Abstract
Myocyte enhancer factor-2 (MEF2) transcription factors control muscle-specific and growth factor-inducible genes. We show that hypertrophic growth of cardiomyocytes in response to phenylephrine and serum is accompanied by activation of MEF2 through a posttranslational mechanism mediated by calcium, calmodulin-dependent protein kinase (CaMK), and mitogen-activated protein kinase (MAPK) signaling. CaMK stimulates MEF2 activity by dissociating class II histone deacetylases (HDACs) from the DNA-binding domain. MAPKs, which activate MEF2 by phosphorylation of the transcription activation domain, maximally stimulate MEF2 activity only when repression by HDACs is relieved by CaMK signaling to the DNA-binding domain. These findings identify MEF2 as an endpoint for hypertrophic stimuli in cardiomyocytes and demonstrate that MEF2 mediates synergistic transcriptional responses to the CaMK and MAPK signaling pathways by signal-dependent dissociation from HDACs.
Myocyte enhancer factor-2 (MEF2) transcription factors (1) participate in diverse gene regulatory programs, including those for muscle and neural differentiation, cardiac morphogenesis, blood vessel formation, and growth factor responsiveness (reviewed in ref. 2). The four MEF2 factors, MEF2A, -B, -C, and -D, share high homology in an amino-terminal MADS (MCMI, Agamous, Deficiens, Serum response factor) domain that mediates DNA-binding and dimerization and an adjacent MEF2-specific domain that influences DNA-binding affinity and interaction with transcriptional cofactors (2). The carboxyl-terminal regions of MEF2 factors, which are more divergent, act as transcription activation domains (TADs).
Studies in T cells and fibroblasts have shown that the mitogen-activated protein kinases (MAPKs) p38 and ERK5 stimulate transcriptional activity of MEF2 factors by phosphorylating conserved sites in their TADs (3–12). Calcium, calmodulin-dependent protein kinase (CaMK), and calcineurin also stimulate MEF2 activity (13, 14), but the underlying mechanisms are unknown. It is also unclear whether MAPK, CaMK, and calcineurin pathways cross-talk or act in parallel to activate MEF2.
Most studies of MEF2 activation by extracellular signaling have focused on cells that are highly responsive to mitogens. Whether MEF2 retains the ability to respond to growth factor signals in terminally differentiated muscle cells, which are permanently postmitotic, and whether MEF2 is activated in muscle cells by the same signaling pathways as in other cell types has not been determined. In cardiac myocytes, growth factor signals evoke a hypertrophic response characterized by cell enlargement, activation of immediate early genes, reactivation of fetal cardiac muscle genes, and sarcomere assembly (reviewed in ref. 15).
To investigate the mechanisms that regulate MEF2 activity in response to extracellular signals and to test whether MEF2 retains its ability to respond to growth factor signals in terminally differentiated muscle cells, we sought to determine whether MEF2 could be activated by hypertrophic signals in cardiomyocytes and, if so, to define the mechanism linking signaling in the cytoplasm to MEF2-dependent transcription. We show that the MEF2 DNA-binding domain confers responsiveness to hypertrophic signals mediated by CaMK. Using this region as bait in a two-hybrid screen for possible CaMK-sensitive transcriptional cofactors, we discovered that MEF2 interacts with histone deacetylases (HDACs) 4 and 5, resulting in repression of the transcriptional activity of MEF2. Activation of CaMK results in dissociation of MEF2 from these HDACs and unmasking of MEF2 transcriptional activity. MAPK signaling pathways also target MEF2, but are directed at the TAD, and require CaMK signaling to the DNA-binding domain for maximal stimulation of MEF2. Thus, synergistic activation of MEF2 involves convergence of CaMK and MAPK signaling pathways on different domains of the protein, providing a potential mechanism for transcriptional cross-talk between these two signaling pathways in hypertrophic cardiomyocytes and other cell types.
Materials and Methods
Transfections.
