Abstract
Stra13 is a transcriptional repressor related within its basic helix–loop–helix domain with the Drosophila Hairy, Enhancer of Split, and the mouse Hes1 proteins that interact with the corepressor Groucho. Because Stra13 lacks the conserved WRPW motif for interaction with Groucho, we examined the function and mechanism of transcriptional repression mediated by Stra13 that exhibits several distinctive features. Here, we report that Stra13 expression is closely associated with cell growth arrest induced by several triggers such as retinoic acid and trichostatin A (TSA; a specific histone deacetylase inhibitor) as well as by serum starvation. Stra13 expression is transcriptionally repressed and maintained at a low level in cells through a negative autoregulatory mechanism that is brought about by its interaction with the corepressor histone deacetylase (HDAC1). This interaction requires the Stra13 C-terminal domain containing three α-helices, which are also functionally critical to its repressive activity. Thus, inhibition of HDAC activity by TSA abrogates Stra13-mediated repression of its promoter, resulting in induction of Stra13 expression that is coincident with TSA-induced growth arrest. Further, once induced, Stra13 strongly represses the expression of the cell proliferation-associated gene c-Myc through an HDAC1-independent pathway that involves its interaction with the basal transcription factor TFIIB. Our studies indicate that Stra13 may play a key role in signaling pathways that lead to growth arrest and terminal differentiation by repression of target genes via HDAC-dependent and HDAC-independent mechanisms.
Transcription factors of the basic helix–loop–helix (bHLH) family are important regulators of cellular growth, differentiation, and apoptosis (1). Stra13 is a novel bHLH gene (2) that exhibits the highest sequence identity in the bHLH domain with the Drosophila Hairy (H), Enhancer of Split [E(Spl)], and mouse Hes1 proteins (3). Members of this subfamily bind to an N-box sequence element and act as transcriptional repressors by recruiting the corepressor Groucho through a highly conserved “WRPW” motif (4). Outside the bHLH domain, Stra13 shares no significant identity with known proteins and is characterized by three putative α-helices in its C terminus. Although Stra13 does not bind the N-box element, it does exhibit a strong transcriptional repression activity that is mediated through the α-helices (2). Moreover, unlike E(Spl), Hairy, and Hes, Stra13 lacks a WRPW motif, suggesting that it may mediate transcriptional repression by interaction with corepressors other than Groucho.
Recent studies have provided molecular evidence that modification of chromatin structure by histone deacetylation is an important mechanism in controlling gene transcription. Several transcriptional repressors such as YY-1, RB, and CBF-1 interact directly with histone deacetylases (HDAC), whereas nuclear hormone receptors, Mad, and PLZF are linked indirectly to HDAC through additional components [silencing mediator of retinoic acid and thyroid hormone receptor (SMRT), nuclear receptor corepressor (NCoR)] of the Sin3-HDAC corepressor complex (5–9). Recruitment of HDAC by these factors results in deacetylation of histone tails and in transcriptional repression.
We demonstrate here that Stra13 expression is associated with growth arrest of cells induced by several different triggers such as all-trans-retinoic acid (RA) or trichostatin A (TSA; a specific HDAC inhibitor) or by serum starvation. Stra13 mediates TSA-induced growth arrest through two distinct mechanisms. One involves alleviation of negative autoregulation of Stra13 expression that is brought about by a physical interaction of Stra13 with HDAC1. In addition, Stra13 strongly represses the expression of the cell proliferation-associated gene c-Myc. This inhibition of c-Myc expression is not dependent on the ability of Stra13 to interact with HDAC1 but, rather, on its ability to interact with the basal transcription factor TFIIB. Thus, Stra13 can use two independent pathways to mediate TSA-induced growth arrest via interactions with distinct regulatory factors. These results not only demonstrate a functional divergence between Stra13 and Hes proteins, but also indicate the existence of a cross-regulatory network involving members of the bHLH family of transcription factors.
Materials and Methods
Cell Culture and FACS Analysis.
NIH 3T3 cells grown in DMEM/10% FCS (GIBCO/BRL) were seeded at a density of 5 × 105/10-cm dishes. On the next day, cells were transferred to medium containing 0.1% FCS or 1 μM TSA. Samples were collected at various time points and analyzed by Western blot and FACS analyses. For FACS analysis, cells were fixed with cold 70% ethanol and treated with 1 mg/ml RNase A for 30 min at 37°C. Cell pellets were resuspended in 50 μg/ml propidium iodide (Sigma) in 30 mM sodium citrate. Cell cycle analysis was performed with 20,000 cells on a FACScan (Becton Dickinson).
