Abstract
The Drosophila nucleosome remodeling factor (NURF) is a protein complex of four distinct subunits that assists transcription factor-mediated chromatin remodeling. One NURF subunit, ISWI, is related to the transcriptional regulators Drosophila brahma and yeast SWI2/SNF2. We have determined peptide sequences and isolated cDNA clones for a second NURF component (the 55-kDa subunit). Immunological studies show that p55 is an integral subunit of NURF and is generally associated with polytene chromosomes. The predicted sequence of p55 reveals a WD repeat protein that is identical with the 55-kDa subunit of the Drosophila chromatin assembly factor (CAF-1). Given that WD repeat proteins related to p55 are associated with histone deacetylase and histone acetyltransferase, our findings suggest that p55 and its homologs may function as a common platform for the assembly of protein complexes involved in chromatin metabolism.
The organization of DNA in nucleosomes presents a barrier to gene transcription and other chromosomal activities. Numerous studies have shown that nucleosomes inhibit both the accessibility of promoter DNA to the general transcriptional machinery and the binding of upstream regulatory proteins (for reviews see refs. 1–4). Recent advances have provided a number of mechanisms by which accessibility to general and sequence-specific transcription factors is facilitated. These include the influences of DNA structure, histone modification, particularly acetylation, and the action of a number of ATP-dependent chromatin remodeling factors (for reviews see refs. 5–8).
The multi-protein SWI–SNF complex uses the energy of ATP hydrolysis to facilitate transcription of a number of yeast promoters in vivo (for reviews see refs. 9–12). The SWI–SNF complex has been conserved in evolution; homologs with related subunits have been purified from yeast, Drosophila, as well as from mammalian cells (8). Biochemical studies from our laboratory have identified an ATP-dependent chromatin remodeling complex, designated nucleosome remodeling factor (NURF), that facilitates a local, transcription factor-mediated nucleosomal disruption within a nucleosome array (13). Purified NURF is composed of four subunits of molecular mass 215, 140, 55, and 38 kDa, assembled in a native complex of ≈500 kDa. Previously, we identified the 140-kDa subunit of NURF as ISWI, a member of the SWI–SNF2 family that is distinguished by a highly conserved ATPase domain (14). ISWI is an abundant nuclear protein that is present throughout Drosophila development. The ATP dependence of NURF activity is likely to be mediated by the action of ISWI, but unlike the DNA-dependent ATPase activity of the SWI–SNF complex, the ATPase activity of NURF is stimulated more by nucleosomes than by free DNA, suggesting a recognition by NURF of both histone and DNA components of the nucleosome.
To elucidate the structure and mechanism of action of NURF, we have sought to identify all the subunits of NURF. Here, we describe the cloning and initial characterization of the 55-kDa NURF component. Our studies reveal that NURF-55 is a WD repeat protein, identical to the 55-kDa subunit of the Drosophila chromatin assembly factor dCAF-1 (15).
MATERIALS AND METHODS
NURF Purification and Peptide Sequencing.
NURF was purified from nuclear extracts of 0–12-h Drosophila embryos as described though the glycerol gradient step (13). Purified fractions were precipitated with acetone and separated by SDS-PAGE. The 55-kDa band was excised and digested with Achromobacter protease. The resulting peptides were eluted, separated by HPLC, and sequenced as previously described (14). The peptide sequences are DYSVHRLILGTHTSDEQ, LMIWDTRNNNTSKP, TVALWDLRNL, LHSFESHK, DEIFQVQWSPHNETILAS, and IGEEQSTEDAEDGPP.
Isolation of p55 cDNA.
Oligonucleotide primers were synthesized based on the peptide sequences of p55 protein and employed for PCR analysis using DNA prepared from a cDNA library derived from 6–14-h Drosophila embryos (Novagen). The sequences are #1, 5′-YTGYTCYTCICCIATYTT-3′; #2, 5′-ATGATCTGGGACACCCGC-3′; #3, 5′-CACAGGGAYGAGATCTTCCAG-3′; #4, 5′-CTGGAAGATCTCRTCCTTGTG-3′; #5, 5′-CTGCTCCTCGCCGATCTT-3′ (I, inosine; R, A/G; Y, C/T). Among five primers synthesized, three combinations (#1/#2, #2/#4, and #2/#5) produced PCR products. The longest PCR product was cloned, sequenced, and found to encode four of the p55 peptides obtained by peptide sequencing. This DNA fragment was used to screen by plaque hybridization a cDNA library derived from 6–14-h Drosophila embryos (Novagen). About 300,000 plaques were screened, and 12 independent positives were obtained. Restriction enzyme digestion showed that all clones had common restriction fragments, suggesting that they were derived from the same gene. A clone containing the longest DNA fragment was completely sequenced for both strands using the dideoxy chain termination method (Sequenase); the predicted ORF contains all the p55 peptide sequences obtained. This ORF was judged to be complete because of the presence of in-frame stop codons 5′ to the presumptive translation initiation codon and in-frame stop codons at the 3′ end of the fragment sequenced.
Preparation of Epitope-Tagged p55 and Polyclonal Antibodies.