Primary neonatal rat cardiomyocytes were prepared as described (16) and plated at a density of 8 × 105 cells per 35-mm dish in DMEM with 10% horse serum and 5% FBS. Twenty-four hours after plating, cells were transfected by using Lipofectamine reagent with 800 ng of luciferase expression plasmids, 100 ng of MEF2 expression plasmid, and 100 ng of CMV-lacZ as an internal control for transfection efficiency. Six hours after transfection, cells were rinsed and refed with serum-free DMEM and incubated overnight. The next morning, phenylephrine (PE) was added to 10 μM, along with KN62 (1 μM) and SB202190 (20 μM), as specified. These concentrations of inhibitors do not affect cell viability. Cells were harvested 36–38 h after transfection, and luciferase assays were performed on cell extracts under conditions of linearity with respect to time and extract concentration. 10T½ cells at 50% confluence in 6-well plates were transfected in duplicate by using FuGENE 6 reagent (Roche Molecular Biochemicals) and were maintained in DMEM with 10% FBS, which accounts for the activity of MEF2 in the absence of exogenous CaMK (see Figs. 3 and 4). Each transfection included 0.3 μg of luciferase reporter, 0.3 μg of MEF2 expression vector, 0.1 μg of HDAC expression vector, and 0.2 μg of CaMK and MAPK kinase (MKK)6 expression vector. CMV-lacZ or SV40-lacZ (0.1 μg) were included for normalization. The MKK6 expression plasmid encoded an activated form of the enzyme with Ser-207 and Thr-211 to Glu mutations (17). The activated CaMKIV plasmid encoded an activated form of the enzyme in which Glu-318 was replaced with a stop codon to remove the autoinhibitory domain (13). The activated CaMKI plasmid encoded an activated form of the enzyme in which isoleucine-294 was replaced with a stop codon. Both of these CaMK mutants function constitutively without a requirement for Ca2+ and calmodulin for activation.
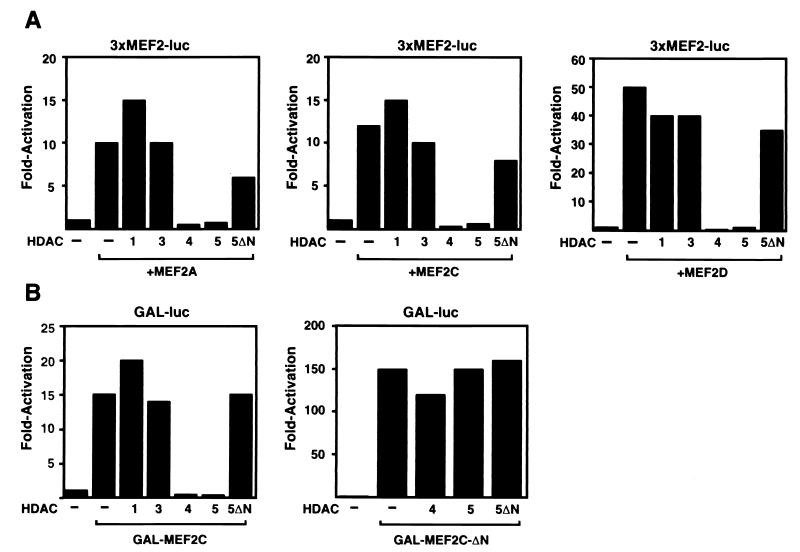
Figure 3.
HDAC4 and -5 inhibit MEF2-dependent transcription. (A) 10T½ cells in serum-containing medium (see Materials and Methods) were transiently transfected with the MEF2-dependent reporter, 3xMEF2-luciferase, along with expression vectors for the indicated HDACs and MEF2 factors. Forty-eight hours later, cells were harvested and luciferase activity was determined. (B) 10T½ cells were transiently transfected with pG5E1b-luciferase reporter and expression vectors for GAL4-MEF2C, GAL4-MEF2C-ΔN, and the indicated HDACs and luciferase activity was determined as in A.
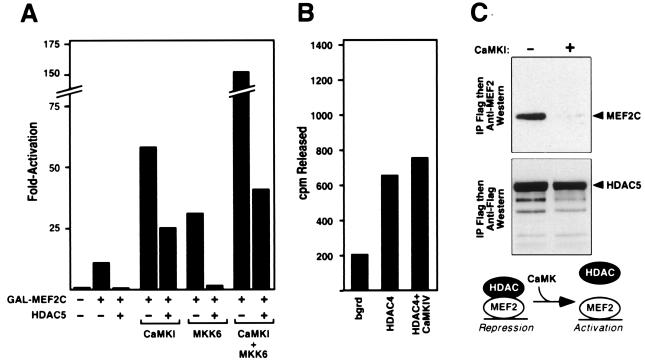
Figure 4.