Northern Blot Analysis.
Twenty micrograms of total RNA from P19 cells grown in the absence or presence of 1 μM RA (Sigma) was run on formaldehyde-agarose gels, transferred onto Hybond N+ membranes, and hybridized with 32P-labeled cDNA probes for Stra13, c-Myc, and 36B4.
Colony-Formation Assay.
NIH 3T3 cells were cotransfected with 4 μg of Stra13 (1–411) or pCS2 along with 0.4 μg of pD503, which contains the puromycin resistance gene. Forty-eight hours after transfection, one-half of the cells were collected for Western blot analysis and the remaining were cultured in the presence of 2 μg/ml puromycin (Sigma). Two weeks later, the colonies were fixed and stained with 0.1% crystal violet.
Plasmid Constructs.
Stra13 promoter constructs were made by cloning a KpnI-NheI fragment spanning −3.5 kb to +101 and a PmlI-NheI fragment spanning −422 to +101 into the promoterless luciferase reporter pGL3 (Promega) from a 129Sv/J BAC genomic library (10). Expression vectors for Stra13 (1–411) or (1–127) were constructed in the EcoRI or EcoRI/XhoI sites of pCS2. His-Stra13 (1–127) was constructed from GAL-Stra13 (1–127) (2) in the BamHI site of HisC (Invitrogen). Glutathione S-transferase (GST)-Stra13 (1–411) construct has been described (2). The GST-Stra13 constructs 1–127, 1–147, and 111–411 were generated from the respective GAL-Stra13 plasmids (2) and cloned into the BamHI site of pGEX-2TK. GST-Stra13 (111–343) was generated from GST-Stra13 (111–411) by intramolecular ligation using KpnI and SmaI. The HDAC1 vector was a gift from S. Schreiber (Howard Hughes Medical Institute, Harvard University); Sin3A was from D. Ayer (Huntsman Cancer Institute, University of Utah); NCoR came from C. Glass (University of California, San Diego); and the c-Myc promoter [P2 (-2489)-luc] and cDNA were from E. Ziff (New York University Medical Center).
Transient Transfection Assays.
COS-7 cells were transfected with the reporter to expression vector in ratios indicated in the figures. Transfection efficiencies were normalized by using 50 ng of the β-galactosidase plasmid pCH110 (Amersham Pharmacia). Forty-eight hours after transfection, cells were assayed for luciferase activity (Promega) and normalized with β-galactosidase activity.
GST Pull-Down Assays and Coimmunoprecipitation Analysis.
GST assays were done as described previously (2). For immunoprecipitation assays, COS-7 cells were cotransfected with GAL-Stra13 (1–411) or (1–127) and expression vectors for HDAC1, Sin3A, or NCoR. Two hundred micrograms of proteins was immunoprecipitated and detected by Western blot with appropriate antibodies as indicated in the figures.
Immunostaining.
COS-7 cells were transfected with His-Stra13 (1–127) and fixed with 4% paraformaldehyde. Cells were incubated with a monoclonal His antibody (1:400 dilution; Qiagen) at 4°C overnight, followed by 1 h at room temperature with a goat anti-rabbit biotinylated antibody (1:1,000 dilution; Vector Laboratories), and visualized by the Vectastain ABC Kit (Vector Laboratories).
Results
RA Induces Stra13 Expression and Down-Regulates c-Myc Expression in P19 Cells.
RA induces growth arrest and differentiation of several cell lines. We have shown previously that Stra13 expression is up-regulated upon RA treatment of P19 cells (2). To determine whether the induction of Stra13 expression by RA related to the early events of growth arrest or a later phase of differentiation, we analyzed the kinetics of Stra13 expression by Northern blot analysis upon RA treatment of P19 cells at various time points (Fig. 1A). Stra13 transcripts were up-regulated within 3 h of RA treatment, and this rapid induction suggested that Stra13 might be associated with cell growth arrest rather than differentiation induced by RA. Interestingly, the up-regulation of Stra13 expression was concomitant with the down-regulation of c-Myc expression levels (Fig. 1A), suggesting that Stra13 may act as a repressor of c-Myc.
Figure 1.
