins have been primarily studied in bacteria, but two-component pathways are also present in archea (13) and eukaryotes (14).
In this report, we demonstrate that in vitro CtrA directly binds the five 9-mer sites within the chromosome replication origin. Further, strains with site-directed mutations to one of the 9-mer sites show increased level of replication, as do strains bearing mutant alleles of ctrA. Overexpression of CtrA blocks replication. Thus, CtrA represses the replication origin in vivo. We propose that cell cycle cues are channeled through CtrA to regulate the initiation of chromosome replication.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, Media, Growth Conditions, and Cell Synchrony.
The synchronizable wild-type strain NA1000 and derivative strains of C. crescentus were grown in PYE complex media or M2G minimal media at 30°C (15). The ctrA401ts strain LS2195 is isogenic to NA1000 (9); the PxyIX::ctrΔ3 strain has a 1.2-kb (BstBI–EcoRI) deletion of chromosomal ctrA complemented by a derivative of the pJS14 high copy number plasmid, pID42HA, carrying a promoterless ctrAΔ3 allele driven by the xylose-inducible PxylX promoter (9, 16). CtrAΔ3 lacks three C-terminal amino acids, causing it to be resistant to proteolysis and thus present at all times in the cell cycle (17). Synchronous swarmer cell cultures were obtained by the method of Evinger and Agabian (18).
Fluorescent Cell Cytometry.
Cultures of C. crescentus wild-type (ctrA+), ctrA401ts, and PxyIX::ctrAΔ3 were fixed in 70% ethanol, and their chromosomal DNA was stained with 10 mg/liter chromomycin A3 (Sigma) and analyzed by a Becton Dickinson FACStar Plus machine as described (20). Data were analyzed by using facs/desk software (Stanford University, Stanford, CA).
Protein Abundance and the Percentage of Replicating Chromosomes.
Pure populations of swarmer cells were obtained by Ludox density centrifugation (18) and allowed to proceed synchronously through the cell cycle (Fig. 1). Cell samples were analyzed by Western blotting with anti-CtrA antibody as described (17). The relative abundance of CtrA protein was measured by laser densitometry of immunoblots (17). Chromosome replication was measured by the previously described methylation state assay at the dnaA locus approximately 4 kb away from Cori (6). This assay is based on the fact that unreplicated DNA is methylated on both strands, but once replicated, DNA remains hemimethylated until the predivisional cell stage (19). DNA was prepared from aliquots of the same cell samples used for Western blot analysis and was assayed for sensitivity to HincII digestion, based on the fact that one HincII recognition site overlaps the Caulobacter CcrM DNA methylation site at the 5′ end of the dnaA gene (19).
Site-Directed DNA Mutagenesis of Cori CtrA Binding Site.
To change CtrA binding site d to the Mut-d sequence shown in Fig. 4, the following oligonucleotides were ligated between unique EcoR47III and EcoRI endonuclease sites at Cori base pair positions 430 and 447 (6). The Mut-d top oligonucleotide GCCTTGAACACACAGGTG was annealed to the Mut-d bottom oligonucleotide AATTCACCTGTGTGTTCAAGGC, and these oligonucleotides were ligated with double-stranded Cori plasmid pGM1270 (7). As a control, oligonucleotides containing the wild-type sequence were likewise annealed and ligated. Both sequences were confirmed by the Sanger dideoxy-sequencing method by using the T7 sequencing kit (Pharmacia).
Figure 4.
Directed mutagenesis at CtrA binding site d alters protein binding and increases Cori plasmid copy number in vivo. (A) Wild-type Cori sequence at site d showing the clustered base pair changes in Mut-d. (B) DNase I protection footprinting experiments were performed as in Fig. 2B (third panel). Outside lanes show the 5′-32P end-labeled DNA fragments cut with EcoRI or HpaI. (C) Cori plasmids per chromosome, as determined by Southern blot hybridization. Caulobacter strain CB15N Δbla was electroplated with pBluescriptII plasmids (Stratagene) bearing otherwise identical wild-type (Wt) or mutant (Mut-d) Cori DNA between BamHI sites at −680 and +998 (see Fig. 1A) and analyzed as described (7). Radioactivity was measured with a PhosphorImager (Molecular Dynamics), and the ratio between Cori plasmid and chromosome band intensities is shown.