CaMK-dependent activation of MEF2 overcomes HDAC-mediated repression. (A) 10T½ cells in serum-containing medium were transiently transfected with the indicated expression vectors, and luciferase activity was determined. (B) 293T cells were transfected with expression plasmids encoding Flag-tagged HDAC4 with and without CaMKIV as indicated. Forty-eight hours after transfection, cell extracts were prepared, immunoprecipitated with anti-Flag antibody, and HDAC activity was determined by release of [3H]acetate from acetylated histones, as described in Materials and Methods. (C) Immunoprecipitations from extracts of cells transfected with MEF2C, HDAC5, and CaMKI expression vectors, as indicated, were performed as described in Fig. 2 B and C. In the presence of activated CaMKI, interaction between MEF2C and HDAC5 was significantly diminished. An illustration of how CaMK may activate MEF2 by dissociating HDAC is shown.
Two-Hybrid Screen.
Rat aortic smooth muscle and mouse C2C12 myotube cDNA libraries were screened with MEF2C baits by the yeast two-hybrid system, as described (16). For each library, approximately 5 × 106 independent clones were screened. We isolated one cDNA encoding HDAC4 and two encoding HDAC5 from the aortic smooth muscle library, and from the C2C12 myotube library we isolated eight HDAC4 cDNAs.
Immunoprecipitations.
For coimmunoprecipitation experiments, 5 × 105 293T cells were transfected by using FuGENE 6 with expression vectors (1 μg each) encoding the indicated Flag epitope-tagged HDAC and MEF2 proteins. Forty-eight hours later, cells were harvested in PBS containing 0.5% Triton X-100, 1 mM EDTA, 1 mM PMSF, and protease inhibitors (Complete; Roche Molecular Biochemicals). Cells were subjected to brief sonication and cellular debris was removed by centrifugation. Flag-tagged HDAC proteins were immunoprecipitated from cell lysates by using anti-Flag M2 affinity resin (Sigma) and washed five times with lysis buffer. Precipitated proteins were resolved by SDS/PAGE, transferred to poly(vinylidene difluoride) membranes, and sequentially immunoblotted with polyclonal antisera raised against the indicated MEF2 protein (3) and an anti-Flag monoclonal antibody (Sigma). Proteins were visualized by using a chemiluminescence system (Santa Cruz Biotechnology).
HDAC Assays.
HDAC assays were performed as described (18) with immunoprecipitates of Flag-tagged HDACs from transiently transfected 293T cells.
Results
Mapping CaMK- and MAPK-Responsive Domains of MEF2.
To determine whether MEF2 was a target for hypertrophic signaling in cardiomyocytes, we tested whether hypertrophy in response to PE and FBS activated a MEF2-dependent reporter (3xMEF2-luciferase), containing three MEF2-binding sites. In cardiomyocytes maintained in serum-free medium, the MEF2 reporter was expressed at a relatively low level. Addition of PE or FBS resulted in stimulation of the reporter (Fig. 1A), whereas a reporter containing a mutant MEF2 site showed no response (data not shown).
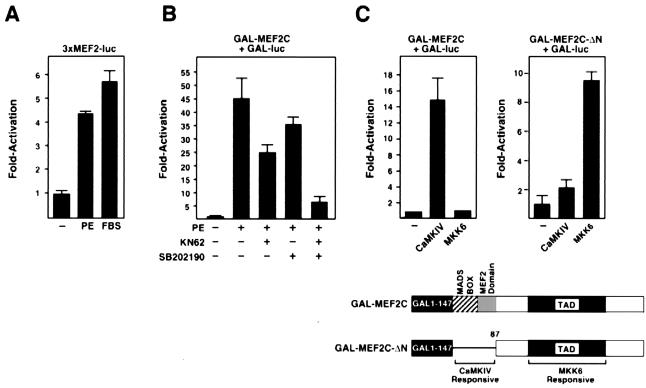
Figure 1.
CaMK and MAPK target different domains of MEF2. Primary neonatal rat cardiomyocytes in serum-free medium were transiently transfected with the indicated expression plasmids, and luciferase activity was determined in cell extracts. (A) Cells were stimulated with PE (10 μM) or 10% FBS, as indicated, and expression of the MEF2-dependent reporter, 3xMEF2-luciferase, was assayed. (B) Cells transiently transfected with pG5E1b-luciferase (Gal-luc) and GAL-MEF2C were stimulated with PE, as in A, in the presence of KN62 or SB202190, as indicated. (C) Cells were transiently transfected with pG5E1b-luciferase and GAL-MEF2C (Left) or GAL-MEF2C-ΔN (Right), along with activated CaMKIV and MKK6, as indicated. A schematic of the GAL4-MEF2 fusions is shown at the bottom.