Up-regulated Stra13 expression is associated with growth arrest. (A) P19 cells were treated with 1 μM RA for 0–24 h, and the expression of Stra13 and c-Myc was analyzed by Northern blot analysis. The amounts of RNA were normalized with 36B4 transcripts that are not altered upon RA treatment. (B) Asynchronized NIH 3T3 cells were treated with 1 μM TSA for 0–24 h, and the expression of Stra13 and c-Myc was analyzed by Western blot analysis. FACS analysis of control and TSA-treated cells (collected 12 h after TSA addition) confirmed that the cells are arrested in the G2/M phase. (C) NIH 3T3 cells grown in 10% FCS were transferred to medium containing 0.1% FCS. Cell lysates were collected at the times indicated and subjected to Western blot analysis. FACS analysis confirmed that the cells are arrested in the G1 and G2/M phases of the cell cycle. (D) NIH 3T3 cells were cotransfected with either the empty vector (pCS2) or a Stra13 expression vector together with a puromycin resistance plasmid. After 14 days of selection with puromycin, the colonies were stained with crystal violet. (Left) Representative plates from two independent experiments. The mean colony numbers obtained with pCS2 vs. Stra13 are shown in the Center. (Right) Western blot analysis of cell lysates from vector- (pCS2) or Stra13-transfected samples collected 48 h after transfection, confirming that Stra13 was expressed in the transfected cells.
Elevated Stra13 Expression Is Associated with Growth Arrest.
We further evaluated whether Stra13 expression is invariably up-regulated during growth arrest induced by different agents. TSA and other histone deacetylase inhibitors such as trapoxin and butyrate induce growth arrest in the G1 and G2/M phases of the cell cycle (11–13) by regulation of largely unknown targets. NIH 3T3 cells treated with TSA for various time points were analyzed for Stra13 expression by Western blot analysis (Fig. 1B). Interestingly, TSA strongly up-regulated the expression of the endogenous Stra13 gene, which was coincident with TSA-induced arrest of the cells in the G2/M phase of cell cycle as measured by FACS analysis, and a decrease in c-Myc expression (Fig. 1B).
We next examined Stra13 expression by Western blot analysis upon serum starvation of NIH 3T3 cells, which also leads to growth arrest. As shown in Fig. 1C, although Stra13 is expressed at a low basal level in exponentially growing cells (time 0), it is up-regulated within 6 h of serum starvation. This again is coincident with repression of c-Myc expression and a cell cycle arrest in serum-starved cells relative to control cells grown in 10% serum as determined by FACS analysis (Fig. 1C). Taken together, these results indicate that Stra13 expression is consistently up-regulated during growth arrest and therefore may be associated with or directly act as a suppressor of cell growth.
To evaluate the potential role of Stra13 as a growth suppressor, colony-formation assays were performed. A eukaryotic expression vector for Stra13 (1–411) or the empty vector (pCS2) was cotransfected with a puromycin selection plasmid pD503 into NIH 3T3 cells. Cells were selected in the presence of puromycin, and the number of resistant colonies was determined after 2 weeks (Fig. 1D Left and Center). NIH 3T3 cells transfected with a Stra13 expression vector showed an ≈5-fold reduction in colony numbers compared with vector-transfected cells. Immunoblot analysis of cell lysates collected 48 h after transfection confirmed that Stra13 was expressed in the transfected cells (Fig. 1D Right). Taken together, these results indicate that Stra13 can function as a growth inhibitor.
Stra13 Negatively Autoregulates Its Expression Through an HDAC-Dependent Pathway.
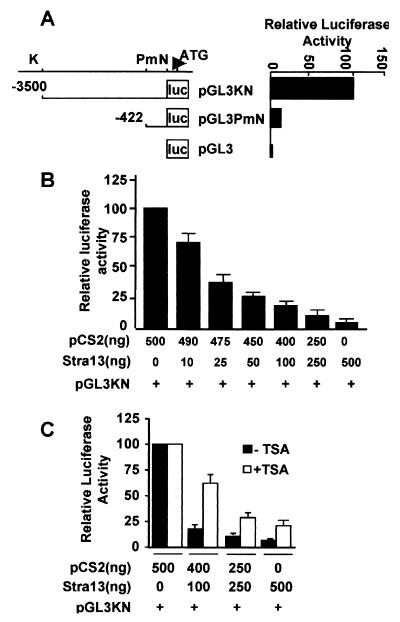
The up-regulation of the endogenous Stra13 gene in the presence of TSA (Fig. 1B) indicated that the Stra13 promoter is repressed by histone deacetylation. To investigate which factors recruit HDAC to repress the Stra13 promoter, we cloned a genomic fragment spanning −3.5 kb to +101 from a 129/SvJ BAC genomic library (10) upstream of the promoterless luciferase reporter pGL3 (pGL3KN, Fig. 2A). In addition, a 5′ deletion mutant was constructed that contains sequences from −422 to +101 (pGL3PmN). Both constructs displayed basal promoter activity in transiently transfected COS-7 cells (Fig. 2A). Because many bHLH factors autoregulate their expression and Stra13 is a strong transcriptional repressor, we then examined whether Stra13 could negatively regulate its own expression. COS-7 cells transfected with pGL3KN in the presence of increasing amounts of a Stra13 expression vector were assayed for luciferase activity relative to pGL3KN alone. The luciferase activity was reduced in a Stra13 expression vector dose-dependent manner, indicating that Stra13 strongly repressed its own promoter activity (Fig. 2B).
