Abstract
Angiotensin-converting enzyme (ACE) is a type I ectoprotein that is cleaved off the cell surface by a plasma membrane-bound metalloprotease. However, CD4, another type I ectoprotein does not undergo such cleavage-secretion. In this study, we investigated the structural determinants of the ACE protein that regulate the cleavage-secretion process. Substitution and deletion mutations revealed that the cytoplasmic domain, the transmembrane domain, and the juxtamembrane region encompassing the major and the minor cleavage sites of ACE do not regulate its cleavage. Moreover, a chimeric protein containing the distal extracellular domain of CD4 and the juxtamembrane, transmembrane, and the cytoplasmic domains of ACE, although transported to the cell surface, was not cleavage-secreted. In contrast, the distal extracellular domain of ACE was shown to be the important determinant: a protein containing the distal extracellular domain of ACE and the juxtamembrane, transmembrane, and cytoplasmic domain of CD4 was efficiently cleaved off the cell surface. The chimeric protein was cleaved within the CD4 sequence and the responsible enzymatic activity was inhibited by Compound 3, a relatively specific inhibitor of the ACE secretase activity. These results demonstrate that, in a chimeric protein, the distal extracellular domain of a cleavable protein, such as ACE, can induce a proteolytic cleavage within the juxtamembrane domain of an uncleaved protein such as CD4.
Many proteins displayed on the surface of a mammalian cell are embedded in the plasma membrane via their hydrophobic transmembrane domains. Some of these ectoproteins undergo regulated proteolytic cleavage at sites near the plasma membrane, and the extracellular domain is released to the medium or circulation. This process has been variously called as solubilization, ectodomain shedding, or cleavage-secretion. Cleavage-secretion appears to be an important and widely used cellular posttranslational regulatory process because a variety of structurally and functionally unrelated cell-surface proteins undergo this process (1, 2). They include membrane-bound growth factors, cytokine and growth factor receptors, β-amyloid precursor protein that is implicated in Alzheimer’s disease, cell adhesion molecules, and enzymes such as angiotensin-converting enzyme (ACE). An appropriate balance between the membrane-anchored and the soluble forms of these proteins is thought to be necessary for their normal physiological role. A perturbation in that balance may lead to a diseased state as suggested for β-amyloid plaque formation in Alzheimer’s disease (3). Genetic studies have shown that the membrane-anchored form of kit ligand is required for normal mouse development (4). Similarly, inhibiting the formation of soluble tumor necrosis factor α (TNF-α) in mouse, without affecting the cell-bound form, inhibits the pathologies associated with this inflammatory cytokine (5–7).
In spite of the biological importance of this process, little is known about the identity of the responsible proteases, their mode of activation, and the structural determinants of the specific ectoproteins that make them susceptible to the cleavage secretion process.
We have been studying the characteristics of cleavage-secretion of ACE in vitro and in vivo (8–11). There are two structurally related isozymes of this enzyme, testicular ACE (ACET) and pulmonary ACE (ACEP) (12–15), that are involved in male fertility and blood pressure regulation (16). Studying the cleavage-secretion process of ACE is particularly significant because tissue-bound ACE and soluble ACE in circulation may have different physiological roles. Studies by us and others have shown that both ectoproteins, ACEP and ACET, undergo specific cleavage-secretion (8–11, 17–19). We have used extensively mouse and human cells in culture to study the cleavage-secretion of transfected rabbit ACET protein. The rabbit ACET protein has 737 residues of which the N-terminal 32 residues are cleaved off, as a signal peptide, during its synthesis. The ectodomain spans up to residue 690 followed by a transmembrane domain of 17 residues and an intracellular domain of 30 residues (12). The protein is cleaved off the cell surface in a regulated fashion. The major cleavage site is between Arg-663 and Ser-664. A minor alternative cleavage occurs between Arg-673 and Val-674 (9). The cleavage activity is resistant to inhibitors of serine, chymotrypsin, trypsin, cysteine, aspartate, and elastase type proteases but it is susceptible to Compound 3 (10, 11) an inhibitor of a specific class of metalloproteases (20). The protease activity was not lost from the plasma membrane by salt wash, indicating that it is carried out by an integral membrane protein (10). Similar to cleavage-secretion of many other ectoproteins the ACE-secretase activity was enhanced by treatment of cells with phorbol ester (9).
The current study was designed to identify the structural determinants in the ACET protein that are recognized by the cleavage-secretion machinery. The available information about other cleavage-secretion proteins suggests that the same rules do not apply to all of them. Although small deletions in the juxtamembrane domain abolished cleavage-secretion (21–25), mutations of residues around the cleavage sites of β-amyloid precursor protein (26), TNF-α receptor (24), interleukin 6 receptor (21), l-selectin (25), and TNF-α (22) did not reveal any strict sequence requirement for the cleavage process. For cleavage-secretion of human ACET in Chinese hamster ovary cells, Ehlers et al. (27) have shown that the sequence around the cleavage site or its specific distance from the plasma membrane are not important determinants. In contrast, Arribas et al. (28) reported that the juxtamembrane domains of pro-TNF-α and β-amyloid precursor protein are the crucial determinants that regulate ectodomain shedding. Here, we present experimental evidence to conclude that, for rabbit ACET, the distal extracellular domain, and not the cytoplasmic, the transmembrane, or the juxtamembrane domain containing the cleavage sites, is the critical determinant for efficient cleavage-secretion. When attached to a uncleaved ectoprotein CD4 (29), the ACET distal extracellular domain converted it to a chimeric protein that was cleavage-secreted very efficiently.
MATERIALS AND METHODS
Materials.