Gel mobility shift assays showed no change in MEF2 DNA-binding activity in response to PE or FBS (data not shown), suggesting that acquisition of MEF2 transcriptional activity was independent of changes in DNA binding. To further investigate this, we tested whether a GAL4-MEF2C fusion, containing the complete ORF of MEF2C (19) fused to the GAL4 DNA-binding domain, could be activated by these stimuli. PE (Fig. 1B) and FBS (data not shown) also stimulated activity of GAL-MEF2C. Because CaMKs (20, 21) and MAPKs (22) are activated by PE in cardiomyocytes and have been shown to stimulate MEF2 activity in other cell types (3–14), we tested whether the CaMK and p38 MAPK inhibitors, KN62 and SB202190, respectively, could block the ability of PE to stimulate transcriptional activity of GAL-MEF2C. At the concentrations used for these inhibitors, they have been shown to selectively inhibit CaMK and MAPK, respectively (23, 24). Either inhibitor alone partially blocked activation of GAL-MEF2C by PE, whereas both inhibitors together blocked almost all activation (Fig. 1B), as well as morphologic hypertrophy (data not shown) (Fig. 1B). This suggested that CaMK and MAPK pathways cooperated to activate MEF2 in hypertrophic cardiomyocytes.
To map the regions of MEF2C that conferred responsiveness to CaMK and MAPK, we compared the effects of these kinases on GAL-MEF2C and a deletion mutant containing the TAD, but lacking the MADS- and MEF2-domains (GAL-MEF2C-ΔN). Whereas activated CaMKIV and CaMKI (data not shown) stimulated activity of GAL-MEF2C, they had little effect on GAL-MEF2C-ΔN (Fig. 1C). In contrast, the MAPK kinase MKK6, which stimulates MEF2 by activating p38 MAPK (25), preferentially activated GAL-MEF2C-ΔN (Fig. 1C). Mutation to alanines of Ser-387, Thr-293, and Thr-300, shown previously to be phosphorylated by MAPK (3), prevented activation of MEF2C by MKK6 and PE (data not shown). All of these GAL-MEF2C constructs were expressed at comparable levels as determined by Western blot with anti-GAL4 antibody (data not shown). Together, these results demonstrated that the CaMK and MAPK pathways targeted the amino- and carboxyl-terminal regions, respectively, of MEF2C and that the MADS/MEF2-region interfered with activation of the TAD by MKK6.
Interaction of MEF2 with HDAC4 and -5.
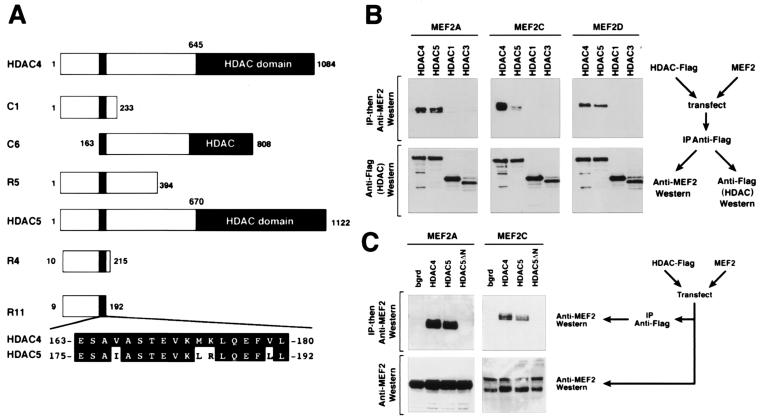
Because activation of MEF2 by CaMK did not appear to involve an effect on DNA binding, we speculated that CaMK might regulate MEF2 activity by means of a CaMK-sensitive cofactor. We therefore used the minimal CaMK-sensitive domain of MEF2 to perform yeast two-hybrid screens of muscle cell cDNA libraries for potential cofactors that might confer CaMK-sensitivity to MEF2. Among 11 strongly positive clones identified using the MADS-MEF2-domain (amino acids 1–86) of MEF2C fused to GAL4 as bait, 9 corresponded to HDAC4 and 2 to HDAC5. These HDACs are expressed at highest levels in heart, brain, and skeletal muscle (26–28), the same tissues in which MEF2 is enriched (29–31).
HDACs 4 and 5 are distinguished from other HDACs by the presence of amino-terminal extensions (Fig. 2A) (26–28). The portions of HDAC4 and -5 rescued from the two-hybrid screens overlapped in a nearly identical 18-aa segment near their amino termini. Association of MEF2 factors with HDAC4 was also reported recently (32–34), but the physiological significance of this association was not determined (see below), nor were MEF2 factors shown to interact with HDAC5.