A full-length p55 cDNA clone was tagged with the 9E10 c-Myc epitope at the 5′ end of the coding region and subcloned between the BamHI and HindIII sites within the multi-cloning site of the pET28a vector (Novagen). The vector supplies the His6 tag at the N-terminal end of the recombinant protein. Recombinant protein was induced in Escherichia coli with 1 mM isopropyl-β-d-thiogalactoside for 3 h at 37°C. p55 was extracted from inclusion bodies with 6 M guanidine·HCl and purified to 90–95% homogeneity using Nickel-NTA-agarose beads (Qiagen). Immunization of rats and rabbits with purified recombinant His6/Myc-tagged p55 protein followed standard protocols using a commercial vendor (Babco, Richmond, CA). Crude and affinity-purified rabbit polyclonal antibodies against p55 were also obtained from J. Tyler and J. Kadonaga. Antibodies against histone H3 were obtained from Michael Bustin.
Baculovirus Expression of p55 Protein.
Full-length p55 cDNA with the 9E10 c-Myc epitope tag was cloned into the pVL1393 vector (PharMingen) and cotransfected with BaculoGold DNA (PharMingen) into Sf9 cells. The recombinant protein lacks the histidine6 tag. Production of recombinant virus and recombinant protein were done according to manufacturer’s instructions (PharMingen).
Affinity Purification of Antisera.
Antibodies against p55 were affinity purified from whole antisera by chromatography on Ni-NTA-agarose affinity resins containing bound His6/Myc-p55 and His6/FLAG-ISWI protein, respectively, using 3 ml bed volume of resin and 1.5 ml crude serum, according to the manufacturer’s (Qiagen) protocols. After washing with 5 bed volumes of equilibration buffer (150 mM NaCl/50 mM Tris⋅HCl, pH 7.4) and 5 bed volumes of wash buffer (2 M NaCl/50 mM Tris⋅HCl, pH 7.4) antibodies were eluted in 1.5 bed volumes of 4 M MgCl2. The eluate was extensively dialyzed against PBS and stored with 1 mg/ml carrier BSA.
SDS-PAGE and Western Blotting.
Proteins were analyzed by SDS-PAGE (8 or 10% polyacrylamide) and Western blotting, using nitrocellulose membranes and chemiluminescence detection (ECL system, Amersham). Primary antibodies were rabbit anti-p55 used at dilutions of 1:1000 (serum) and 1:500 (affinity purified), rat anti-p55 at 1:10,000, rabbit anti-ISWI at 1:1000 (serum antibodies raised against ISWI residues 1–880; T.T. and M.M., unpublished data) and 1:500 (affinity purified), and anti-histone H3 at 1:1000 (gift of M. Bustin). Secondary antibodies were goat anti-rabbit IgG conjugated to peroxidase (Amersham) at 1:20,000 dilution, goat anti-mouse IgG conjugated to peroxidase (Amersham) at 1:10,000 dilution, or goat anti-rat IgG (Jackson ImmunoResearch) conjugated to peroxidase at 1:10,000 dilution.
Gel Filtration Chromatography.
Drosophila embryo S-150 extracts were prepared as described previously (16, 17). 200 μl of extract (0.5 mg of protein) was applied to a Superose 6 (Pharmacia) column and eluted with HEMGN-0.3 (25 mM Hepes-KOH, pH 7.6, 0.1 mM EDTA/12.5 mM MgCl2/10% glycerol/0.1% Nonidet P-40/1 mM DTT/0.1 mM 4(2-aminoethyl)benzenesulfonyl fluoride·hydrochloride (AEBSF)/1 μg/ml leupeptin/1 μg/ml aprotinin/1 μg/ml pepstatin/25 mM NaF/5 mM sodium orthovanadate/1 mM sodium pyrophosphate/5 μM microcystin). Fractions (0.5 ml) were collected and assayed for p55 and ISWI proteins by Western blotting.
Indirect Immunofluorescence.
Drosophila Schneider line 2 (SL2) cells were cultured in 1-ml slide flasks (Nunc). Cells were fixed in 4% formaldehyde in PBS for 20 min at room temperature, followed by methanol treatment for 10 min. After blocking with 3% BSA in PBS, 0.1% Tween 20 for 30 min, the slides were incubated with a 1:50 affinity-purified anti-p55 in PBS, 3% BSA for 2 h, followed by incubation for 1 h with fluorescein isothiocyanate-conjugated goat anti-rabbit IgG (1:250 dilution in PBS, 3% BSA). After each incubation with antibody, slides were extensively washed with PBS, 0.05% Tween 20 twice for 10 min at room temperature and stained with Hoechst 33342. Polytene chromosome squashes and chromosome staining were performed with the additional prefixing step as described (18) and with dry milk substituting for BSA. A 1:200 dilution of rat anti-p55 and a 1:150 dilution of the affinity-purified rabbit anti-ISWI antibody were used as primary antibodies. Fluorescein isothiocyanate-conjugated goat anti-rat IgG (Cappel) and rhodamine-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) were used as secondary antibodies at 1:200 dilution.
Immunoprecipitation Assays.