Compound 3 {N-[d,l-[2-(hydroxyaminocarbonyl)methyl]-4-methylpentanoyl]-l-3(tert-butyl)alanyl-l-alanine, 2-aminoethyl amide} was provided by Roy A. Black (Immunex Research and Development). Anti-FLAG M2 affinity gel and anti-FLAG antibody (FLAG-Probe D-8) were purchased from Eastman Kodak and Santa Cruz Biotechnology, respectively. Anti-CD4 antibodies, OKT4 and T4-4, were purchased from Ortho Diagnostics and the National Institutes of Health AIDS Research and Reference Reagent Program, respectively.
Construction of Expression Plasmids.
Rabbit ACET cDNA cloned in pGEM7Zf (8, 30) was the starting material for all constructs. Cytoplasmic deletion mutants (see Fig. 1) were generated by introducing translational stop codons after residue 729, 718, and 708. To generate the mutants with deletions in the cleavage region (see Fig. 2B), advantage was taken of the unique SfiI site at nt 1,748 in the ACET cDNA. Desired mutations were first introduced into the SfiI/BamHI (BamHI cuts in the plasmid) fragment, and the mutated fragments were subcloned into pGEM-ACET by using appropriate restriction sites. Five chimeric constructs (see Fig. 3) were generated that will encode for proteins containing portions of ACET and portions of human CD4 molecule. Human CD4 (29) was used as a template to generate the relevant CD4 fragments by PCR. Constructs containing the cytoplasmic, transmembrane, and increasing number of membrane proximal residues of CD4 were generated and ligated to the desired length of the extracellular domain of ACET. For CD4/ACET-1 (see Fig. 5), the unique AvaI site in CD4 cDNA was used to generate the CD4 fragment that was ligated to the appropriate ACET fragment with engineered AvaI site. The other chimeras (Table 1) were also constructed by using similar PCR technology.
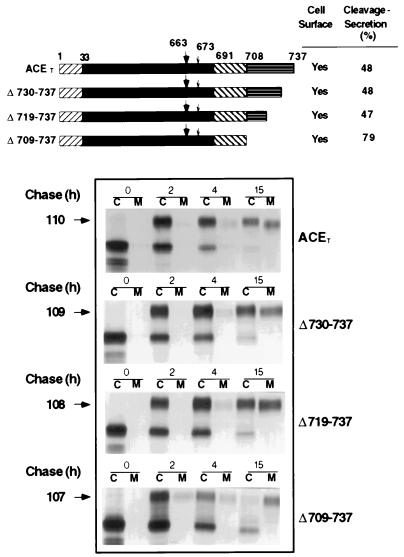
Figure 1.
Cleavage-secretion of cytoplasmic deletion mutants. (Upper) Schematic representation of ACET and the ACET mutants from which 8 residues (Δ730–737), 19 residues (Δ719–737), or 29 residues of the cytoplasmic domain (Δ709–737) have been deleted. The numbers above the bar indicate ACET residue numbers as described by Kumar et al. (12). ▨, cleaved signal sequence, residues 1–32; ▪, extracellular domain, residues 33–690; ▧, membrane anchoring domain, residues 691–707; ▤, cytoplasmic domain, residue 708–737. The large (▾‖) and small (↓) arrows indicate the position of the major and the minor cleavage site in ACET, respectively (9). (Lower) The synthesis, intracellular processing, and cleavage-secretion of the ACET proteins in HeLa cells. HeLa cells, infected with the recombinant vaccinia virus expressing T7 RNA polymerase, were transiently transfected with wild-type or mutant ACET-expression vectors. The cells were pulse-labeled with [35S]methionine for 30 min followed by incubation without the labels for different periods of time as indicated by chase (h). Labeled detergent lysates of cells (C) and medium (M) were immunoprecipitated with ACET antibody and analyzed by SDS/PAGE. Arrows on the left indicate the position of the mature glycosylated ACET proteins and the numbers correspond to their estimated molecular weights. Cleavage-secretion (top panel) is estimated by PhosphorImager analysis of the dried gels. The amount (arbitrary PhosphorImager units) of mature ACET proteins in cell extract and medium combined at 15 h is taken as 100%. Cell surface expression of ACET or the mutant proteins was assessed by indirect immunofluorescence analysis as described.
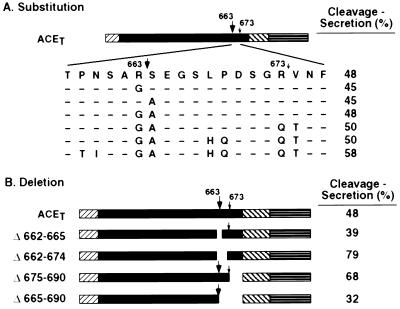
Figure 2.
Influence of substitution and deletion mutagenesis at and around the cleavage sites of ACET on cleavage-secretion. (A) Schematic representation of point mutations at and around the cleavage site in ACET. The amino acid sequence and the numberings are for the cleavage domain of wild-type ACET (12). Sequence of the cleavage region of different mutants is indicated below this with identical amino acids indicated by −, and changed amino acids are indicated by the single letter code. All mutants were expressed in HeLa cells, analyzed by pulse–chase experiments (data not shown), and cleavage-secretion was quantitated by PhosphorImager analysis (numbers on the right) as described in the legend of Fig. 1. (B) Schematic representation of the wild-type ACET and the deletion mutants. Δ662–665 indicates an in-frame deletion of four amino acids, A662 to E665, which includes the major cleavage site. Δ662–674 indicates a deletion of 13 amino acids that encompass both major and the minor cleavage sites. Δ675–690 and Δ665–690 indicate deletion of 16 or 26 residues of the membrane proximal region of the ectodomain that includes none or only the minor cleavage site respectively. The mutant proteins were expressed in HeLa cells (data not shown), and cleavage-secretion was quantitated as described above.