CtrA Protein Footprinting Experiments.
DNase I protection footprinting experiments (21) were performed with a purified His6–CtrA fusion protein as described (9). For the experiment described in Fig. 2, the following 32P end-labeled DNA fragments from Cori were employed: first panel, CtrA binding sites a, b, and c were assayed by end-labeling pGM1070 (7) at an artificial HindIII (+221) and by using gel electrophoresis to isolate the short fragment cut at EcoRI (+452); second panel, sites a, b, and c were assayed by end-labeling pGM1070 at EcoRI (+452) and by using gel electrophoresis to isolate the short fragment cut at HindIII; third panel, pGM1070 (7) was 5′ end-labeled at the BglII site (+254); fourth panel, pGM1022 (7) was 5′ end-labeled at an artificial BamHI linker (+714).
RESULTS
Kinetics of CtrA Disappearance Coincides with the Initiation of DNA Replication in a Synchronized Cell Population.
We have shown that a strong promoter within the origin of replication (Fig. 2A) is essential for the temporal control of Cori plasmid replication (7). These studies also suggested that the promoter is bound in vivo by a swarmer cell-specific repressor (7). Consensus binding sites for the CtrA protein overlap the Cori strong promoter (Fig. 2A), and in a strain bearing a ctrA ts allele, origin promoter activity is significantly increased (9). Furthermore, we have recently shown that CtrA is present only in the cell type that is unable to initiate DNA replication (17). To assess the temporal relationship of CtrA disappearance and the initiation of chromosome replication during the swarmer-to-stalked cell transition, samples of a synchronized population were assayed by immunoblotting with CtrA antibody and in the same samples, by monitoring the replication state of the chromosome, as described in Materials and Methods (Fig. 1). As swarmer cells differentiated into stalked cells, CtrA protein levels decreased coincident with the initiation of chromosome replication. Thus, CtrA is likely to be a swarmer cell-specific repressor of chromosome replication. To directly test this hypothesis, we have assayed both in vitro binding of CtrA to the origin promoter and the in vivo effect on DNA replication of genetically lowering or raising levels of CtrA.
CtrA Protein Binds to Five Sites within the Caulobacter Replication Origin.
To test for CtrA binding to Cori, we performed DNase I footprinting experiments (21) throughout the Cori region by using purified His6-tagged CtrA protein (Fig. 2B). We observed a CtrA-dependent 24- to 26-bp zone of protection, flanked by hypersensitive sites, centered over each of the five 9-mer motifs (Fig. 2, sites a–e). No additional DNase I protection in the ≈1-kb Cori region was seen, demonstrating that CtrA selectively binds the 9-mer motifs.
Genetic Manipulation of CtrA Demonstrates Repression of Chromosome Replication.
To test whether CtrA represses replication in vivo, we genetically altered CtrA activity and examined chromosome replication. CtrA activity was decreased by shifting the ctrA401 temperature-sensitive (ts) mutant to a restrictive temperature or increased by overexpressing the CtrAΔ3 protein from the xylose-inducible promoter PxylX (17). CtrAΔ3 lacks a C-terminal degradation signal and, unlike wild-type CtrA (Fig. 1), is present in all cell types (17). To monitor the changes in CtrA activity, we measured Cori strong promoter transcription from a lacZ transcription fusion (Fig. 3A). This promoter is coregulated with chromosome replication (7) and is repressed by CtrA in vivo (9). Thus, it provides a physiologically relevant measure of CtrA activity at Cori.