40 μl of a slurry of protein G-Sepharose CL-4B (Pharmacia) was incubated for 1 h at 4°C with affinity-purified anti-ISWI (25 μg protein) or with affinity-purified anti-p55 (15 μg protein), or preimmune serum in interaction buffer (IB; 50 mM Tris⋅HCl, pH 7.5, 120 mM KCl/0.1% Nonidet P-40/0.1 mM EDTA/3 mM MgCl/0.5 mM DTT/0.1 mM AEBSF/1 μg/ml leupeptin/1 μg/ml aprotinin/1 μg/ml pepstatin). The anti-ISWI or anti-p55 beads (20-μl bed volume) were washed extensively (four times each with 5–10 volumes of IB for 5 min at 4°C, with rotation) and incubated with 50 μl of nuclear extract prepared from 0–12-h Drosophila embryos as described by Wampler et al. (19) (Figs. 2B and 3D) or with 100 μl of Drosophila S-150 assembly extract or reconstituted chromatin (Fig. 5A). Beads were washed extensively as above with IB, 0.3 M KCl prior to pelleting. After mixing with SDS-PAGE sample buffer (pellet, 30 μl; supernatant, 1 volume of 2 × buffer), samples were boiled for 3–5 min before SDS-PAGE and Western blotting. For quantitative immunodepletion of NURF activity (Fig. 2 C and D), 100 μg of control antibody (rabbit anti-chicken IgG, Jackson ImmunoResearch) or affinity-purified rabbit anti-p55 antibody were coupled to 20 μl of Affi-Prep protein-A beads (Bio-Rad) overnight at 4°C. The antibody-coupled beads were then used to immunodeplete NURF (equivalent to P-11 fraction; ref. 13). Forty microliters of a 1:10 NURF dilution in HEGN-0.15 containing 1 mM AEBSF and 0.2 mg/ml insulin were incubated with the antibody-coupled beads for 1 h at 4°C. The supernatant was then collected and used for the GAGA factor-mediated nucleosome disruption assay. The beads were washed twice in interaction buffer for 5 min at 4°C and then boiled in SDS-PAGE sample buffer. 10-μl equivalents of bead and supernatant samples were analyzed by SDS-PAGE and Western blotting.
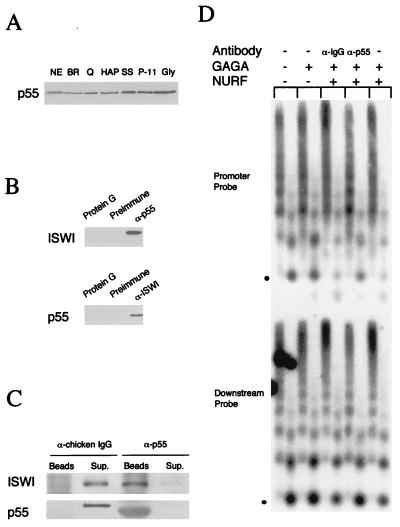
Figure 2.
p55 is a subunit of NURF. (A) p55 copurifies with active NURF fractions. Western blot of chromatographic fractions containing equivalent NURF activity. NE, nuclear extract; BR, DE52/BioRex; Q, Q Sepharose; HAP, hydroxylapatite; SS, single-stranded DNA cellulose; P11, phosphocellulose P-11; Gly, glycerol gradient. The blot was probed with antibodies to p55. Antibody specificity was demonstrated by the inhibition of immunoreactivity by the inclusion of purified recombinant p55 protein with anti-p55. (B) p55 coimmunoprecipitates with ISWI. Western blot of proteins coimmunoprecipitating with p55 and ISWI. Nuclear extracts were incubated with affinity-purified anti-ISWI (α-ISWI) (Left) or anti-p55 serum (α-p55) (Right), preimmune sera, or protein G beads alone. Immunoprecipitates were analyzed by SDS-PAGE and Western blotting using α-p55 and α-ISWI, respectively. (C) Quantitative immunodepletion of ISWI and p55 from a NURF fraction (P-11 equivalent; ref. 13) by affinity-purified anti-p55 and control anti-chicken IgG. Western blot analysis of the supernatants (Sup) and pellet (Beads) showing ISWI and p55. The SDS-PAGE mobility of p55 immunoprecipitated by α-p55 beads is slightly increased by the presence of comigrating IgG. (D). GAGA transcription factor-mediated nucleosome disruption on reconstituted hsp70 plasmid chromatin, using NURF (P-11 fraction) and the same fraction immunodepleted with α-p55 and control anti-chicken IgG (α-IgG). Southern blot showing nucleosome disruption as assayed by MNase digestion (3- and 15-min digestion points for each chromatin sample); the blot was hybridized with radiolabeled oligonucleotide corresponding to the hsp70 promoter (−113 to −142) or downstream sequences (+1803 to +1832) as described (13). The position of mononucleosomal DNA is indicated by the dot. NURF activity is gauged by the relative loss of the mononucleosomal DNA fragment and concomitant appearance of subnucleosomal DNA fragments.