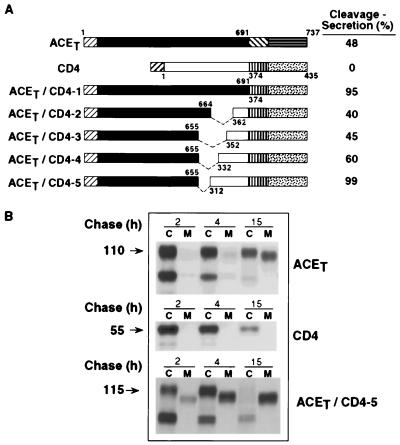
Figure 3.
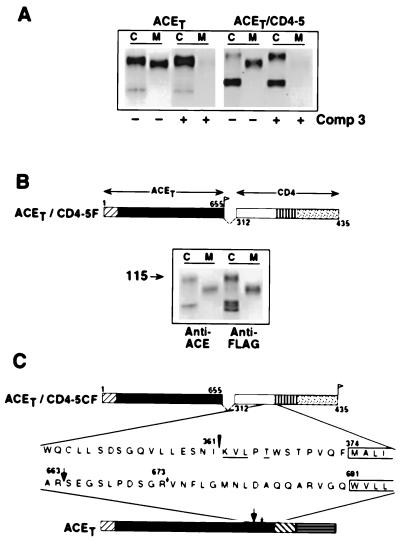
Cleavage-secretion of ACET, CD4, and ACET/CD4-chimeric molecules. (A) Schematic representation of ACET, CD4, and the five chimeric molecules containing the cytoplasmic, transmembrane, and increasing lengths of the membrane proximal region of the extracellular domain of CD4 molecule joined with the extracellular domain of ACET. Numbers above and below the bars indicate ACET or CD4 residue numbers, respectively. The top bar represents the 737-amino acid polypeptide chain of ACET. The numberings start from the signal sequence (12) and amino acid 691 indicates the beginning of the predicted transmembrane domain. ▨, cleaved signal sequence of CD4 molecule; □, extracellular domain; ▥, membrane anchoring domain; and ░⃞, cytoplasmic domain of CD4. Amino acids in the CD4 molecule are numbered according to Maddon et al. (29). Thus, 1 corresponds to the first amino acid after the cleaved signal sequence. Residues 374 and 435 mark the beginning of the membrane anchoring domain and the C-terminal amino acid of CD4, respectively. The deleted membrane proximal sequences of the ACET extracellular domain are indicated by ---. (B) Synthesis and cleavage secretion in HeLa cells. ACET, CD4, and ACET/CD4-5 were transiently expressed in HeLa cells and pulse–chase analysis performed as described in the legend of Fig. 1. Cell lysates (C) and media (M) were immunoprecipitated by using anti-ACE antibody (ACET and ACET/CD4-5 proteins) or anti-CD4 antibody (CD4 proteins) and analyzed by SDS/PAGE. Arrows on the left indicate the position of mature proteins in the cell extract and the numbers correspond to their estimated molecular weight.
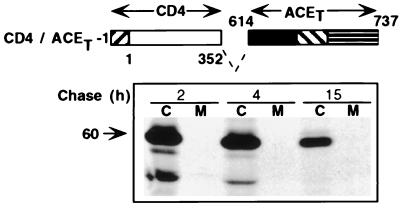
Figure 5.
Lack of cleavage-secretion of CD4/ACET-1. (Upper) Schematic representation of CD4/ACET-1 containing the distal extracellular domain of CD4 (residues 1–352) and proximal extracellular, transmembrane, and cytoplasmic domains of ACET (residues 614–737). Symbols for different domains and residue numberings are as described in the legend of Fig. 3. (Lower) Pulse–chase analysis of synthesis and cleavage-secretion of CD4/ACET-1 in HeLa cells.
Table 1.
ACET-CD4 chimeric proteins not transported to the cell-surface
| N-terminal region | C-terminal region |
|---|---|
| ACET 1–342 | CD4352–435 |
| ACET 1–426 | CD4352–435 |
| ACET 1–536 | CD4352–435 |
| CD41–97 | ACET 343–737 |
| CD41–187 | ACET 427–737 |
| CD41–277 | ACET 537–737 |
Absence of surface immunofluorescence was taken as an index for lack of transport to the cell surface.
For adding the FLAG epitope (DYKDDDDK) to the construct ACET/CD4-5, double-stranded oligonucleotide encoding the FLAG epitope was prepared by PCR by using pairs of primers designed to encode residues upstream of the SfiI site and the FLAG epitope with a BamHI site engineered in it. The amplified products were digested with SfiI and BamHI and ligated to similarly digested ACET/CD4-5. Thus ACET/CD4-5F (see Fig. 6B) will encode for a protein having residues 1–655 of ACET followed by the FLAG epitope at the junction and CD4 residues 312–435. ACET/CD4-5CF, which has the FLAG epitope at the C terminal, was generated by utilizing a primer that encodes for the FLAG epitope followed by a stop codon.
Figure 6.