Figure 2.
Stra13 negatively regulates its own expression. (A) COS-7 cells were transfected with 0.5 μg of Stra13 promoter constructs pGL3KN and pGL3PmN. Forty-eight hours after transfection, luciferase activity was measured by using pGL3 as a control. The activity of pGL3PmN was plotted relative to pGL3KN, which was given an arbitrary value of 100. The abbreviations for the restriction enzyme sites used to generate the constructs are: K, KpnI; Pm, PmlI; and N, NheI. (B) COS-7 cells were transfected with the reporter pGL3KN in the presence of increasing amounts of a Stra13 expression vector (1–411). The extent of repression in the presence of Stra13 was measured relative to the activity of pGL3KN in the presence of equivalent amounts of the empty vector (pCS2) alone. (C) pGL3KN reporter (0.5 μg) was cotransfected into COS-7 cells with varying amounts of Stra13 expression vector or equivalent concentrations of an empty vector (pCS2), as indicated. TSA (100 nM) was added 24 h after transfection, and the cells were harvested 20 h later. The luciferase activity of pGL3KN in the absence or presence of TSA was given a value of 100. Note, however, that treatment with TSA alone resulted in a 2- to 3-fold increase in the promoter activity of pGL3KN.
To determine whether Stra13-mediated repression of its promoter required HDAC activity, the effect of TSA, a specific HDAC inhibitor (13), was tested in cotransfection assays. COS-7 cells were cotransfected with the Stra13 reporter pGL3KN along with varying concentrations of the Stra13 expression vector in the absence or presence of TSA treatment (Fig. 2C). The addition of TSA strongly reversed Stra13-mediated repression of its own promoter, indicating that Stra13 negatively regulates its expression through an HDAC-dependent mechanism.
Stra13 Interacts with Components of the HDAC Corepressor Complex Through Its C-Terminal α-Helices.
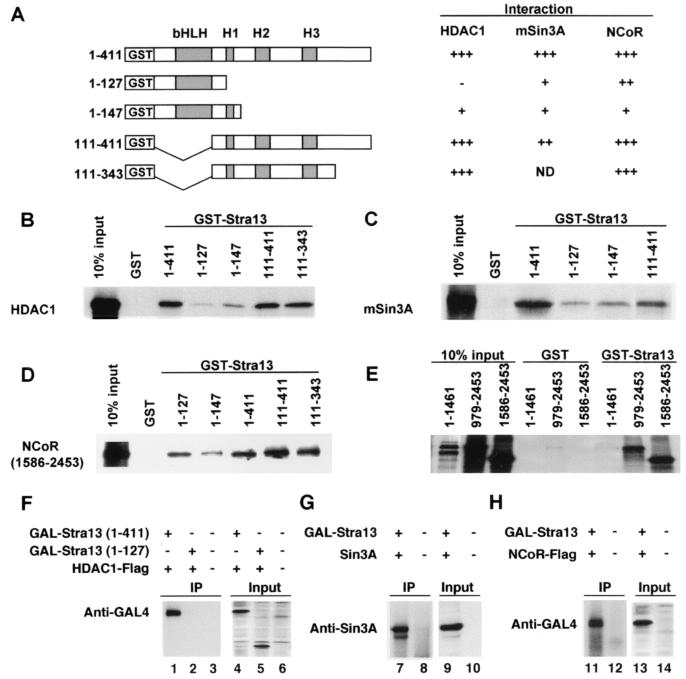
To determine whether Stra13 can physically interact with components of the HDAC-Sin3A corepressor complex and to define the domains in Stra13 that are required for these interactions, we performed GST pull-down assays. Equivalent amounts of GST fusion proteins containing various domains of Stra13 (Fig. 3A) were used for interaction with in vitro translated HDAC1, Sin3A, and NCoR. As shown in Fig. 3 B and C, GST-Stra13 (1–411) interacted with HDAC1 and Sin3A, whereas GST-Stra13 (1–127), which contains only the bHLH domain, and GST-Stra13 (1–147), which contains the bHLH domain and Helix 1 (H1), exhibited little or no interaction. In contrast, GST-Stra13 (111–411) and GST-Stra13 (111–343) exhibited an interaction indicating that the association occurs primarily with the region containing α-helices 2 and 3. Stra13 also interacted with NCoR through the domain containing the α-helices (Fig. 3D). The apparently stronger interaction of Stra13 (1–127) relative to Stra13 (1–147) with NCoR is surprising, but may reflect a conformation dependency for interaction. Interestingly, only the C-terminal NCoR constructs (979–2453 and 1586–2453) interacted with GST-Stra13 (1–411), whereas an N-terminal construct (1–1461) as well as 758-1817 (data not shown) failed to interact in GST