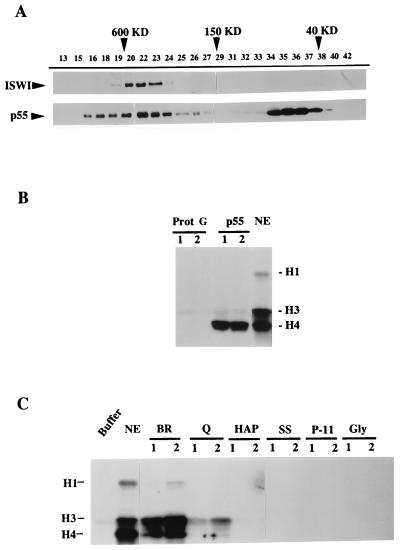
Figure 3.
p55 is associated with other proteins in Drosophila extracts. (A) Superose 6 gel filtration chromatography of Drosophila embryo S-150 extract. Fractions as indicated by numbers were analyzed by Western blotting for ISWI (Upper) and p55 (Lower). (B) Recombinant p55 interacts with a HAT activity. 20 μl of anti-Myc protein G-Sepharose beads, unbound or bound to saturating amounts of Myc epitope-tagged p55, were incubated for 3 h with Drosophila nuclear extract (NE) (19), pelleted and washed, and resuspended in 30 μl of HAT assay buffer. Bead suspension (lane 1, 5 μl; lane 2, 15 μl) and the NE (1 μl) were analyzed for HAT activity, as monitored by the transfer of the [3H]acetyl group to exogenously added histones and detection by SDS-PAGE and fluorography. Histones H1, H3, and H4 are indicated. Myc beads alone did not associate with HAT activity. (C) NURF has no HAT activity. Active NURF fractions through purification steps as in the Fig. 2A legend were assayed for HAT activity, as monitored by transfer of the [3H]acetyl group to histones and detection by SDS-PAGE and fluorography.
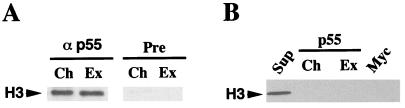
Figure 5.
Chromatin associates with native but not recombinant p55. (A) Coimmunoprecipitation of p55 with reconstituted chromatin. 100 μl of S-150 chromatin assembly extract alone (Ex) or chromatin (Ch) reconstituted with S-150 extract and hsp70 promoter-containing plasmid DNA (and purified by spin-column chromatography) were analyzed by immunoprecipitation with α-p55 and preimmune serum (Pre). Immunoprecipitates were analyzed for histones by SDS-PAGE and Western blotting using antibodies to histone H3. (B) p55 alone does not bind to nucleosomes. Baculovirus-expressed p55 immobilized on beads (20-μl bed volume) was incubated with 100 μl (∼20 mg/ml protein) of S-150 extract (Ex) and with hsp70 promoter-containing plasmid chromatin (Ch) reconstituted in the same extract and purified by spin-column chromatography. After washing in extract buffer, 0.1 M KCl, the pelleted beads and 10% of the supernatant (Sup) were boiled in 30 μl SDS-PAGE sample buffer. 10-μl samples were analyzed for histone H3 by Western blotting.
Immobilized p55 for Interaction Assays.
Anti-Myc antibody (Myc 9E-10 epitope, Santa Cruz Biotechnology) was bound to protein G-Sepharose beads and further bound with a saturating amount of baculovirus-expressed His6/Myc epitope-p55. Beads were equilibrated in extract buffer (16) and then incubated with the Drosophila S-150 assembly extract or reconstituted chromatin for 3 h at 4°C. After pelleting, beads were washed in extract buffer, and the pellets and supernatant were analyzed by SDS-PAGE and Western blotting with anti-histone H3 antibody. For histone acetyltransferase (HAT) assays, after incubation with extract, beads were pelleted and washed three times for 5 min at 4°C in IB and resuspended in 60 μl of buffer A (see HAT assay).
Nucleosome Assembly and Disruption Assay for NURF Activity.
Regularly spaced arrays of nucleosomes were assembled in vitro on the plasmid pdHspXX3.2 using the S-150 Drosophila embryo extract as described (16, 17). [For co-immunoprecipitation experiments, the assembled plasmid chromatin was purified away from unassembled histones by gel filtration spin-column chromatography (Pharmacia Sepharose CL4B). This was verified by Western analysis of the spin-column eluate of an S-150 extract alone, using anti-H3]. Sarkosyl treatment to inactivate NURF in preparations of plasmid chromatin assembled with the S-150 extract, spin-column chromatography to remove detergent, and GAGA transcription factor-mediated disruption of hsp70 promoter nucleosomes were performed as described previously (13).
Histone Acetylase Assay.
Liquid histone acetyltransferase assays were performed according to Sobel et al. (20) in a volume of 50 μl. Reactions contained 0.1 μM [3H]acetyl-CoA (6 Ci/mmol) and 1 mg/ml histone (Sigma) in buffer A (50 mM Tris⋅HCl, pH 8.0, 0.1 mM EDTA, pH 8.0, 100 mM NaCl/1 mM DTT/1 mM phenylmethylsulfonyl fluoride). After incubation at 30°C for 30 min, samples were precipitated with 20% trichloroacetic acid (final), washed in acetone, air-dried, and analyzed by SDS-PAGE (15% acrylamide). 3H-labeled acetyl histones were visualized by fluorography.
RESULTS
Cloning of NURF-55 Reveals a WD Repeat Protein Identical to CAF-1 p55.