Characteristics of ACET/CD4-5 cleavage-secretion. (A) Compound 3 blocks the process. Transfected HeLa cells were pulse-labeled for 30 min and chased for 4 h with (+) or without (−) the addition of 50 μM Compound 3 in the culture medium. Immunoprecipitation with anti-ACE antibody and SDS/PAGE was performed as described elsewhere. (B) ACET/CD4-5 is cleaved within the CD4 domain. Schematic representation of ACET/CD4-5F that is similar to ACET/CD4-5 chimera (see Fig. 3) with a FLAG epitope introduced at the junction of ACET and the CD4 sequences. Following pulse–chase analysis for 4 h the cell lysates and the culture medium were divided into two parts and immunoprecipitated with either anti-ACE antibody or anti-FLAG affinity gel as indicated. (C) Cleavage site in ACET/CD4-5. The top bar schematically represents ACET/CD4-5CF, which is similar to ACET/CD4-5 chimera with a FLAG epitope introduced at the C terminal. The bottom bar is a schematic illustration of wild-type ACET, with arrows representing the major (▾‖) and the minor (↓) cleavage site; the putative transmembrane domains (only partial sequence shown) are boxed. The amino acid sequences of the indicated regions are expanded and aligned from the respective membrane anchoring domain for comparison. The N-terminal residues of the purified C-terminal tail of ACET/CD4-5CF determined by sequence analysis is underlined; the inverted triangle (▾) and the number indicate the position of the determined cleavage site.
The point mutants described in Fig. 2A were generated as described before (30). R-663 was mutated to G by changing the codon CGC to GGC; S664 to A by changing TCG to GCG; R673 to Q by changing CGC to CAA; V674 to T by changing GTC to ACC; L668 to H by changing CTC to CAC; P669 to Q by changing CCA to CAA; P659 to T by changing CCA to ACA; and N660 to I by changing AAC to ATC. All constructs were verified by restriction mapping and sequencing of the entire PCR-amplified fragments. The sequences of all oligonucleotide primers used are available from the authors on request.
Transient Expression of ACET and Chimeric Proteins, Pulse–chase Analysis, and Immunoprecipitation.
ACET and chimeric proteins were transiently expressed in HeLa cells by using the vaccinia virus T7RNA polymerase system as described (30). The transfected cells were pulse-labeled with [35S]methionine for 30 min, and the label was chased for the indicated time. ACET-related proteins were immunoprecipitated from the cell extracts and medium by using anti-ACE or anti-CD4 (T4-4, for CD4/ACET-1 proteins) antibody and analyzed by SDS/PAGE, as described (30). The FLAG-tagged chimeric proteins were immunoprecipitated by using anti-FLAG M2 affinity gel according to manufacturers instructions. For quantitating cleavage-secretion, the dried gels were subjected to PhosphorImager (Molecular Dynamics) analysis by using Image Quant software. The amount of mature ACET or chimeric proteins (measured in arbitrary PhosphorImager units) present in the cell extract and medium combined after 15 h of chase, is taken as 100%, and the amount present in the culture medium at that time is represented as cleavage-secreted. The intra- and interassay variability for PhosphorImager analysis was 2–3% and 4–5%, respectively. Secretion measured by PhosphorImager analysis correlates very well with that measured by ACE enzyme activity assay. Transfection and pulse–chase analysis of each construct was repeated three times.
Purification and Amino Terminal Sequencing of the Cleaved C-Terminal Peptide of ACET/CD4–5.
HeLa cells (48 × 106), transiently transfected with ACET/CD4–5CF, were harvested 20 h after transfection and suspended in 2 ml of 50 mM Tris⋅HCl buffer (pH 8.0) and 1 mM phenylmethylsulfonyl fluoride. After three cycles of rapid freezing, and thawing at 37°C, the suspension was homogenized in glass–glass homogenizer and centrifuged at 25,000 × g for 1 h. The pellet was resuspended in 1 ml of homogenization buffer containing 150 mM NaCl and 1% Triton X-100. After 1 h on ice, the samples were similarly centrifuged and the supernatant containing the solubilized C-terminal peptide was added to 300 μl of an anti-FLAG M2 affinity gel, mixed for 16 h at 4°C, centrifuged, washed with the homogenization buffer, and eluted with 0.1 M glycine (pH 3.5). The eluate was immediately adjusted to pH 7 by 1 M Tris⋅HCl (pH 8.0), concentrated, and analyzed by 20% SDS/PAGE, electroblotted onto ProBlott membranes (Applied Biosystems), and detected by staining with Coomassie brilliant blue as described (9). A duplicate membrane was subjected to Western blot analysis by using anti-FLAG antibody, which was detected by an enhanced chemiluminescence detection method (Amersham). The Coomassie brilliant blue-stained peptide band, corresponding to the C-terminal peptide detected by Western blot analysis, was cut out and used for N-terminal sequence analysis after Edman degradation by using Procise, model 492 protein sequencer (Applied Biosystems) attached to 140C microgradient system and 610A Version 2.1 data analysis system.
Immunodetection of ACET, CD4, ACET/CD4-5, and CD4/ACET-1 in Transfected HeLa Cells by Indirect Immunofluorescence.
HeLa cells grown on glass coverslips were transfected with appropriate constructs, fixed, permeabilized for internal staining, and treated with anti-ACE antibody (for ACET and ACET/CD4-5 transfected cells) or anti-CD4 antibody (OKT4, for CD4 and CD4/ACET-1 transfected cells), and fluorescein conjugated appropriate IgGs as described (31).
RESULTS
Effects of Mutations in the Cytoplasmic and the Juxtamembrane Domains of ACET on its Cleavage-Secretion.