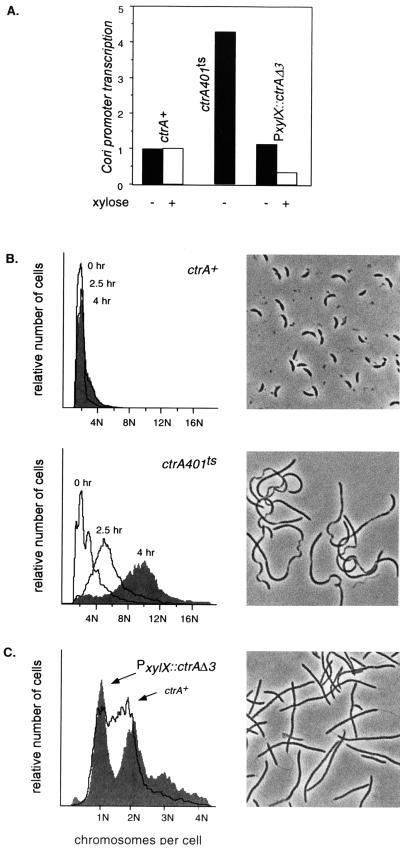
Figure 3.
CtrA is necessary and sufficient to repress chromosome replication in vivo. (A) Results of decreasing and increasing CtrA activity on transcription from the Cori strong promoter (Fig. 1A) measured as relative β-galactosidase units (normalized to wild-type cells) from a lacZ reporter plasmid as described (9). Transcription was assayed in wild-type cells (ctrA+), in a temperature-sensitive mutant (ctrA401ts) and in a strain (PxylX::CtrΔ3) capable of oversupplying a nonproteolyzable CtrAΔ3 protein (17) controlled by a xylose-inducible promoter (16). (B) Loss of CtrA activity derepresses chromosome replication. Wild-type (ctrA+) and mutant ctrA401ts strains were grown at 30°C and then shifted to 37°C for the times indicated; DNA content (N chromosomes per cell) was then measured by fluorescent flow cytometry. Phase contrast photomicrographs correspond to the shaded histograms. (C) Oversupplying CtrA represses chromosome replication. Wild-type (ctrA+) and PxylX::ctrAΔ3 cultures were shifted to 0.2% xylose minimal medium for 10 h and analyzed as in B.
Our genetic experiments demonstrate that CtrA is both necessary and sufficient to repress chromosome replication initiation (Fig. 3 B and C). The ctrA401 ts allele decreased CtrA activity, as evidenced by a 4-fold increase in Cori strong promoter transcription (Fig. 3A), and caused ctrA401 cells to progressively accumulate an average of 10 chromosomes per cell (10n) within 4 h after the temperature shift (Fig. 3B). Conversely, overexpression of CtrAΔ3 (in a chromosomal CtrA null background) increased CtrA activity, as evidenced by a 3-fold decrease in Cori strong promoter transcription (Fig. 3A), and caused cells to arrest with primarily one or two complete chromosomes (1n or 2n) even after 10 h of induction by xylose. Few cells had DNA contents intermediate between 1n and 2n (Fig. 3C), indicating that they could complete but not initiate chromosome replication (20). Overexpressing wild-type CtrA caused no discernible replication block (data not shown), presumably because it was efficiently removed by proteolysis in stalked cells (ref. 17; Fig. 1). Despite the large differences in DNA content, both decreasing and increasing CtrA activity blocked cell division and caused a comparable filamentous cell morphology (Fig. 3 B and C). This argues that CtrA represses DNA replication independent of its effects on cell division (9), most likely because of altered transcription of cell division genes (10).
Direct Repression of Chromosome Replication by Binding CtrA to Origin DNA.