To identify the 55-kDa subunit of NURF (NURF-55), we purified the NURF-55 polypeptide from Drosophila nuclear extracts and isolated peptides derived from protease digestion as previously described (13). Microsequence analysis yielded the sequences of six NURF-55 peptides, allowing the design of oligonucleotide primers for PCR analysis. The resulting DNA fragments were used to probe a Drosophila cDNA library, identifying several overlapping cDNAs, the longest of which contained an ORF of 430 amino acids with a predicted molecular weight of 48,600 and an acidic pI of 4.76. The deduced sequence of the encoded protein completely matched with the sequences of all six NURF-55 peptides (Fig. 1). We therefore conclude that this ORF encodes NURF-55.
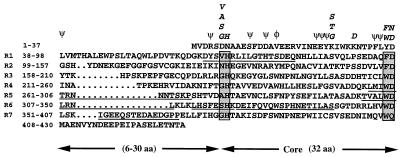
Figure 1.
Identification of the NURF-55. Predicted amino acid sequence of NURF-55 in single letter code, presented as the WD repeat motifs. The six NURF-55 peptides sequenced are underlined. Each WD repeat contains a variable region of 6–30 residues and a 28-residue core. The core regions start with GH and end with WD. φ, conserved aromatic residues; ψ, conserved hydrophobic residues.
Computer comparisons revealed that cDNA sequence of NURF-55 is identical to the sequence of the 55-kDa subunit of the Drosophila chromatin assembly factor CAF-1 reported by Tyler et al. (15). In agreement with these workers, Southern blot analysis and in situ hybridization to polytene chromosomes showed that p55 is a single copy gene that maps to chromosomal locus 88E (data not shown). NURF-55 and Drosophila CAF-1 p55 (both referred to as “p55” hereafter) are homologous to the related mammalian proteins, RbAp48 and RbAp46, initially discovered by virtue of their association with the human retinoblastoma protein in vitro (21). RbAp48 is also associated with mammalian histone deacetylase (22–24) and is a component of the human CAF-1 complex (25). In addition, there is 20–40% amino acid identity between p55 and the Saccharomyces cerevisiae proteins Hat2p, a subunit of the histone acetyltransferase B (26), and MSI1p, a negative regulator of Ras (27) and a subunit of yeast CAF-1 (28).
The amino acid sequence of p55 shows that it is a member of the large family of WD (or WD40) repeat proteins confined to eukaryotes and involved in numerous regulatory functions, including signal transduction, RNA processing, gene regulation, vesicular trafficking, and cell division (29). These proteins are composed of repeating units that occur four to eight times in the polypeptide, and each repeat comprises a region of variable length preceding a conserved core of ∼30 amino acids, bordered by characteristic Gly-His (GH) and Trp-Asp (WD) dipeptide residues (see Fig. 1 boxes). p55 has seven WD repeats with the variable regions ranging between 6 and 30 residues. All of the seven WD repeats found in p55 are well matched to the consensus motifs determined for this protein family.
p55 Is a Subunit of NURF.
Previous studies from our laboratory showed that all four NURF polypeptides copurifed with nucleosome disruption activity through seven chromatographic and centrifugal separation steps (13). However, no further biochemical evidence was available to support the proposal that these four polypeptides were an integral part of the NURF complex. The identification and cloning of NURF-55 as well as ISWI/NURF-140 (14) has permitted the generation of polyclonal antibodies for immunological analyses.
As shown in Fig. 2A, Western blotting indicates that roughly equivalent amounts of NURF-55 copurify with the NURF activity at all stages of the seven-step purification; similar results were also obtained for ISWI (13). We were also able to use antibodies suitable for immunoprecipitation to demonstrate interactions between these two NURF components. As shown in Fig. 2B, antibodies against p55 were able to immunoprecipitate ISWI from unfractionated nuclear extracts. Likewise, antibodies against ISWI were able to immunoprecipitate p55. Moreover, quantitative immunodepletion of p55 (and ISWI) from a partially purified NURF fraction with antibodies against p55 resulted in substantially reduced NURF activity as revealed by the ATP-dependent, GAGA factor-mediated nucleosome disruption assay (Fig. 2 C and D). These results strongly support the hypothesis that both p55 and ISWI are integral components of the NURF complex.
p55, but Not NURF, Associates with Histone Acetyltransferase.
Because Drosophila p55 is a subunit of NURF, CAF-1, and probably other protein complexes, we investigated the size distribution of p55 in Drosophila embryo extracts by gel filtration analysis and Western blotting. As shown in Fig. 3A, p55 was found in two broad fractions ranging in apparent molecular mass of 40–80 kDa and 150–850 kDa (Fig. 3A). This distribution is inclusive of the native size of the NURF complex, previously found to be approximately 500 kDa, as indicated by the chromatographic profile of the ISWI protein (Fig. 3A and ref. 14). However, the gel filtration profile of p55 is broader than ISWI and indicates that it may be also found in complexes both larger and smaller than NURF.