assays (Fig. 3E). We then tested whether Stra13 can interact with HDAC1 in mammalian cells by using coimmunoprecipitation assays. A Stra13 expression vector, GAL4-Stra13 (1–411), was cotransfected into COS-7 cells with a HDAC1 expression vector. Forty-eight hours after transfection, cell extracts were prepared, and the immunoprecipitates revealed a significant interaction of Stra13 with HDAC1 (Fig. 3F). Consistent with GST assays, GAL4-Stra13 (1–127) failed to immunoprecipitate with HDAC1 (Fig. 3F), indicating that the interaction between Stra13 and HDAC1 requires the C-terminal region containing the α-helices of Stra13. We further investigated whether Stra13 also associates with Sin3A and NCoR. Expression vectors for Sin3A or NCoR (1586–2453) were cotransfected with GAL-Stra13 (1–411). As shown in Fig. 3 G and H, both Sin3A and NCoR coimmunoprecipitated with Stra13. Untransfected cells, which were used as controls, showed no interaction.
Figure 3.
Stra13 interacts with HDAC1, Sin3A, and NCoR. (A) Schematic representation of the GST-Stra13 fusion proteins tested for interaction with HDAC1, Sin3A, and NCoR. The various domains of Stra13 are shown. H1, helix 1; H2, helix 2; H3, helix 3. The strength of interaction between Stra13 and HDAC1 and Sin3A and NCoR was scored from − (background level), + (weak), ++ (fair), to +++ (strong). (B– E) GST-Stra13 constructs were tested for interaction with 35S-labeled in vitro translated HDAC1, Sin3A, or NCoR as indicated. Ten percent of input was run on the gels as controls. (F– H) COS-7 cells were transfected with expression vectors for HDAC1 and Stra13 (1–411 or 1–127) as indicated in F, Sin3A and Stra13 (1–411) as indicated in G, or NCoR (1586–2453) and Stra13 (1–411) as indicated in H. Cell lysates were immunoprecipitated and analyzed for interaction by Western blot with appropriate antibodies as indicated. The inputs (50 μg) were analyzed with anti-GAL antibodies in lanes 4–6 and 13–14 and with the Sin 3A antibody in lanes 9 and 10.
A Stra13 Mutant Lacking the HDAC–Corepressor Interaction Domain Fails to Repress Transcription.
We examined the functional importance of HDAC1, Sin3A, and NCoR interaction for Stra13-mediated repression of its promoter. Expression vectors for either full-length Stra13 (1–411) or a mutant lacking the HDAC-interaction domains [Stra13 (1–127)] were cotransfected with Stra13 reporter construct pGL3KN or pGL3PmN in COS-7 cells (Fig. 4A). In contrast to Stra13 (1–411), which repressed the basal activity of both pGL3KN as well as pGL3PmN, Stra13 (1–127) had no significant effect on the basal activity of either reporter construct. To ascertain that the inability of Stra13 (1–127) to repress transcription was not due to its lack of appropriate localization in the nucleus, we determined its subcellular localization by immunostaining. An epitope-tagged construct, His-Stra13 (1–127), was transfected into COS-7 cells and detected with an antibody directed against the His-epitope. As shown in Fig. 4B, Stra13 (1–127) is correctly targeted to the nucleus. Taken together, these results indicate that Stra13-mediated repression of its promoter requires the C-terminal domain that encompasses the α-helices.
Figure 4.