Although direct CtrA binding to Cori in vitro (Fig. 2) implies direct binding in vivo (Fig. 3), CtrA could indirectly repress replication. CtrA regulates the transcription of several genes, and one of these might be a repressor of chromosome replication. To address this issue, we mutated CtrA binding site d as shown in Fig. 4A. This mutation (Mut-d) altered CtrA binding in vitro such that only the unaltered TTAA half site was footprinted (Fig. 4B). To assay the effects of altered CtrA binding to the replication origin, plasmids bearing a wild-type origin or an origin with a Mut-d mutation were moved into a wild-type Caulobacter strain, and plasmid replication activity was monitored (Fig. 4C). Altered binding of CtrA to binding site d increased the in vivo number of Cori plasmids per chromosome approximately 7-fold. These results indicate that wild-type Cori plasmid replication is repressed by CtrA binding in vivo and that the Mut-d Cori base pair changes, shown in Fig. 4A, are by themselves sufficient to relieve this repression.
Our data, in its entirety, therefore demonstrate that CtrA controls chromosome replication during the cell cycle by directly binding and repressing the replication origin in swarmer cells.
DISCUSSION
How Might CtrA Repress Replication?
CtrA could repress replication by binding sites a and b, which completely overlap the strong promoter in the origin of replication (Fig. 2), and blocking RNA polymerase binding. This is a key regulatory step because the Cori strong promoter is essential for replication; it is only transcribed in stalked cells, and its inappropriate transcription abolishes the temporal control of Cori plasmid replication (7). However, the function of the other CtrA binding sites are not explained by this model. Site e lies only 3 bp away from a DnaA box that is essential for autonomous replication (6). CtrA footprinting experiments clearly demonstrate that CtrA overlaps this DnaA box (Fig. 2B). Therefore, CtrA may also act by inhibiting DnaA interaction with the origin. In Escherichia coli, chromosome replication is controlled by a balance between positively acting proteins, such as RNA polymerase (22, 23) and DnaA (8, 24), and negatively acting ones such as SeqA (25, 26) and an inhibitor of DnaA (27). In Caulobacter, an analogous antagonism may restrict replication to the stalked cells.
At present, the role of CtrA binding site c is unknown, although site d is clearly involved in repression (Fig. 4). How might site d repress replication? The presence of five CtrA binding sites within the minimal Cori sequences suggests that CtrA and Cori form a multimeric nucleoprotein complex. Cooperative protein–protein interactions within such a complex could allow CtrA bound at site d to repress RNA polymerase and/or DnaA at distant sites by a DNA looping mechanism. Such cooperative interactions allow the homologous response regulator OmpR to repress transcription (28, 29), and similar interactions regulate plasmid R6K replication (30).
Negative regulation of chromosome replication seems to be required to prevent premature initiation of chromosome replication. The Caulobacter cell cycle provides a striking example of the “once and only once” principle of chromosome replication, a concept normally applied to the eukaryotic S phase, that accounts for exactly one round of chromosome replication per cell cycle. In E. coli, negative regulation of chromosome replication is accomplished, at least in part by the SeqA protein binding origin DNA that becomes hemimethylated by new replication through the Dam methylation sites (25, 26). Although both SeqA and CtrA repress replication, they act by different mechanisms, and they clearly act at different times of the cell cycle. In E. coli, SeqA blocks replication immediately after the initiation of chromosome replication to prevent a premature repetition of the replication cycle (25). CtrA is absent at the time of replication initiation (Fig. 1), and it is not resynthesized until mid S phase (9, 17). The kinetics of CtrA synthesis and its segregation to swarmer cells imply that CtrA acts late in the cell cycle to establish a repressed state. Caulobacter may use a second repression system that, like E. coli SeqA/Dam, functions at the beginning of S phase. Conversely, E. coli SeqA releases the origin well in advance of chromosome replication (25), implying that E. coli may use a second repression system that, like Caulobacter CtrA, functions during later stages of the cell cycle.
How Might CtrA Communicate Cell Cycle and Developmental Cues?