Figure 2.
Interaction of MEF2 and HDACs 4 and 5 in yeast and mammalian cells. (A) Schematic diagrams of HDACs 4 and 5 and the different regions of the proteins encoded by cDNAs rescued as “prey” in two-hybrid screens are shown. The rescued HDAC cDNAs overlap in the 18-aa segment shown at the bottom. The HDAC catalytic domain is located at the extreme C termini of the proteins. (B and C) Coimmunoprecipitation of MEF2 factors and HDACs 4 and 5 from transfected cells. HDACs with Flag epitopes at their carboxyl termini and MEF2 factors were expressed in transiently transfected 293 T cells. Forty-eight hours after transfection, cell extracts were prepared and immunoprecipitated with anti-Flag antibody. Immunoprecipitates were then separated by SDS/PAGE and sequentially immunoblotted with anti-MEF2 or anti-Flag antibodies. (Upper) The results of anti-MEF2 Western and specific interaction of HDAC 4 and 5 with MEF2A, -C, and -D. (Lower) The results of anti-Flag (HDAC) Western blot show that comparable amounts of each HDAC were expressed in transfected cells. A schematic of the experiment is shown at the side. (C) Cell extracts were immunoprecipitated with anti-Flag antibody followed by Western blot with anti-MEF2 (Upper) or were probed by anti-MEF2 Western without prior immunoprecipitation (Lower). Deletion of the HDAC5 amino terminus prevents interaction with MEF2A, MEF2C, and MEF2D (not shown).
Immunoprecipitation with an antibody against a Flag-epitope tag on HDACs, followed by Western blot with anti-MEF2 antibody, showed that HDACs 4 and 5 formed a complex with MEF2A, -C, and -D in transiently transfected 293T cells (Fig. 2B). In contrast, there was no interaction between MEF2 and HDACs 1 or 3, which lack a MEF2-interacting region. Consistent with the requisite role of the 18-aa segment near the amino termini of HDACs 4 and 5 for association with MEF2, a HDAC5 deletion mutant lacking amino acids 22–488 (HDAC5-ΔN), eliminating the MEF2-interacting region, failed to coimmunoprecipitate with MEF2 factors (Fig. 2C).
Repression of MEF2 Transcriptional Activity by HDAC4 and -5.
HDACs are thought to inhibit transcription by deacetylating histones (35). Thus, interaction of HDACs 4 and 5 with the region of MEF2 that represses the TAD suggested that this interaction played a role in regulation of MEF2 function. Indeed, in the presence of HDACs 4 and 5, MEF2A, -C, and -D were unable to transactivate 3xMEF2-luciferase in 10T½ fibroblasts, whereas in the presence of HDACs 1 or 3, these MEF2 factors were fully active (Fig. 3A). Transcriptional activity of GAL-MEF2C was also completely inhibited by HDACs 4 and 5, but not by HDACs 1 or 3 or HDAC5ΔN (Fig. 3B). In contrast, GAL-MEF2C-ΔN, which lacks the HDAC-binding region, was insensitive to HDAC4 and -5 (Fig. 3B).
CaMK Signaling Overcomes HDAC-Mediated Repression of MEF2 Activity.
The finding that HDACs 4 and 5 interacted with the same region of MEF2 required for CaMK activation suggested that signal-dependent activation of MEF2 might involve derepression by HDACs. To test this, we assayed the responsiveness of GAL-MEF2C to HDAC5 and CaMKs. Whereas HDAC5 completely repressed activity of GAL-MEF2C in serum-containing medium, activated CaMKI (Fig. 4A) and CaMKIV (data not shown) restored transcriptional activity to GAL-MEF2C in the presence of HDAC5 (Fig. 4A). In contrast, MKK6 was unable to overcome inhibition by HDAC5, in agreement with the earlier conclusion that activation of the carboxyl-terminal TAD by MKK6 required signaling to the DNA-binding domain by CaMK. Together, MKK6 and CaMKI synergistically activated MEF2-dependent transcription greater than 100-fold and potently interfered with the inhibitory effect of HDAC on MEF2 activity.
Dissociation of HDAC5 from MEF2 by CaMK Signaling.