Although cleavage of ACET occurs at extracellular sites in the juxtamembrane domain of the protein, the cytoplasmic tail could influence this process by helping to recruit specific proteases to the site of cleavage. This possibility was tested by introducing progressive deletions to the cytoplasmic tail of ACET (Fig. 1). Residues 708–737 of the ACET are thought to constitute the intracellular domain of the protein. Three deletion mutants missing 8, 19, and 29 residues from the C terminus were expressed in HeLa cells by using the transient vaccinia virus system described before (30). All these mutant proteins were transported to the plasma membrane as judged by cell surface immunofluorescence (data not shown). They were also cleavage-secreted efficiently. The cleavage-secretion rates of two mutants were similar to that of the wild-type protein, whereas the protein missing all of the intracellular domain was cleaved even more efficiently. That the ACET proteins in the medium were indeed cleaved-off was confirmed by the absence of the hydrophobic transmembrane domain as judged by detergent-partitioning test (10). These results demonstrate that the cytoplasmic domain of the ACET protein is not required for the cleavage-secretion process.
In the next series of experiments we investigated the nature of mutations that can be tolerated at or around the cleavage sites in the extracellular juxtamembrane domain of ACET. The major site of ACET cleavage is between residues 663 and 664. We carried out various substitution mutations at the two cleavage sites to examine if the nature of the flanking residues influence the process. All substitution mutants were cleaved as efficiently as the wild-type protein (Fig. 2A). In the most mutated protein, 8 of 16 residues in this region were substituted without any effect. These results suggest that either the specific mutations introduced in our mutants can be tolerated by the cleavage-secretion machinery or the specific sequence at or near the cleavage sites are not important. The latter possibility was tested in the next experiments described below.
Several deletion mutants carrying specific deletions around the two natural cleavage sites were tested for cleavage-secretion (Fig. 2B). Elimination of the major cleavage site (Δ662–665) or of both major and minor cleavage sites (Δ662–674) did not affect the process. Similarly, deletions of the region between the minor cleavage site and the transmembrane domain (Δ675–690) and the region between the major cleavage site and the transmembrane domain (Δ665–690) were also without any effect. These results strongly suggested that the specific sequences present in the juxtamembrane domain encompassing the two cleavage sites are unimportant for the cleavage-secretion of ACET.
Cleavage-Secretion of ACET/CD4 Chimeras.
Because the cytoplasmic and the juxtamembrane domains of the ACET protein appeared to be unimportant for the cleavage-secretion process, we hypothesized that its distal extracellular region may be influencing this process. To test this hypothesis, we constructed chimeric proteins containing portions of ACET proteins and portions of CD4 protein. Like ACET, CD4 is a type I ectoprotein expressed on the cell surface. However, unlike ACET, CD4 is not cleavage-secreted. The first chimera tested, ACET/CD4-1, contained the transmembrane and the intracellular domains of CD4 (Fig. 3). ACET/CD4-1 was cleavage-secreted efficiently (data not shown), thus demonstrating that the transmembrane and the intracellular domains of ACET are irrelevant for the process. A series of additional chimeras, containing increasing portions of the CD4 protein and the distal extracellular region of ACET, were constructed. All of these chimeric proteins were enzymatically active (data not shown), transported to the cell surface and cleavage secreted (Fig. 3A). ACET/CD4-5 contained 62 residues of the juxtamembrane domain of CD4 in addition to its transmembrane and cytoplasmic domains. It also contained 655 residues of the distal extracellular domain of ACET. This protein was cleavage-secreted extremely efficiently. Almost all of the protein was in the culture medium 15 h after its synthesis (Fig. 3B). That the secreted protein was a cleavage product was apparent from its lower molecular weights. In contrast, CD4 was not secreted at all and about 50% of the ACET protein was cleavage-secreted after 15 h.
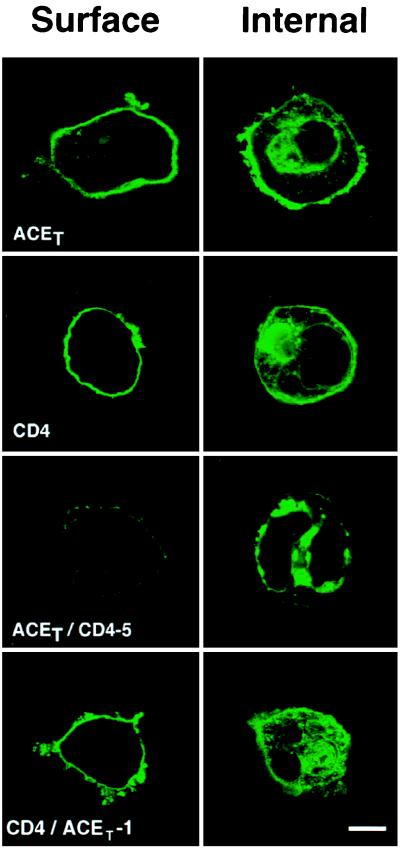
The above conclusions drawn from pulse–chase experiments were confirmed by immunofluorescence experiments shown in Fig. 4. ACET, CD4, and ACET/CD4-5 proteins were expressed in HeLa cells and the presence of the expressed protein on its cell surface and inside the cells was examined by indirect immunofluorescence by using appropriate antibodies. All these proteins could be detected intracellularly, but only CD4 and ACET were detected on the cell surface. Little ACET/CD4-5 was on the plasma membrane presumably because it was cleaved off the surface very efficiently. The results demonstrated that the distal extracellular domain of ACET could regulate cleavage-secretion of a chimeric protein.
Figure 4.
Cell surface expression of ACET, CD4, ACET/CD4-5, and CD4/ACET-1. HeLa cells transfected with ACET, CD4, ACET/CD4-5F, or CD4/ACET-1 expression vectors were processed for indirect immunofluorescence analysis as described. Anti-ACE or anti-CD4 antibody and appropriate fluorescein conjugated IgGs were used to detect the proteins expressed intracellularly (Internal) or on the cell surface (Surface). (Bar = 10 μm.)
