The perceived phobia (fear) of some apolar substances for aqueous (hydro) environments does not imply lack of attraction to water. Rather, it originates from the strength of this attraction, which, similar to that between the constituents of apolar substances, is smaller than the force between water molecules. Fear of those who dominate the scene (in this case, water hydrogen bonds) is hardly surprising. The concept of hydrophobicity, intuitively associated with the demixing of oil and water, appears in most biophysics and biochemistry textbooks, and it is generally accepted that hydrophobic interactions are a major driving force of fundamental biological processes, for example, protein folding, molecular recognition, and the formation of membranes (1,2). Yet, the molecular theory of the hydrophobic effect and the microscopic understanding of the hydration of hydrophobic species (or hydrophobic hydration), are incomplete. The article by Li et al. (3) in this issue of PNAS presents the first investigation of hydrophobic association for a model system (two methane molecules in water) entirely based on first principles, i.e., on a quantum mechanical description of electronic and electron–ion interactions (4) of the combined solute–solvent system. The potential of mean force between the two methane molecules computed in ref. 3 is consistent with the data extracted from several hydrocarbon solubility experiments, but it is substantially different from what is found in simulations based on empirical classical force fields. These simulations represent the majority of computer simulations carried out in the last 30 years to investigate hydrophobic effects at the microscopic level. The results of ref. 3 indicate that these empirical simulations may lead to an underestimation of the hydrophobic effect in many instances, and they point to the need for an accurate, ab initio description of hydrophobic interactions, capable of accounting for subtle, yet key, interfacial effects between the solute and water. Similar conclusions on the discrepancy between empirical potentials and ab initio descriptions have also been reported by Grossman et al. (5) and Allesch et al. (6), who studied isolated methane and benzene molecules in water, respectively.
The first use of the hydrophobic concept dates back (7) to I. Traube, who, in 1891, noted that many organic solutes absorbed at a water–air interface have polar ends in the solvent but nonpolar ends “repelled” by water and sticking out. This type of interface was the subject of intense studies by I. Langmuir, and, in 1938, he was probably the first scientist to discuss the possible relationship between hydrophobicity and protein structures (8). However, it was only in 1959 that W. Kautzmann (9) clearly identified hydrophobic interactions as a primary source of protein stability. It was recognized very early that the antipathy between oily substances and water is a consequence of the great strength of water hydrogen bonds. As clearly stated by D. Chandler (10), “oil and water molecules actually attract each other, but not nearly as strongly as water attracts itself.” Quantifying this concept in different situations has been difficult, however, because hydrophobicity manifests itself in different manners (e.g., from a thermodynamics standpoint), depending on whether a small solute, such as the methane molecules studied by Li et al. (3), or an extended hydrophobic surface is in contact with water (11, 12). Indeed, reordering of water hydrogen bonds (thus entropic effects) is mainly responsible for the behavior of solvation-free energies as a function of temperature and pressure in the small solute limit, whereas it is the breaking of these bonds (thus enthalpic effects) that plays a key role for large solutes. Interestingly, the small solute limit study of ref. 3 highlights the importance of a volume entropy term in distinguishing commonly used potential mean effective forces from the potential of mean force. Whether in the case of mere reordering or of breaking of hydrogen bonds, the ability to understand the nature of hydrophobic interfaces at the microscopic level is crucial for building a molecular theory of hydrophobicity. The work of Li et al. (3) shows that unraveling the properties of these interfaces requires an accurate description of electronic and structural properties of the combined solute–solvent system, and this description may not be provided by potentials fitted to water bulk properties. In addition, accurate simulations give insight into the spatial distribution functions of water around the solute (see Fig. 1) and allow one to understand in detail both steric and caging effects.
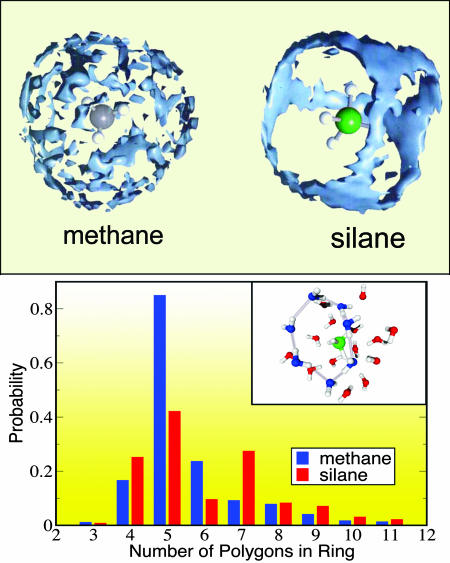
Fig. 1.
(Upper) The spatial distribution functions of water molecules around two hydrophobic solutes of different size, methane and silane, as obtained by using ab initio molecular dynamics simulations. (Lower) Distribution of hydrogen bond pathways for the first solvation shell of the two solutes. Although the water molecules surrounding these two small hydrophobic solutes are oriented in a similar fashion, the water spatial distribution function is determined by steric effects (modified from ref. 5).
In recent years, the progress in molecular statistical thermodynamic theories of the hydrophobic effect (1) has been nicely complemented by atomistic simulations with empirical potentials. These simulations have been instrumental in qualitative investigations of some microscopic properties of hydrophobic assemblies in water, as predicted by F. Stillinger ≈35 years ago (11), in the conclusions of his paper on scale-particle theory for nonpolar solutes in water. However, to make atomistic simulations a robust and predictive tool for biological processes involving hydropho-bic interactions, as well as in materials science, we face the challenge of performing accurate and rather complex quantum simulations. Acquiring the ability to perform such simulations is particularly desirable because experiments to probe hydrophobic interfaces are still very difficult to perform, and quantitative studies are not yet available. Do we have all of the ingredients and tools to proceed and perform quantum simulations for various systems involving hydrophobic interactions? Not quite yet, although the progress reported in refs. 3, 5, and 6 is definitely encouraging.