To begin to determine the mechanism whereby CaMK signaling was able to overcome HDAC-mediated repression of MEF2 activity, we tested whether CaMK inhibited HDAC enzymatic activity in vivo. Cells were transiently transfected with expression vectors for Flag-tagged HDAC4 with and without activated CaMKI or CaMKIV, followed by immunoprecipitation of HDAC and an assay for HDAC enzymatic activity. We observed no reduction in HDAC activity in cells expressing activated CaMKI or IV (Fig. 4B), suggesting that overall inhibition of enzyme activity was not the mechanism for CaMK-dependent activation of MEF2.
We next tested whether CaMK signaling might detach HDACs from MEF2, resulting in transcriptional activation. As shown in Fig. 4C, activated CaMKI prevented association of HDAC5 and MEF2C, as detected by coimmunoprecipitation assays in transfected COS cells (Fig. 4C). Similar results were obtained for HDAC4 (data not shown). We conclude that CaMK signaling disrupts the MEF2/HDAC complex, which unmasks the transcriptional activity of MEF2.
Discussion
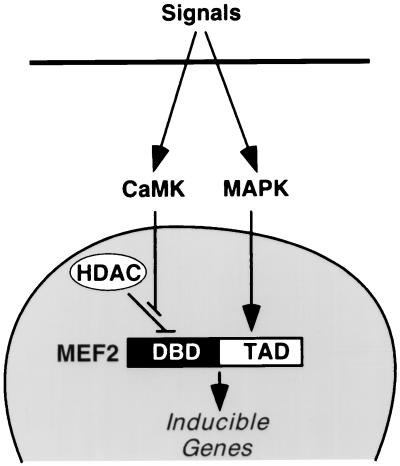
Numerous studies have implicated CaMK and MAPK signaling in cardiac hypertrophy (15, 22), but little is known of the transcriptional targets for these pathways or whether they are independent or interdependent. Our results demonstrate that MEF2 acts as a bipartite target for CaMK and MAPK signaling pathways in hypertrophic cardiomyocytes. PE appears to activate both of these pathways. However, one could also envision other stimuli that might preferentially activate one pathway or the other. A model consistent with our results is shown in Fig. 5.
Figure 5.
A model for the regulation of MEF2 activity by CaMK signaling. Hypertrophic signals that activate CaMK and MAPK lead to MEF2 activation by different mechanisms. Some stimuli, such as PE, may activate both pathways, whereas other stimuli may preferentially activate one pathway or the other. Association of HDACs 4/5 with the DNA-binding domain (DBD) of MEF2 represses MEF2 transcriptional activity. CaMK activates MEF2 by preventing association of HDACs 4/5 with MEF2. MAPK stimulates MEF2 activity by direct phosphorylation of the TAD. Together, the CaMK and MAPK pathways synergize to activate MEF2.
The DNA-binding domain of MEF2 confers sensitivity to CaMK signaling and also mediates repression of MEF2-dependent transcription through interaction with HDACs 4 and 5. CaMK signaling unmasks the transcriptional potential of MEF2 by inducing the release of these HDACs. In contrast, activation of MEF2 by MAPK is mediated by phosphorylation of the TAD (3–12) and can be prevented by association of HDAC with the DNA-binding domain. Thus, full activation of MEF2 depends on costimulatory pathways activated by CaMK and MAPK, which target different domains of MEF2 to synergistically activate transcription. These findings reveal a potential form of cross-talk between the CaMK and MAPK signaling pathways and demonstrate a molecular basis for synergistic transcriptional activation by these pathways.
Repression of MEF2 activity is specific for HDACs 4 and 5 and was not observed for other HDACs that lack the MEF2-interacting region. HDACs 4 and 5, classified as class II HDAC enzymes, have been shown to deacetylate all four core histones in vitro (26–28), which would be predicted to result in suppression of gene expression. In principle, the association of MEF2 with HDACs allows MEF2 to act as a transcriptional activator or repressor, depending on intracellular signaling and combinatorial associations with other transcription factors. Thus, in cells such as cardiomyocytes that express high levels of class II HDACs, MEF2 would be expected to repress transcription in the absence of CaMK signaling, whereas, in other cell types that express lower levels of these HDACs, MEF2 would be expected to show higher basal activity and less responsiveness to CaMK signaling.
The specific target for CaMK in the MEF2-HDAC complex remains to be identified. We do not believe the HDAC-interacting region of MEF2 is a direct substrate for CaMK phosphorylation because in vitro phosphorylation experiments have failed to demonstrate efficient phosphorylation of this region by purified CaMK and mutation of potential phosphorylation sites in this region does not alter HDAC-mediated repression of MEF2 (unpublished results). Thus, we favor the possibility that HDAC, or possibly another nuclear factor that controls MEF2–HDAC interactions, is the target for CaMK.