Because it has been shown that a p55 homolog, yeast Hat2p, is an integral component of a histone acetyltransferase enzyme (26), we explored the potential interaction between Drosophila p55 and HAT activity by incubating nuclear extracts with recombinant p55 tagged with the Myc epitope and overexpressed in Sf9 insect cells. We found that recombinant p55 could bind specifically to a histone acetylase activity in the nuclear extract, as revealed by the preferential incorporation of [3H]acetate in free histone H4 (Fig. 3B). This interaction of p55 with HAT activity prompted us to examine whether NURF itself was physically associated with HAT by analyzing chromatographic fractions at each stage of NURF purification using the liquid histone acetyltransferase assay. As shown in Fig. 3C, the Drosophila nuclear extract and the DE-52/BioRex (BR) fractions were found to exhibit strong HAT activity, but this activity was reduced substantially after the Q-Sepharose step and was undetectable when NURF was subjected to subsequent steps of purification. Hence, we conclude that whereas p55 is associated with active HAT enzyme(s) in nuclear extracts, this interaction is not shared by the subpopulation of p55 protein present in the NURF complex.
Immunofluorescent Localization of p55 on Polytene Chromosomes.
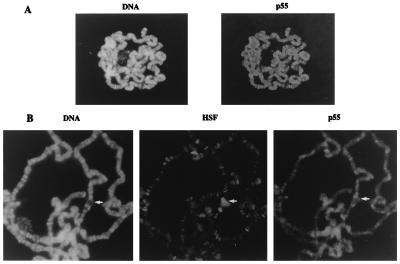
In accord with previous studies of Drosophila CAF-1, which localized the p55 protein to the embryonic cell nuclei (15), we have found that the p55 was predominantly localized in nuclei of Schneider line 2 tissue culture cells (data not shown). To further investigate the chromosomal localization of p55, we visualized the staining pattern of p55 on Drosophila polytene chromosomes by indirect immunofluorescence. As shown in Fig. 4A, p55 was found to be associated with polytene chromosomes, consistent with its presence in multiple complexes involved in chromatin metabolism. However, the immunostaining pattern of p55 revealed a general, nonspecific association with polytene chromosomes that followed, to a first approximation, the staining intensity for DNA in bands, interbands, and puffs (Fig. 4B). In general, p55 staining was not found to be focused at specific loci as has been frequently observed for sequence-specific transcription factors. During heat shock, activation of heat shock transcription factor HSF leads to a redistribution of this protein to numerous specific loci, including the chromosomal puffs at 87A/87C that contain the hsp70 genes; however, these heat shock puffs show no preferential accumulation of p55 protein (Fig. 4B). Our results indicate that the bulk of the p55 found in the multiple protein complexes associates with chromatin in a nonspecific manner, although targeting to specific loci is not excluded for a small fraction of the molecules.
Figure 4.
Immunofluorescent localization of p55 on polytene chromosomes. (A) Indirect immunofluorescent staining for p55 in a polytene nucleus (Left) and DNA staining (Right). Antibody specificity was demonstrated by the inhibition of immunoreactivity by the inclusion of purified recombinant p55 protein with anti-p55. (B) Indirect immunofluorescent staining for p55, HSF, and DNA on polytene chromosomes prepared from heat-shocked third instar larvae. The heat shock puff at 87C is indicated by the arrow. We have noticed several instances where p55 appears more localized to a few specific sites above the general nonspecific staining; the reproducibility and cytological location of this staining require further study for clarification.
Native, but Not Recombinant, p55 Associates with Reconstituted Chromatin.
The occurrence of p55 and its homologues in several protein complexes involved in chromatin metabolism suggested that this subunit could have a chromatin targeting or “escort” function for the protein complexes (26, 30). We explored physical interactions between reconstituted chromatin and the endogenous p55 in the S-150 chromatin assembly extract by immunoprecipitation assays. Antibodies directed against p55 (α-p55) were found to immunoprecipitate partially purified chromatin reconstituted with the S-150 chromatin assembly extract (Ch), as revealed by Western blot analysis using an antibody against one representative histone, H3 (Fig. 5A). α-p55 was also capable of immunoprecipitating unassembled histone H3 from the S-150 chromatin assembly extract. The latter coimmunoprecipitation is likely to be due to CAC, a complex of histones H3 and H4 and the human chromatin assembly factor CAF-1, of which the p55 homolog, RbAp48, is a subunit (25). Interestingly, recombinant p55 produced from a baculovirus expression system failed to interact with reconstituted chromatin. As shown by Western blotting (Fig. 5B), recombinant p55 did not exhibit detectable interactions with reconstituted chromatin (Ch) nor with unassembled histones in the S-150 extract (Ex). These results suggest that the recombinant p55 polypeptide, despite detectable binding to HAT activity (Fig. 3B), may be unable to associate with nucleosomal or unassembled histones. It is possible that the observed interaction of native p55 with chromatin could occur through conformational changes that confer histone-binding properties to p55 when it is complexed with partner proteins. Alternatively, p55 could just provide a platform for the assembly of other protein subunits that have the capacity to directly bind chromatin.