In the above constructs, 655 residues of ACET were present. Our attempts to perform further deletions of the ACE extracellular domain were ineffective because the chimeric proteins containing 342, 426, or 536 ACET residues and residues 352–435 of CD4 were not transported to the cell surface and consequently their cleavage-secretion could not be tested (Table 1). The same was true for three other proteins containing various N-terminal regions of the CD4 protein and C-terminal regions of ACET (Table 1). Similarly, a series of ACET derivatives carrying deletions from residue 35 to residues 79, 342, 426, or 536 was also not transported to the plasma membrane (data not shown). These proteins were arrested in the endoplasmic reticulum, as judged by their prolonged endoglycosidase H-sensitivity, and eventually degraded (data not shown).
The conclusion that the distal extracellular domain of ACET, and none of its other domains, dictates its cleavage-secretion process, was confirmed by the results of the next experiment. CD4/ACET-1, a chimeric protein containing the distal extracellular domain of CD4 (residues 1–352) and the juxtamembrane, transmembrane, and cytoplasmic domains of ACET (residues 614–737) was not cleavage-secreted as shown by the lack of appearance of this protein in the culture medium (Fig. 5). However, this chimeric protein was transported to the cell surface as efficiently as CD4 (Fig. 4 Lower).
Characteristics of ACET/CD4-5 Cleavage-Secretion.
The membrane associated protease activity responsible for ACET cleavage-secretion has been partially characterized (10). One of its characteristics is its susceptibility to Compound 3, an inhibitor of a specific class of metalloproteases. In the experiment shown in Fig. 6A, we examined the properties of the protease activity responsible for cleaving the chimeric protein, ACET/CD4-5. Compound 3 inhibited cleavage-secretion of both ACET and ACET/CD4-5 completely, thus indicating that the same protease is responsible for cleaving both proteins.
The results reported above indicated that the distal extracellular domain of ACET is capable of directing cleavage-secretion of a chimeric protein such as ACET/CD4-5. The size of the secreted derivative of ACET/CD4-5 suggested that the cleavage was taking place within the CD4 juxtamembrane domain. Whether that was indeed true, was tested in the experiment shown in Fig. 6B. We designed a construct, ACET/CD4-5F, which contained a FLAG epitope at the junction of the ACET and the CD4 sequences. Cell-bound and secreted proteins were analyzed by both anti-ACET and anti-FLAG antibodies. The secretion product was immunoprecipitated with either antibody, thus establishing that the cleavage had occurred in the CD4 region and not in the ACET region.
In the next experiment, the exact cleavage site was determined by protein sequencing (Fig. 6C). For this purpose, a derivative of ACET/CD4-5 protein was designed. It contained the FLAG epitope at the C terminus. After expression and cleavage-secretion of ACET/CD4-5CF, the cell-bound C-terminal fragment was purified by affinity chromatography on anti-FLAG affinity gel and the purified protein was subjected to N-terminal sequencing. The sequence obtained was KVLXT which corresponds to residues 362–366 of CD4. Thus, the chimeric protein had been cleaved between residues 361 and 362 of CD4. The cleavage site was 12 residues upstream from the transmembrane domain and 49 residues downstream from the ACET-CD4 junction. For comparison, the major cleavage site in ACET is 27 residues upstream from the transmembrane domain (Fig. 6C).
DISCUSSION
In this study, we sought to identify the structural determinants of the ACET protein that regulates its cleavage-secretion. In the past, such determinants have not been clearly defined for any protein that undergoes cleavage-secretion, although studies from several laboratories indicated that there are clear distinctions among different ectoproteins with respect to the specific domains that regulate their cleavage-secretion. We have been studying the characteristics of cleavage-secretion of rabbit ACET (8–10) and ACEP (11) in vitro and in vivo. These studies have shown that ACET expressed in mouse or human cells undergo specific and regulated cleavage secretion (9). The extracellular domain is cleaved off by a membrane-associated metalloprotease activity that is inhibited by Compound 3 but not by a number of other inhibitors of specific proteases (9–11). A major and a minor site of cleavage in the juxtamembrane domain of ACET have been identified (9). It was also shown that ACET expressed in the yeast is cleaved at the same site by a similar secretase (32).
Results presented here clearly showed that the cytoplasmic domain of ACET is not required for its cleavage-secretion. In this respect, ACET is similar to the kit ligand, interleukin 6 receptor, and β-amyloid precursor protein (21, 33, 34). Moreover, deletion of the cytoplasmic domain (Fig. 1) or exchanging the cytoplasmic and the transmembrane domain with the corresponding domains of CD4 (Fig. 3) resulted in increased secretion, indicating that these domains might slow down the process of cleavage-secretion. In contrast, the cytoplasmic domains of pro-transforming growth factor α and the TNF-α receptor are needed for their cleavage-secretion (35, 36). We observed that juxtamembrane domain of ACET, which contains both the major and the minor cleavage sites, is also dispensable for the secretion process (Fig. 2B). Substitution mutations of the residues at or around the cleavage site could not block the cleavage process indicating a lack of sequence specificity at the cleavage site (Fig. 2A). Similar observations have also been made for β-amyloid precursor protein (26), TNF-α receptor (24), interleukin 6 receptor (21), TNF-α (22), and l-selectin (25). Total deletion of the juxtamembrane region containing the two cleavage sites did not affect ACET cleavage-secretion. These results are consistent with the observations made by Ehlers et al. (27) who studied cleavage-secretion of human ACET in Chinese hamster ovary cells. They concluded that the cleavage activity present in those cells was not constrained by a specific cleavage site motif or by a specific distance from the transmembrane domain. Thus, cleavage-secretion of rabbit ACET in human HeLa cells had the same properties as cleavage-secretion of human ACET in Chinese hamster ovary cells as far as the role of the juxtamembrane was concerned. However, this is not true for pro-transforming growth factor α or β-amyloid precursor protein. Arribas et al. (28) have reported that the short juxtamembrane region of the two proteins are the primary determinants of their cleavage-secretion. These domains, when transplanted to a noncleaved ectoprotein, rendered it susceptible to cleavage-secretion.