Stra13-mediated repression requires the C-terminal α-helices. (A) COS-7 cells were transfected with 0.25 μg of the Stra13 reporter constructs pGL3KN or pGL3PmN and the same amount of Stra13 expression vectors Stra13 (1–411) or Stra13 (1–127). Luciferase activity was measured 48 h after transfection, and the activity of pGL3PmN was expressed relative to pGL3KN, which was given an arbitrary value of 100. (B) COS-7 cells were transfected with His-Stra13 (1–127). Forty-eight hours after transfection, cells were fixed and stained with anti-His mAb (Qiagen).
Stra13 Acts as a Repressor of c-Myc Expression by an HDAC-Independent Mechanism That Involves Interaction with TFIIB.
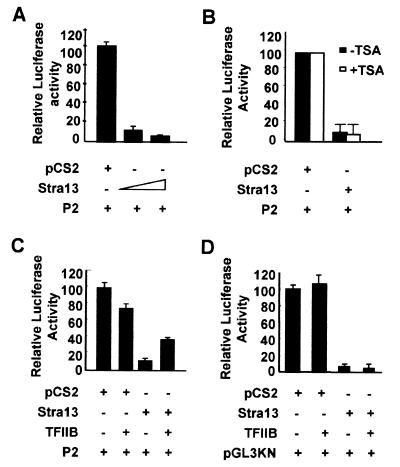
The inverse correlation between Stra13 and c-Myc expression (Fig. 1 A–C) suggested that one mechanism by which Stra13 may mediate growth arrest is by negatively regulating the expression of c-Myc. To test this possibility, a c-Myc promoter construct (14) was transfected into COS-7 cells in the presence of increasing amounts of Stra13 (1–411). As shown in Fig. 5A, Stra13 strongly repressed c-Myc promoter activity in a dose-dependent manner. We then analyzed whether Stra13 recruited HDAC/Sin3A corepressors to inhibit c-Myc promoter activity. COS-7 cells were cotransfected with the c-Myc promoter construct and a Stra13 expression vector in the absence or presence of TSA. Interestingly, TSA had no significant effect on Stra13 repression of the c-Myc promoter, indicating that an HDAC-independent pathway is required for Stra13-mediated repression of the c-Myc promoter (Fig. 5B).
Figure 5.
Stra13 negatively regulates c-Myc expression. (A) The c-Myc promoter was transfected in COS-7 cells with increasing concentrations of a Stra13 expression vector. Cells were harvested 48 h after transfection and assayed for luciferase activity. (B) The c-Myc promoter construct was cotransfected with Stra13 expression vector in COS-7 cells as described in A in the absence or presence of 100 nM TSA. The luciferase activity in the absence and presence of TSA was given a value of 100. (C) COS-7 cells were transfected with c-Myc promoter construct in the absence or presence of a Stra13 expression vector as well as a TFIIB expression vector. Cell lysates were assayed for luciferase activity after 48 h. (D) COS-7 cells were transfected with pGL3KN in the absence or presence of a Stra13 and TFIIB expression vector as indicated. Cell extracts were assayed for luciferase activity 48 h after transfection.
We have shown previously that Stra13 physically interacts with TBP and TFIIB (2). To examine whether the interaction with the basal transcription machinery was required for Stra13-mediated repression of the c-Myc promoter, we tested whether cotransfection of a TFIIB expression vector could reverse this repression. Although an expression vector for TFIIB on its own was slightly inhibitory to c-Myc transcription possibly because of squelching effects, its addition along with a Stra13 expression vector strongly alleviated Stra13-mediated repression of the c-Myc promoter (Fig. 5C). Because transfection of a TFIIB expression vector alone causes a moderate inhibition of c-Myc promoter activity, the reversal of Stra13 inhibition by cotransfection of TFIIB is likely to be a specific functional and physiologically relevant interaction. These results indicated that Stra13 represses c-Myc transcription by interacting with basal transcription factors. We also tested the effect of TFIIB on Stra13-mediated repression of its own promoter. As shown in Fig. 5D, the addition of a TFIIB expression vector had little effect on Stra13-mediated repression of its promoter activity. The effect of overexpression of TBP could not be tested, because TBP strongly inhibited transcription in our assays (data not shown).