It is conceivable that the release of HDAC from MEF2 in response to CaMK signaling depends on, or is accompanied by, displacement by another factor that is CaMK-sensitive. In this regard, the transcriptional coactivator CBP/p300, previously shown to interact with MEF2 (36, 37) and to be activated by CaMKIV (38), might be recruited to MEF2-dependent promoters in response to CaMK signaling, resulting in transcriptional activation. Because CBP/p300 possesses histone acetyltransferase activity, its recruitment to MEF2 following CaMK activation could also account for the signal-dependent activation of MEF2.
The finding that MEF2 is activated in cardiomyocytes by hypertrophic signals raises the question whether MEF2 activation is essential for hypertrophic growth. Consistent with this possibility are recent studies showing that a dominant negative MEF2 mutant prevents postnatal cardiac growth (39). Cardiac hypertrophy has also been shown to be controlled by a signaling pathway involving calcineurin and the transcription factor NFAT3 (16), but there is evidence for alternate pathways (15). Hypertrophic activation of MEF2 by CaMK-mediated dissociation of HDAC may constitute such an alternate pathway for cardiac growth. Given the essential roles of MEF2 in muscle and neural development (2, 40), HDAC and CaMK signaling may also play a role in these processes.
Acknowledgments
We thank S. Schreiber, T. Soderling, A. Means, and R. Prywes for reagents and J. Page and A. Tizenor for help with the manuscript. This work was supported by grants from National Institutes of Health, the Texas Advanced Technology Program, and the Robert A. Welch Foundation (to E.N.O.). T.A.M. is a Pfizer Fellow of the Life Sciences Foundation, and R.L.N. was supported by a postdoctoral fellowship from the National Institutes of Health.
Abbreviations
- CaMK
calcium, calmodulin-dependent protein kinase
- HDAC
histone deacetylase
- MAPK
mitogen-activated protein kinase
- MEF2
myocyte enhancer factor-2
- MKK
MAPK kinase
- PE
phenylephrine
- TAD
transcription activation domain
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.080064097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.080064097
References
- 1.Gossett L A, Kelvin D J, Sternberg E A, Olson E N. Mol Cell Biol. 1989;9:5022–5033. doi: 10.1128/mcb.9.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black B L, Olson E N. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 3.Han T H, Prywes R. Mol Cell Biol. 1995;15:2907–2915. doi: 10.1128/mcb.15.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han J, Jiang Y, Li Z, Kravchenko V V, Ulevitch R J. Nature (London) 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 5.Kato Y, Kravchenko V V, Tapping R I, Han J, Ulevitch R J, Lee J D. EMBO J. 1997;16:7054–7066. doi: 10.1093/emboj/16.23.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zetser A, Gredinger E, Bengal E. J Biol Chem. 1999;274:5193–5200. doi: 10.1074/jbc.274.8.5193. [DOI] [PubMed] [Google Scholar]
- 7.Zhao M, New L, Kravchenko V V, Kat Y, Gram H, Di Padova F, Olson E N, Ulevitch R J, Han J. Mol Cell Biol. 1999;19:21–30. doi: 10.1128/mcb.19.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang C C, Ornatsky O, McDermott J C, Cruz T F, Prody C A. Nucleic Acids Res. 1998;26:4771–4777. doi: 10.1093/nar/26.20.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke N, Arenzana N, Hai T, Minden A, Prywes R. Mol Cell Biol. 1998;18:1065–1973. doi: 10.1128/mcb.18.2.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ornatsky O I, Cox D M, Tangirala P, Andreucci J J. Nucleic Acids Res. 1999;27:2646–2654. doi: 10.1093/nar/27.13.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marinissen M J, Chiariello M, Pallante M, Gutkind J S. Mol Cell Biol. 1999;19:4289–4301. doi: 10.1128/mcb.19.6.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang S, Galanis A, Sharrocks A D. Mol Cell Biol. 1999;19:4028–4038. doi: 10.1128/mcb.19.6.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S, Liu P, Borras A, Chatila T, Speck S H. EMBO J. 1997;16:143–153. doi: 10.1093/emboj/16.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woronicz J D, Lina A, Calnan B J, Szychowski S, Cheng L, Winoto A. Mol Cell Biol. 1995;15:6364–6376. doi: 10.1128/mcb.15.11.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKinsey T A, Olson E N. Curr Opin Genet Dev. 1999;9:267–274. doi: 10.1016/s0959-437x(99)80040-9. [DOI] [PubMed] [Google Scholar]