The most significant observations in the current study came from the experiments done with ACET/CD4 chimeric proteins. Both ACET and CD4 are type I ectoproteins that are anchored in the plasma membrane and displayed on the cell surface. However, unlike ACET, CD4 is not cleavage-secreted. Although this cell-surface protein is internalized and recycled, it is not shed into the extracellular medium by cleavage from the cell surface. Results reported in this paper clearly showed that in the context of a chimeric protein, CD4 can be cleavage-secreted very efficiently. When the distal extracellular domain of CD4 was replaced by the corresponding region of ACET, the chimeric protein was cleaved off the cell surface almost completely. The cleavage occurred exclusively in the CD4 juxtamembrane region, and the responsible protease had the same characteristics as the ACET cleavage-secretase. Thus, we succeeded in transferring the property of a cleavable protein, ACET, to an uncleaved protein, CD4, by swapping the latter’s distal extracellular domain with the corresponding domain of the former protein.
In contrast to the above chimeras, a chimeric protein consisting of the distal extracellular domain of CD4 and the C-terminal one-sixth of ACET was not cleavage-secreted (Fig. 5), although it contained the proximal extracellular domain of ACET (residues 614–690). These results strongly suggest that the distal extracellular domain of ACET is the primary determinant of inducing its cleavage-secretion. Future experiments will be required to explore further the nature of the minimal determinant present within that domain. As mentioned in Results, our efforts in that direction was hampered by the fact that many chimeric proteins failed to be transported to the plasma membrane. This experimental limitation has been encountered frequently by other workers who attempted to express different chimeric proteins on the cell surface for various experimental needs. The nature of the protein primary structure, which dictates its successful transport to the plasma membrane, is not yet fully understood. Thus, it is not possible to rationally design chimeric proteins that will appear on the cell surface; they have to be empirically tested. In another ectoprotein, amyloid precursor protein, the alternatively spliced Kuniz protease inhibitor domain, which is part of its distal extracellular domain, was also shown to influence its cleavage-secretion (37).
Given the limited information available currently, it is tempting to speculate that the distal extracellular domain of ACET is capable of obtaining a specific structure that folds back and initiate a proteolytic cleavage process at the plasma membrane. Once activated, this secretase activity seems not to care about the specific sequence at or around the cleavage site, nor is it very specific about the distance of the cleavage site from the plasma membrane. These observations suggest that the crucial element that determines whether a specific protein will be cleaved or not are dictated by the higher order structure of the protein and they cannot be readily predicted from the primary sequence of the protein.
Acknowledgments
We are grateful to Roy A. Black (Immunex Research and Development) for providing Compound 3. This work was supported in part by the National Institutes of Health Grants HL-48258 and HL-54297 and by a fellowship from the American Heart Association, Northeast Ohio Affiliate.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: ACE, angiotensin-converting enzyme; ACET, testicular ACE; ACEP, pulmonary ACE; TNF-α, tumor necrosis factor α; Compound 3, N-[d,l-[2-(hydroxyaminocarbonyl)methyl]-4-methylpentanoyl]-l-3(tert-butyl)alanyl-l-alanine, 2-aminoethyl amide.
References
- 1.Ehlers M R W, Riordan J F. Bochemistry. 1991;30:10065–10074. doi: 10.1021/bi00106a001. [DOI] [PubMed] [Google Scholar]
- 2.Massagué J, Pandiella A. Annu Rev Biochem. 1993;62:515–541. doi: 10.1146/annurev.bi.62.070193.002503. [DOI] [PubMed] [Google Scholar]
- 3.Haass C, Selkoe D J. Cell. 1993;75:1039–1042. doi: 10.1016/0092-8674(93)90312-e. [DOI] [PubMed] [Google Scholar]
- 4.Flanagan J G, Chen D C, Leder P. Cell. 1991;64:1025–1035. doi: 10.1016/0092-8674(91)90326-t. [DOI] [PubMed] [Google Scholar]
- 5.Mohler K M, Sleath P R, Fitzner J N, Cerretti D P, Alderson M, Kerwar S S, Torrance D S, Otten-Evans C, Greenstreet T, Weerawarna K, Kronheim S R, Petersen M, Gerhart M, Kozlosky C J, March C J, Black R A. Nature (London) 1994;370:218–220. doi: 10.1038/370218a0. [DOI] [PubMed] [Google Scholar]