Discussion
Transcriptional repression by several factors including nuclear hormone receptors, Mad, Rb, and p53 is mediated by direct or indirect interactions with HDAC (5–9). Inhibition of HDAC activity by TSA can release repression mediated by these factors. TSA also induces growth arrest at the G1 and G2/M phases of the cell cycle (11). However, the molecular basis of TSA-induced growth arrest is largely unknown. One of the few target genes whose expression is up-regulated in response to TSA-mediated growth arrest is p21, a key cell cycle regulatory gene (15, 16). In this paper we demonstrate that Stra13 expression is up-regulated and coincident with TSA-induced growth arrest. These results indicated to us that not only is Stra13 expression repressed by histone deacetylation, but also that its up-regulated expression may be important to achieve growth arrest, and, therefore, Stra13 may function as a growth suppressor. Consistent with this, overexpression of Stra13 strongly suppressed colony formation in NIH 3T3 cells, indicating that increased Stra13 levels are associated with growth inhibition.
Transcriptional repression of Stra13 expression occurs through a negative autoregulatory mechanism and requires recruitment of HDAC1. The interaction of Stra13 with HDAC1, Sin3A, and NCoR occurs primarily, but not exclusively, through a large C-terminal region in Stra13 that contains α-helical structures, although the interaction with NCoR also requires the N-terminal bHLH domain, suggesting that these interactions may be modulated by conformational changes in Stra13. Whether Stra13 can interact simultaneously with HDAC1, Sin3A, and NCoR and whether these interactions occur through different α-helices require further investigation. Nevertheless, these interactions are physiologically important for Stra13-mediated repression of its own promoter because a mutant construct lacking the region containing the α-helices does not interact with HDAC1 and also fails to act as a repressor. The repression of Stra13 promoter by HDAC1 may be important to maintain a low level of Stra13 expression that would permit proliferation to occur, because inhibition of HDAC activity by TSA results in increased Stra13 expression and concomitant growth arrest. Thus, Stra13-HDAC1 interaction may be important for maintenance of the proliferative state of cells. It is interesting in this regard to note that the interaction of retinoid receptors with the HDAC complex also appears to be required for maintenance of cells in a proliferative capacity, because ligand-induced activation of the receptor abrogates this interaction and induces growth arrest and differentiation (17).
Notably, Stra13 expression can be up-regulated not only by TSA but also with RA as well as serum starvation, each of which results in growth arrest of cells. Thus, additional HDAC-independent pathways also may require Stra13 function to mediate growth arrest. In this regard, Stra13 is strikingly similar to p21, whose expression also is induced by TSA (15, 16), RA (18), and serum starvation (19), and, thus, Stra13, like p21, may have a general function in growth arrest.
The inverse expression patterns of Stra13 and c-Myc led us to hypothesize that Stra13 may act as a growth suppressor by repressing the expression of c-Myc. Consistent with this, Stra13 strongly inhibits c-Myc promoter activity in transfection assays. However, HDAC recruitment is not required for this repression, because TSA does not relieve Stra13-dependent repression of the c-Myc promoter. In contrast, overexpression of TFIIB is sufficient to alleviate this repression, indicating that the interaction of Stra13 with TFIIB results in repression of c-Myc promoter activity.
It is not surprising that Stra13 utilizes different mechanisms to repress itself and c-Myc, as many repressors have been shown to mediate repression through multiple mechanisms. For instance, repression of target genes by Rb occurs through both HDAC-dependent and HDAC-independent pathways (20). It is, however, interesting that Stra13 and c-Myc, which are involved in biologically opposite functions, are targets for Stra13-mediated repression through distinct pathways. This probably provides an efficient mechanism for growth arrest, because up-regulation of Stra13 expression by TSA (or other regulators) still would allow it to repress c-Myc expression through an HDAC-independent mechanism. Because Stra13 is up-regulated rapidly by several growth arrest triggers including RA, TSA, and serum starvation, it is likely that Stra13 is a key target in each of these signaling pathways. Once up-regulated through these pathways, Stra13 could function as a growth suppressor by repression of target genes such as c-Myc, which then would cause cellular quiescence leading to terminal differentiation.
It is intriguing that Stra13 and Hes1, although closely related, have biologically opposite functions. Whereas Stra13 expression is induced upon differentiation and growth arrest, Hes1 expression is high in proliferating cells and is down-regulated upon differentiation, and, in fact, sustained Hes1 expression inhibits neural differentiation (3, 21). Moreover, Hes1 interacts with the corepressor Groucho (22), although the role of Groucho in Hes-mediated repression of its own promoter (23) or that of Mash1 (24) has not been documented. The ability of Stra13 and Hes1 to interact with different corepressors and repress expression of distinct target genes might underlie their differential biological activities.