Domian et al. (17) have recently shown that CtrA activity is controlled by temporally regulated phosphorylation as well as by proteolysis. The ability of CtrA to repress the replication origin is therefore likely to be modulated by both of these mechanisms. However, phosphorylation is not strictly required for selective DNA binding in vitro (ref. 9; Figs. 2B and 4B) because we used unphosphorylated protein in our assays. Instead, phosphorylation may increase the affinity and/or cooperativity of CtrA binding, as is the case for OmpR (31, 32). Specific but low affinity binding of unphosphorylated CtrA to Cori probably accounts for the ability of CtrAΔ3 to block chromosome replication in Fig. 3C because CtrAΔ3 is not phosphorylated in stalked cells (17), and overexpression of CtrAΔ3 is required to induce this phenotype (Fig. 3C). Interestingly, CtrAΔ3 overexpression cannot repress replication in a ctrA+ background (ref. 17, unpublished data). The recessive nature of this phenotype may be due to the formation of mixed CtrA+/CtrAΔ3 complexes at Cori that become unable to repress replication once the wild-type protein is targeted for degradation. This idea is also consistent with our hypothesis that CtrA acts cooperatively at Cori.
Relevance to Other Chromosome Replication Systems.
Despite much progress in understanding cell cycle control and DNA replication in both eukaryotes and prokaryotes, the signals that couple these two processes remain obscure. For example, in budding yeast, the timed phosphorylation and proteolysis of the Sic1 protein coordinates DNA replication with the cell cycle (33–36), but how Sic1p-mediated signals are ultimately communicated to replication origins is unclear. The identification of CtrA as a global regulator of cell cycle and developmental events (9), as a cell-type specific protein controlled by proteolysis and phosphorylation (Fig. 1; refs. 9 and 17), and as a repressor of the chromosome replication origin (Figs. 2–4) suggests that CtrA directly communicates cell cycle cues to the Caulobacter replication origin. These observations extend the functions of the versatile two-component regulators to the direct control of DNA replication. Considering the ubiquity and versatility of two-component regulatory systems, it would be surprising if other organisms did not also adapt them to regulate chromosome replication.
Acknowledgments
We thank Ann Reisenauer and Rachel Wright for critical reading of this manuscript. Partial support was received from The Medical Research Council of Canada (MRC) Grant MT-13453 (to B.Y. and G.T.M.), MRC Scholarship Award 273-46 (to G.T.M.), and National Institutes of Health Grant GM51426 (to L.S.).
Footnotes
A commentary on this article begins on page 93.
References
- 1.Brun Y V, Marczynski G T, Shapiro L. Ann Rev Biochem. 1994;63:419–450. doi: 10.1146/annurev.bi.63.070194.002223. [DOI] [PubMed] [Google Scholar]
- 2.Gober J W, Marques M V. Microbiol Rev. 1995;59:31–47. doi: 10.1128/mr.59.1.31-47.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu J, Newton A. Mol Microbiol. 1997;24:233–239. doi: 10.1046/j.1365-2958.1997.3281691.x. [DOI] [PubMed] [Google Scholar]
- 4.Degnen S T, Newton A. J Mol Biol. 1972;64:671–680. doi: 10.1016/0022-2836(72)90090-3. [DOI] [PubMed] [Google Scholar]
- 5.Marczynski G T, Dingwall A, Shapiro L. J Mol Biol. 1990;212:709–722. doi: 10.1016/0022-2836(90)90232-B. [DOI] [PubMed] [Google Scholar]
- 6.Marczynski G T, L. Shapiro L. J Mol Biol. 1992;226:959–977. doi: 10.1016/0022-2836(92)91045-q. [DOI] [PubMed] [Google Scholar]
- 7.Marczynski G T, Lentine K, Shapiro L. Genes Dev. 1995;9:1543–1557. doi: 10.1101/gad.9.12.1543. [DOI] [PubMed] [Google Scholar]