DISCUSSION
Previous studies from our laboratory defined NURF as a complex of four polypeptides that acts in combination with the GAGA transcription factor to disrupt nucleosome structure at a heat shock gene promoter in vitro (13). One NURF subunit, NURF-140, has been identified as the ISWI protein (14). Here, we have identified a second NURF subunit, NURF-55, and found that it is identical to the 55-kDa subunit of Drosophila CAF-1 (p55) (15). p55 is a WD repeat protein whose human homologs RbAp48 and RbAp46 were initially found as proteins that bind to the retinoblastoma protein in vitro (21), although the physiological significance of this interaction is unclear. RbAp48 was also identified as both a component of human CAF-1 (25) and mammalian histone deacetylase complexes (22–24). The related yeast proteins, Hat2p and MSI1p, were found as a regulatory subunit of the cytoplasmic B-type histone acetyltransferase (26) and a subunit of yeast CAF-1, respectively (28). Thus, p55 and its homologs are common components of four different protein complexes involved in histone metabolism, providing an intriguing molecular link between nucleosome disruption, chromatin assembly, histone acetylation, and histone deacetylation.
The presence of WD repeat motifs in p55 suggests how this polypeptide might associate with multiple partners. Structural studies have shown that the prototypical WD repeat protein, the β subunit of heterotrimeric G proteins, is folded in a seven-bladed β propeller structure that provides a scaffold for distinct interactions with Gα and Gγ subunits (31). Many of the residues involved in stabilizing the propeller fold are conserved in the WD repeats of p55 (see Fig. 1). These include the central aromatic and the C-terminal defining tryptophan of each repeat, which, in Gβ, are positioned such that the aromatic rings form the hydrophobic lower face of each propeller blade. Also found in p55 is the structural triad of aspartate (not the aspartate in the WD motif), histidine (of the GH motif), and serine/threonine residues of the WD repeat, which forms inter- and intrablade hydrogen bonds. The conservation of these residues in p55 suggests that the protein is likely to assume a propeller structure, providing a broad platform for multiple, distinct interactions with the remaining subunits of NURF, or for interactions to assemble the other protein complexes involved in histone metabolism. The presence of a common p55 subunit among these complexes invites speculation that this propeller protein may also have a specialized affinity for histones. However, we have been unable to detect substantial interactions between recombinant p55 immobilized on beads and histones in the chromatin assembly extract or in reconstituted chromatin, whereas interactions were readily detected for the endogenous p55 in native complexes. Such interactions could perhaps be allosterically induced by the association of p55 with other NURF subunits or, alternatively, could involve other NURF subunits besides p55. In this respect, it is of interest that only the complex of yeast Hat1p/Hat2p is able to interact productively with histone H4 tails (26).
In an earlier study, the four subunits of NURF were assigned on the basis of copurification with nucleosome disruption activity through seven chromatographic steps. We have obtained additional support for these assignments by coimmunoprecipitation studies with antibodies against p55 and ISWI, which show an interaction between these two polypeptides in crude extracts. Further evidence for a close association of p55 and ISWI with NURF activity is indicated by the ability to immunodeplete nucleosome disruption activity with anti-p55 antibodies.
The role of p55 as a subunit of Drosophila CAF-1 has been the subject of an extensive study (15), which also found an association between p55 and histone deacetylase (HD) activity in Drosophila nuclear extracts, in agreement with studies of the human p55 homolog RbAp48 (25). We have extended these findings by showing that recombinant p55 associates with HAT activity, consistent with the presence of a related protein Hat2p as a regulatory subunit of the major cytoplasmic histone acetyltransferase in yeast (26). Interestingly, the observation of steady-state HAT activity associating with p55 implies that this activity is able to counteract to some extent the opposing histone deacetylase activity presumably also associating with p55. The preferential acetylation of H4 over H3 in the p55-associated HAT activity is consistent with the properties of yeast Hat2p, although the presence of such an activity in the Drosophila nuclear rather than cytoplasmic fraction is puzzling and requires further investigation.
Are the interactions between p55 and the components of NURF, CAF-1, HAT, and HD mutually exclusive? We have found that NURF fractionates away from HAT activity very early in the purification process, suggesting that p55 within the NURF complex is not free to bind to components of HAT. Coimmunoprecipitation assays show little or no interaction between ISWI and Drosophila Rpd3 in nuclear extracts, suggesting that p55 within NURF is generally not available for binding to histone deacetylase components (M.M., F. DeRubertis, P. Spierer, and C.W., unpublished observations). We have not explored direct interactions between NURF and CAF-1 in crude extracts, although an exchange of reagents has indicated that purified NURF and CAF-1 fractions have no significant cross-activities and hence no tight interactions through the common p55 subunit (J. Kadonaga, personal communication and our unpublished observations). It is therefore likely that binding of NURF subunits to the p55 propeller protein excludes binding to components of CAF-1, HAT, and HD. Such an exclusionary interaction is reminiscent of the binding of the regulatory subunit phosducin to the βγ subunits of the G protein transducin, which excludes binding of Gα (32).
Immunolocalization studies show that p55 is in general distributed in a pattern that parallels the content of DNA throughout polytene chromatin. This distribution is consistent with the unlocalized distribution in nuclei previously observed in Drosophila embryos (15). However, because this protein is a component of multiple complexes involved in chromatin metabolism and because the relative prevalence of each complex has not been determined quantitatively, the general distribution we have observed does not exclude the possibility of a more specific pattern of localization for any one complex containing p55. Studies to clarify this issue are in progress with ISWI, which shows a distinct pattern of association on polytene chromosomes (J. Tamkun and S. Pimpinelli, personal communication). It will be of interest to extend the immunolocalization studies to the remaining NURF subunits to fully ascertain whether the NURF complex is targeted to specific loci or is generally associated with chromatin.