Acknowledgments
We are grateful to P. Chambon, S. Schreiber, D. Ayer, C. Glass, M. Boudjelal, Ed Ziff, and K. Calame for various reagents and thank R. Gopalkrishnan, J. Bieker, and R. Mira-y-Lopez for discussions and comments on the manuscript. This work was supported by grants from the National Institutes of Health and the Waxman Cancer Research Foundation. R.T. is a Basil O'Connor Research Scholar.
Abbreviations
- TSA
trichostatin A
- RA
all-trans-retinoic acid
- HDAC
histone deacetylase
- NCoR
nuclear receptor corepressor
- bHLH
basic helix–loop–helix
- GST
glutathione S-transferase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.070526297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.070526297
References
- 1.Murre C, Bain G, Dijk M A, Engel I, Furnari B A, Massari M E, Matthews J R, Quong M W, Rivera R R, Stuiver M H. Biochim Biophys Acta. 1994;1218:129–135. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 2.Boudjelal M, Taneja R, Matsubara S, Bouillet P, Dollé P, Chambon P. Genes Dev. 1997;11:2052–2065. doi: 10.1101/gad.11.16.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kageyama R, Nakanishi S. Curr Opin Genet Dev. 1997;7:659–665. doi: 10.1016/s0959-437x(97)80014-7. [DOI] [PubMed] [Google Scholar]
- 4.Paroush Z, Finley R L, Kidd T, Wainwright S M, Ingham P W, Brent R, Ish Horowicz D. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 5.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 6.Struhl K. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 7.Torchia J, Glass C, Rosenfeld M G. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 8.Maldonado E, Hampsey M, Reinberg D. Cell. 1999;99:455–458. doi: 10.1016/s0092-8674(00)81533-0. [DOI] [PubMed] [Google Scholar]
- 9.Knoepfler P S, Eisenman R N. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- 10.Sun H, Mattei M G, Taneja R. Cytogenet Cell Genet. 2000;87:211–212. doi: 10.1159/000015470. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida M, Beppu T. Exp Cell Res. 1988;177:122–131. doi: 10.1016/0014-4827(88)90030-4. [DOI] [PubMed] [Google Scholar]
- 12.Kijima M, Yoshida M, Sugita K, Horinouchi S, Beppu T. J Biol Chem. 1993;268:22429–22435. [PubMed] [Google Scholar]
- 13.Yoshida M, Horinouchi S, Beppu T. BioEssays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 14.Lee T C, Ziff E B. J Biol Chem. 1999;274:595–606. doi: 10.1074/jbc.274.2.595. [DOI] [PubMed] [Google Scholar]
- 15.Sowa Y, Orita T, Minamikawa S, Nakano K, Mizuno T, Nomure H, Sakai T. Biochem Biophys Res Commun. 1997;241:142–150. doi: 10.1006/bbrc.1997.7786. [DOI] [PubMed] [Google Scholar]
- 16.Archer S Y, Meng S, Shei A, Hodin R A. Proc Natl Acad Sci USA. 1998;95:6791–6796. doi: 10.1073/pnas.95.12.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kouzarides T. Curr Opin Genet Dev. 1999;9:40–48. doi: 10.1016/s0959-437x(99)80006-9. [DOI] [PubMed] [Google Scholar]
- 18.Liu M, Iavarone A, Freedman L P. J Biol Chem. 1996;271:31723–31728. doi: 10.1074/jbc.271.49.31723. [DOI] [PubMed] [Google Scholar]
- 19.Macleod K F, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, Vogelstein B, Jacks T. Genes Dev. 1995;9:935–944. doi: 10.1101/gad.9.8.935. [DOI] [PubMed] [Google Scholar]
- 20.Luo R X, Postigo A A, Dean D C. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 21.Ishibashi M, Moriyoshi K, Sasai Y, Shiota K, Nakanishi S, Kageyama R. EMBO J. 1994;13:1799–1805. doi: 10.1002/j.1460-2075.1994.tb06448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grbavec D, Stifani S. Biochem Biophys Res Commun. 1996;223:701–705. doi: 10.1006/bbrc.1996.0959. [DOI] [PubMed] [Google Scholar]
- 23.Takebayashi K, Sasai Y, Sakai Y, Watanabe T, Nakanishi S, Kageyama R. J Biol Chem. 1994;269:5150–5156. [PubMed] [Google Scholar]
- 24.Chen H, Thiagalingam A, Chopra H, Borges M W, Feder J N, Nelkin B D, Baylin S B, Ball D W. Proc Natl Acad Sci USA. 1997;94:5355–5360. doi: 10.1073/pnas.94.10.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]