- 6.McGeehan G M, Becherer J D, Bast R C, Jr, Boyer C M, Champion B, Connolly K M, Conway J G, Furdon P, Karp S, Kidao S, McElroy A B, Nichols J, Pryzwansky M, Schoenen F, Sekut L, Truesdale A, Verghese M, Warner J, Ways J P. Nature (London) 1994;370:558–561. doi: 10.1038/370558a0. [DOI] [PubMed] [Google Scholar]
- 7.Gearing A J H, Beckett P, Christodoulou M, Churchill M, Clements J, et al. Nature (London) 1994;370:555–557. doi: 10.1038/370555a0. [DOI] [PubMed] [Google Scholar]
- 8.Sen I, Samanta H, Livingston W, III, Sen G C. J Biol Chem. 1991;266:21985–21990. [PubMed] [Google Scholar]
- 9.Ramchandran R, Sen G C, Misono K, Sen I. J Biol Chem. 1994;269:2125–2130. [PubMed] [Google Scholar]
- 10.Ramchandran R, Sen I. Biochemistry. 1995;34:12645–12652. doi: 10.1021/bi00039a021. [DOI] [PubMed] [Google Scholar]
- 11.Ramchandran R, Kasturi S, Douglas J G, Sen I. Am J Physiol. 1996;271:H744–H751. doi: 10.1152/ajpheart.1996.271.2.H744. [DOI] [PubMed] [Google Scholar]
- 12.Kumar R S, Thekkumkara T J, Sen G C. J Biol Chem. 1991;266:3854–3862. [PubMed] [Google Scholar]
- 13.Thekkumkara T J, Livingston W, III, Kumar R, Sen G C. Nucleic Acids Res. 1992;20:683–687. doi: 10.1093/nar/20.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howard T E, Shai S-Y, Langford K G, Martin B M, Bernstein K E. Mol Cell Biol. 1990;8:4294–4302. doi: 10.1128/mcb.10.8.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hubert C, Houot A M, Corvol P, Soubrier F. J Biol Chem. 1991;266:15377–15383. [PubMed] [Google Scholar]
- 16.Krege J H, John S W M, Langenbach L L, Hodgin J B, Hagaman J R, Bachman E S, Jennette J C, O’Brien D A, Smithies O. Nature (London) 1995;375:146–148. doi: 10.1038/375146a0. [DOI] [PubMed] [Google Scholar]
- 17.Wei L, Alhenc-Gelas F, Soubrier F, Michaud A, Corvol P, Clauser E. J Biol Chem. 1991;266:5540–5546. [PubMed] [Google Scholar]
- 18.Ehlers M R W, Chen P Y-N, Riordan J F. Proc Natl Acad Sci USA. 1991;88:1009–1013. doi: 10.1073/pnas.88.3.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beldent V, Michaud A, Wei L, Chauvet M T, Corvol P. J Biol Chem. 1993;268:26428–26434. [PubMed] [Google Scholar]
- 20.Black R A, Rauch C T, Kozlosky C J, Peschon J J, Slack J L, et al. Nature (London) 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 21.Müllberg J, Oberthür W, Lottspeich F, Mehl E, Dittrich E, Graeve L, Heinrich P C, Rose-John S. J Immunol. 1994;153:4958–4968. [PubMed] [Google Scholar]
- 22.Tang P, Hung M C, Klostergaard J. Biochemistry. 1996;35:8226–8233. doi: 10.1021/bi952183l. [DOI] [PubMed] [Google Scholar]
- 23.Deng P, Rettenmier C W, Pattengale P K. J Biol Chem. 1996;271:16338–16343. doi: 10.1074/jbc.271.27.16338. [DOI] [PubMed] [Google Scholar]
- 24.Brakebusch C, Varfolomeev E E, Batkin M, Wallach D. J Biol Chem. 1994;269:32488–32496. [PubMed] [Google Scholar]
- 25.Migaki G I, Kahn J, Kishimoto T K. J Exp Med. 1995;182:549–557. doi: 10.1084/jem.182.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sisodia S S. Proc Natl Acad Sci USA. 1992;89:6075–6079. doi: 10.1073/pnas.89.13.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ehlers M R W, Schwager S L V, Scholle R R, Manji G A, Brandt W F, Riordan J F. Biochemistry. 1996;35:9549–9559. doi: 10.1021/bi9602425. [DOI] [PubMed] [Google Scholar]
- 28.Arribas J, López-Casillas F, Massagué J. J Biol Chem. 1997;272:17160–17165. doi: 10.1074/jbc.272.27.17160. [DOI] [PubMed] [Google Scholar]
- 29.Maddon P J, Littman D R, Godfrey M, Maddon D E, Chess L, Axel R. Cell. 1985;42:93–104. doi: 10.1016/s0092-8674(85)80105-7. [DOI] [PubMed] [Google Scholar]
- 30.Sen I, Kasturi S, Jabbar M A, Sen G C. J Biol Chem. 1993;268:25748–25754. [PubMed] [Google Scholar]
- 31.Sadhukhan R, Sen I. J Biol Chem. 1996;271:6429–6434. doi: 10.1074/jbc.271.11.6429. [DOI] [PubMed] [Google Scholar]
- 32.Sadhukhan R, Sen G C, Sen I. J Biol Chem. 1996;271:18310–18313. doi: 10.1074/jbc.271.31.18310. [DOI] [PubMed] [Google Scholar]
- 33.Cheng H J, Flanagan J G. Mol Biol Cell. 1994;5:943–553. doi: 10.1091/mbc.5.9.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.da Cruz e Silva O A, Iverfeldt K, Oltersdorf T, Sinha S, Lieberburg I, Ramabhadran T V, Suzuki T, Sisodia S S, Gandy S, Greengard P. Neuroscience. 1993;57:873–877. doi: 10.1016/0306-4522(93)90031-a. [DOI] [PubMed] [Google Scholar]
- 35.Crowe P, VanArsdale T L, Goodwin R G, Ware C F. J Immunol. 1993;151:6882–6890. [PubMed] [Google Scholar]
- 36.Bosenberg M W, Pandiella A, Massagué J. Cell. 1992;71:1157–1165. doi: 10.1016/s0092-8674(05)80064-9. [DOI] [PubMed] [Google Scholar]
- 37.Ho L, Fukuchi K-i, Younkin S G. J Biol Chem. 1996;271:30929–30934. doi: 10.1074/jbc.271.48.30929. [DOI] [PubMed] [Google Scholar]