- 8.Skarstad K, Boye E. Biochim Biophys Acta. 1994;1217:111–130. doi: 10.1016/0167-4781(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 9.Quon K C, Marczynski G T, Shapiro L. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 10.Quardokus E, Din N, Brun Y V. Proc Natl Acad Sci USA. 1996;93:6314–6319. doi: 10.1073/pnas.93.13.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parkinson J S, Kofoid E C. Annu Rev Genet. 1992;26:71–83. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 12.Swanson R V, Alex L A, Simon M I. Trends Biochem Sci. 1994;19:485–490. doi: 10.1016/0968-0004(94)90135-x. [DOI] [PubMed] [Google Scholar]
- 13.Rudolph J, Tolliday N, Schmitt C, Schuster S C, Oesterhelt D. EMBO J. 1995;14:4249–4257. doi: 10.1002/j.1460-2075.1995.tb00099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alex L A, Borkovich A, Simon M I. Proc Natl Acad Sci USA. 1996;93:3416–3421. doi: 10.1073/pnas.93.8.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ely B. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- 16.Meisenzahl A, Shapiro L, Jenal U. J Bacteriol. 1997;179:592–600. doi: 10.1128/jb.179.3.592-600.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domian I J, Quon K C, Shapiro L. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 18.Evinger M, Agabian N. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zweiger G, Marczynski G T, Shapiro L. J Mol Biol. 1994;235:472–485. doi: 10.1006/jmbi.1994.1007. [DOI] [PubMed] [Google Scholar]
- 20.Winzeler E, Shapiro L. J Mol Biol. 1995;251:346–365. doi: 10.1006/jmbi.1995.0439. [DOI] [PubMed] [Google Scholar]
- 21.Galas D, Schmitz A. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker T A, Kornberg A. Cell. 1988;55:113–123. doi: 10.1016/0092-8674(88)90014-1. [DOI] [PubMed] [Google Scholar]
- 23.Theisen P W, Grimwade J E, Leonard A C, Bogan J A, Helmstetter C E. Mol Microbiol. 1993;10:575–584. doi: 10.1111/j.1365-2958.1993.tb00929.x. [DOI] [PubMed] [Google Scholar]
- 24.Crooke E, Castuma C E, Kornberg A. J Biol Chem. 1992;267:16779–16782. [PubMed] [Google Scholar]
- 25.Lu M, Campbell J L, Boye E, Kleckner N. Cell. 1994;77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 26.Brendler T, Abeles A, Austin S. EMBO J. 1995;14:4083–4089. doi: 10.1002/j.1460-2075.1995.tb00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katayama T, Crooke E. J Biol Chem. 1995;270:9265–9271. doi: 10.1074/jbc.270.16.9265. [DOI] [PubMed] [Google Scholar]
- 28.Harlocker S L, Bergstrom L, Inouye M. J Biol Chem. 1995;270:26849–26856. doi: 10.1074/jbc.270.45.26849. [DOI] [PubMed] [Google Scholar]
- 29.Huang K J, Igo M M. J Mol Biol. 1996;262:615–628. doi: 10.1006/jmbi.1996.0540. [DOI] [PubMed] [Google Scholar]
- 30.Murherjee S, Erickson H, Bastia D. Cell. 1988;52:375–383. doi: 10.1016/s0092-8674(88)80030-8. [DOI] [PubMed] [Google Scholar]
- 31.Rampersaud A, Harlocker S L, Inouye M. J Biol Chem. 1994;269:12559–12566. [PubMed] [Google Scholar]
- 32.Huang K J, Lan C Y, Igo M M. Proc Natl Acad Sci USA. 1997;94:2828–2832. doi: 10.1073/pnas.94.7.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nugroho T T, Mendenhall M D. Mol Cell Biol. 1994;14:3320–3328. doi: 10.1128/mcb.14.5.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tyers M. Proc Natl Acad Sci USA. 1996;93:7772–7776. doi: 10.1073/pnas.93.15.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knapp D, Bhoite L, Stillman D J, Nasmyth K. Mol Cell Biol. 1996;16:5701–5707. doi: 10.1128/mcb.16.10.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider B L, Yang Q H, Futcher A B. Science. 1996;272:560–562. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]