Size and simulation times attainable in ab initio simulations are certainly issues to be carefully considered because, even for moderate size systems [e.g., an ≈100-molecule solution of benzene in water (6)], the longest simulation times reached so far amount to 150 ps, and have been achieved by using rigid water models. (Typical time scales of simulations employing empirical potentials are at least 10 times longer.) However, some of these shortcomings might be solved by fitting ab initio simulation data obtained for small systems, to derive effective interaction potentials. In this respect, results such as those obtained in refs. 3, 5, and 6, benchmarking several different classical potentials, are extremely valuable. More importantly, fundamental issues regarding the theory used in ab initio calculations [density functional theory (DFT) (13) with gradient corrected approximations] remain open, despite the coming of age of quantum simulation techniques and of the fundamental progress brought about by their use in the fields of condensed matter physics and materials science. Accurate assessments of the parameters used in ab initio simulations of water (ref. 14 and references therein) indicate that the performance of a quasilocal DFT (13) in describing hydrogen bonding is not fully understood. The theory accounts for all of the major structural properties of water, some of its spectroscopic signatures (15) (in a qualitative manner), and several dynamical properties (16); however, it is still unclear to what extent the theory can reproduce the known phase diagram and thermodynamic properties of water at and close to ambient conditions. In addition, the incorporation of “weak” (compared with hydrogen bonding) van der Waals forces acting between apolar molecules and water, into functionals used in ab initio molecular dynamics, is in its early stages (17). We anticipate that it will take several years before an assessment of the validity of DFT and thus of ab initio simulations to investigate thermodynamic properties of hydrophobic solutions can be made. Progress may come, for example, from simulations of model systems using many body approaches, such as quantum Monte Carlo (18), and from systematic comparisons of these results with those obtained by using DFT. In the meantime, studies such as those of ref. 3 for specific systems, and possibly for more complex hydrophobic solutes in the near future, will continue to elucidate the nature of hydrophobic hydration structure and dynamics. These are important prerequisites to gain insight into the influence of hydrophobic hydration on the structure and functions of specific amino acids, as well as into materials science problems such as water in zeolites, fluid flow in small channels (e.g., carbon nanotubes), or evaporation and hydration of nano-particles.
Footnotes
The author declares no conflict of interest.
See companion article on page 2626.
References
- 1.Pratt LR. Annu Rev Phys Chem. 2002;53:409–436. doi: 10.1146/annurev.physchem.53.090401.093500. [DOI] [PubMed] [Google Scholar]
- 2.Dill KA, Truskett TM, Vlachy V, Hribar-Lee B. Annu Rev Biophys Biomol Struct. 2005;34:173–199. doi: 10.1146/annurev.biophys.34.040204.144517. [DOI] [PubMed] [Google Scholar]
- 3.Li J-L, Car R, Tang C, Wingreen NS. Proc Natl Acad Sci USA. 2007;104:2626–2630. doi: 10.1073/pnas.0610945104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Car R, Parrinello M. Phys Rev Lett. 1985;55:2471–2474. doi: 10.1103/PhysRevLett.55.2471. [DOI] [PubMed] [Google Scholar]
- 5.Grossman JC, Schwegler ER, Galli G. J Phys Chem B. 2004;108:15865–15872. [Google Scholar]
- 6.Allesch M, Schwegler ER, Galli G. J Phys Chem B. 2007;111:1081–1089. doi: 10.1021/jp065429c. [DOI] [PubMed] [Google Scholar]
- 7.Edsall JT. Proc Am Phil Soc. 1985;129:371–407. [PubMed] [Google Scholar]
- 8.Tanford C. Protein Sci. 1997;6:1358–1366. doi: 10.1002/pro.5560060627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kautzmann W. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- 10.Chandler D. Nature. 2002;417:491. doi: 10.1038/417491a. [DOI] [PubMed] [Google Scholar]
- 11.Stillinger FH. J Solution Chem. 1973;2:141–158. [Google Scholar]
- 12.Chandler D. Nature. 2005;437:640–647. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- 13.Kohn W, Sham L. Phys Rev. 1965;137:A1697–A1705. [Google Scholar]
- 14.Schwegler ER, Grossman JC, Gygi F, Galli G. J Chem Phys. 2003;121:5400–5409. doi: 10.1063/1.1782074. [DOI] [PubMed] [Google Scholar]
- 15.Prendergast D, Galli G. Phys Rev Lett. 2006;96:215502-1–215502-4. doi: 10.1103/PhysRevLett.96.215502. [DOI] [PubMed] [Google Scholar]
- 16.Sharma M, Resta R, Car R. Phys Rev Lett. 2005;95:187401-1–187401-4. doi: 10.1103/PhysRevLett.95.187401. [DOI] [PubMed] [Google Scholar]
- 17.Dobson J, White A, Rubio A. Phys Rev Lett. 2006;95:073201-1–073201-4. doi: 10.1103/PhysRevLett.96.073201. [DOI] [PubMed] [Google Scholar]
- 18.Pierleoni C, Ceperley DM, Holzmann M. Phys Rev Lett. 2004;93:146402-1–146402-4. doi: 10.1103/PhysRevLett.93.146402. [DOI] [PubMed] [Google Scholar]