Acknowledgments
We thank C. Klee for performing microsequence analysis on NURF peptides, R. Kobayashi for protocols on in-gel proteolytic cleavage and HPLC of peptides, R. Sandaltzopoulos for experimental assistance, J. Kadonaga and J. Tyler for communicating results before publication and for anti-p55 antibodies, M. Bustin for anti-histone H3 antibodies, J. Tamkun for anti-ISWI, T. Jones and M. Mortin for determination of the chromosomal location of the p55 gene, G. Marchler and A. Marchler-Bauer for WD-repeat analysis of p55, F. De Rubertis, P. Spierer, J. Tamkun, and S. Pimpinelli for communication of unpublished observations, and members of our laboratory for helpful discussions. This work was supported by the Intramural Research Program of the National Cancer Institute.
ABBREVIATIONS
- NURF
Drosophila nucleosome remodeling factor
- CAF
chromatin assembly factor
- HAT
histone acetyltransferase
- HD
histone deacetylase
References
- 1.Felsenfeld G. Nature (London) 1992;355:219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- 2.Workman J L, Buchman A R. Trends Biochem Sci. 1993;18:90–95. doi: 10.1016/0968-0004(93)90160-o. [DOI] [PubMed] [Google Scholar]
- 3.Paranjape S M, Kamakaka R T, Kadonaga J T. Annu Rev Biochem. 1994;63:265–297. doi: 10.1146/annurev.bi.63.070194.001405. [DOI] [PubMed] [Google Scholar]
- 4.Owen-Hughes T, Workman J L. Crit Rev Eukaryotic Gene Expression. 1994;4:403–441. [PubMed] [Google Scholar]
- 5.Becker P B. BioEssays. 1994;16:541–547. doi: 10.1002/bies.950160807. [DOI] [PubMed] [Google Scholar]
- 6.Kingston R E, Bunker C A, Imbalzano A N. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 7.Felsenfeld G. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- 8.Tsukiyama T, Wu C. Curr Opin Genet Dev. 1997;7:182–191. doi: 10.1016/s0959-437x(97)80127-x. [DOI] [PubMed] [Google Scholar]
- 9.Henikoff S. Trends Biochem Sci. 1993;18:291–292. doi: 10.1016/0968-0004(93)90037-n. [DOI] [PubMed] [Google Scholar]
- 10.Carlson M, Laurent B C. Curr Opin Cell Biol. 1994;6:396–402. doi: 10.1016/0955-0674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 11.Peterson C L, Tamkun J W. Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 12.Petersen C L. Curr Opin Genet Dev. 1996;6:171–175. doi: 10.1016/s0959-437x(96)80047-5. [DOI] [PubMed] [Google Scholar]
- 13.Tsukiyama T, Wu C. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 14.Tsukiyama T, Daniel C, Tamkun J, Wu C. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 15.Tyler J K, Bulger M, Kamakaka R T, Yashi R, Kadonaga J T. Mol Cell Biol. 1996;16:6149–6159. doi: 10.1128/mcb.16.11.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becker P B, Wu C. Mol Cell Biol. 1992;12:2241–2249. doi: 10.1128/mcb.12.5.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becker P B, Tsukiyama T, Wu C. Methods Cell Biol. 1994;44:207–223. doi: 10.1016/s0091-679x(08)60915-2. [DOI] [PubMed] [Google Scholar]
- 18.Westwood J T, Clos J, Wu C. Nature (London) 1991;353:822–827. doi: 10.1038/353822a0. [DOI] [PubMed] [Google Scholar]
- 19.Wampler S L, Tyree C M, Kadonaga J T. J Biol Chem. 1990;265:21223–21231. [PubMed] [Google Scholar]
- 20.Sobel R E, Cook R G, Allis D. J Biol Chem. 1994;269:18576–18582. [PubMed] [Google Scholar]
- 21.Qian Y-W, Wang Y-C J, Hollingsworth R E, Jones D, Jr, Ling N, Lee E Y-H P. Nature (London) 1993;364:648–652. doi: 10.1038/364648a0. [DOI] [PubMed] [Google Scholar]
- 22.Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 23.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 25.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 26.Parthun M R, Widom J, Gottschling D E. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 27.Ruggieri R, Tanaka K, Nakafuku M, Kaziro Y, Toh-e A, Matsumoto K. Proc Natl Acad Sci USA. 1989;86:8778–8782. doi: 10.1073/pnas.86.22.8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaufman P D, Kobayashi R, Stillman B. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 29.Neer E J, Schmidt C J, Nambudripad R, Smith T F. Science. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 30.Roth S Y, Allis D. Cell. 1996;87:5–8. doi: 10.1016/s0092-8674(00)81316-1. [DOI] [PubMed] [Google Scholar]
- 31.Neer E J, Smith T F. Cell. 1996;84:175–178. doi: 10.1016/s0092-8674(00)80969-1. [DOI] [PubMed] [Google Scholar]
- 32.Gaudet R, Bohm A, Sigler P B. Cell. 1996;87:577–588. doi: 10.1016/s0092-8674(00)81376-8. [DOI] [PubMed] [Google Scholar]