- 16.Molkentin J D, Lu J, Antos C L, Markham B, Richardson J, Robbins J, Grant S, Olson E N. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Y, Chen C, Li Z, Guo W, Gegner J A, Liu S, Han J. J Biol Chem. 1996;271:17920–17926. doi: 10.1074/jbc.271.30.17920. [DOI] [PubMed] [Google Scholar]
- 18.Wu G, Roeder R G. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 19.Martin J F, Schwarz J J, Olson E N. Proc Natl Acad Sci USA. 1993;90:5282–5286. doi: 10.1073/pnas.90.11.5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDonough P M, Stella S L, Glembotski C C. J Biol Chem. 1994;269:9466–9472. [PubMed] [Google Scholar]
- 21.Ramirez M T, Zhao X-L, Schulman H, Brown J H. J Biol Chem. 1997;272:31203–31208. doi: 10.1074/jbc.272.49.31203. [DOI] [PubMed] [Google Scholar]
- 22.Clerk A, Michael A, Sugden P H. J Cell Biol. 1998;142:523–535. doi: 10.1083/jcb.142.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clerk A, Sugden P H. FEBS Lett. 1998;426:93–96. doi: 10.1016/s0014-5793(98)00324-x. [DOI] [PubMed] [Google Scholar]
- 24.Enslen H, Sun P, Brickley D, Soderling S H, Klamo E, Soderling T R. J Biol Chem. 1994;269:15520–15527. [PubMed] [Google Scholar]
- 25.Enslen H, Raingeaud J, Davis R J. J Biol Chem. 1998;273:1741–1748. doi: 10.1074/jbc.273.3.1741. [DOI] [PubMed] [Google Scholar]
- 26.Grozinger C M, Hassig C A, Schreiber S L. Proc Natl Acad Sci USA. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischle W, Emiliani S, Hendzel M J, Nagase T, Nomura N, Voelter W, Verdin E. J Biol Chem. 1999;274:11713–11720. doi: 10.1074/jbc.274.17.11713. [DOI] [PubMed] [Google Scholar]
- 28.Verdel A, Khochbin S. J Biol Chem. 1999;274:2440–2445. doi: 10.1074/jbc.274.4.2440. [DOI] [PubMed] [Google Scholar]
- 29.Edmondson D G, Lyons G E, Martin J F, Olson E N. Development (Cambridge, UK) 1994;120:1251–1263. doi: 10.1242/dev.120.5.1251. [DOI] [PubMed] [Google Scholar]
- 30.Yu Y, Breitbart B, Smoot L B, Lee Y, Mahdavi V, Nadal-Ginard B. Genes Dev. 1992;6:1783–1789. doi: 10.1101/gad.6.9.1783. [DOI] [PubMed] [Google Scholar]
- 31.Martin J F, Miano J M, Hustad C M, Copeland N G, Jenkins N A, Olson E N. Mol Cell Biol. 1994;14:1647–1656. doi: 10.1128/mcb.14.3.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sparrow D B, Miska E A, Langley E, Reynaud-D S, Kotecha N T, Spohr G, Kouzarides T, Mohun T J. EMBO J. 1999;18:5085–5098. doi: 10.1093/emboj/18.18.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miska E A, Karlsson C, Langley E, Nielson S J, Pines J, Kouzarides T. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang A H, Bertos N R, Vezmar M, Pelletier N, Crosato M, Heng H H, Thng J, Han J, Yang X-J. Mol Cell Biol. 1999;19:7816–7827. doi: 10.1128/mcb.19.11.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strahl B D, Allis D. Nature (London) 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 36.Sartorelli V, Huang J, Hamamori Y, Kedes L. Mol Cell Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eckner R, Yao T P, Oldread E, Livingston D M. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 38.Chawala S, Hardingham G E, Quinn D R, Bading H. Science. 1998;281:1505–1509. doi: 10.1126/science.281.5382.1505. [DOI] [PubMed] [Google Scholar]
- 39.Kolodziejczyk S M, Wang L, Balazsi K, DeRepentigny Y, Kothary R, Megeney L A. Curr Biol. 1999;9:1203–1206. doi: 10.1016/S0960-9822(00)80027-5. [DOI] [PubMed] [Google Scholar]
- 40.Lilly B, Zhao B, Ranganayakulu G, Paterson B M, Schultz R A, Olson E N. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]