In the early 2000s, the realization that a significant fraction of the so-called heterotrophic marine bacterioplankton is capable of phototrophy has challenged our views of the carbon and energy budgets in the oceans (1) and, consequently, the biosphere. Because of their widespread occurrence and high abundance, bacterioplankton represent large pools of major elements and are catalysts in many biogeochemical cycles of global significance. In 2000–2002, a new picture of marine aerobic anoxygenic phototrophs (AAnPs) in the oceans was revealed. Using biophysical techniques, Kolber et al. (2, 3) detected a significant signal originating from these organisms in different regions of the World Ocean. These findings came as a big surprise because this type of photosynthesis was believed to be limited to a few organisms living in specialized ecological niches (4, 5). Based on cultivation studies (3–8), these newly discovered AAnPs were thought to be limited to members of Alphaproteobacteria. This notion was challenged in 2002 when a study based on environmental genomics (9) suggested that some marine AAnPs genes likely originated from members of Gammaproteobacteria (10). The work of Fuchs et al. (11) in this issue of PNAS is the first to unequivocally assign these elusive AAnP genes to a cultured member of Gammaproteobacteria.
The results offer a glimpse of the physiological adaptations and ecology of organisms that we now consider as typical marine bacterioplankton. Before the application of cultivation-independent technique analyses in the early 1990s, marine bacterioplankton was studied mainly after cultivation in rich media, and most isolated organisms were members of a limited set of copiotrophic (i.e., organisms adapted to growth in high substrate concentrations) Gammaproteobacteria and Bacteroidetes. Cloning and sequencing of 16S rRNA genes (12–15) and direct probing for these genes in the environment (16, 17) challenged the importance of these organisms and also led to the discovery of novel environmentally significant groups (18, 19). The findings also propelled the development and application of alternative cultivation techniques for bacterioplankton (20–23) that resulted in the cultivation of many organisms that were previously known only from cultivation-independent studies. The KT71 strain described by Fuchs et al. (11) represents an environmentally significant clade (NOR5/OM60) that has been brought to cultivation by using low-nutrient media (24) and that, like Pelagibacter ubique (25) and Silicibacter pomeroyi (26), now has its full genomic sequence uncovered.
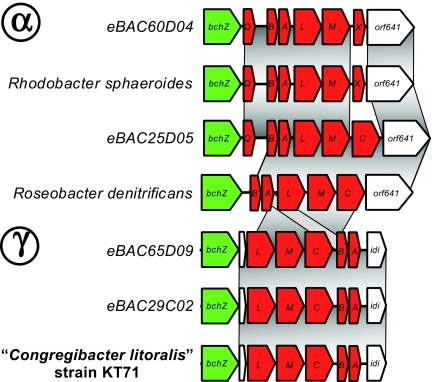
The sequencing of KT71 was part of the Gordon and Betty Moore Foundation's marine microbiology initiative launched in 2004 (www.moore.org/microgenome), in which >200 marine bacteria have or are in the process of full genome sequencing aiming to “produce a better understanding of the ocean's basic biological and chemical processes.” Full genome sequencing analysis revealed that KT71 is, in fact, closely related to the mysterious gammaproteobacterial AAnP group detected in 2002 because its photosynthetic operon is identical to that of environmental BAC clones from Monterey Bay in California (Fig. 1). Based on physiological tests and genomic information, these organisms are likely photoheterotrophs, organisms that use light as an energy source while exploiting organic compounds as their carbon and energy source. Genomic evidence for photoheterotrophy includes the presence of genes involved in bacteriochlorophyll-based phototropy but the lack of key genes for autotrophic carbon fixation of the Calvin cycle, the reductive citrate cycle, or the reductive acetyl-CoA pathway. Despite the fact that these organisms do not fix significant amounts of carbon dioxide, photoheterotrophs still have the potential to affect carbon budgets in the ocean because part of the carbon necessary for energy production could be saved by the cells, and bacterial growth efficiency could be higher under illuminated conditions. However, whether significant differences in growth efficiency occur in the environment, and what the significance of photoheterotrophy is to global carbon budgets are currently not well understood.
Fig. 1.
Diversity of puf operons. Homologous regions are connected by dark gray areas. Puf genes and other reaction center genes are marked in red; green identifies bchZ genes, and white indicates nonphotosynthetic or hypothetical proteins with unknown function. Alphaproteobacteria (Upper) and Gammaproteobacteria (Lower) groups are indicated. Examples are from puf operons of cultured bacteria and environmental BAC sequences currently available in GenBank.
In addition, the genome contains several indications of putative niches occupied by the NOR5/OM60 clade, particularly because of its microaerophily, suggested by the presence of genes involved in oxygen detoxification and confirmed by empirical observations. The presence of several genes associ-ated with exopolymer production and cell aggregation in culture indicates an association with particles such as marine snow that has been shown to contain suboxic microenvironments (27). On the other hand, the presence of mercury-resistance genes was used as evidence for adaptation to costal sediments. Although annotation and experimental observations were not sufficient to determine the main substrates used by the organism in the environment, there were indications of auxotrophic growth (i.e., growth dependent on substrates not synthesized by the organism) and an inability to utilize of glucose or polysaccharides such as chitin, which is commonly present in the marine environment. Finally, the presence of genes coding for a nitrogen storage compound is remarkable and probably reflects an adaptation to life in environments with fluctuating nitrogen availability.
It should be pointed out that it is unclear at this point how widespread these phenotypic characteristics are among organisms in the NOR5/OM60 clade. The within-clade 16S rRNA sequence similarity ranges from 93% to 95% in the clade originally described by Eilers et al. (24), and a BLAST search using a frequently sequenced stretch of the rRNA gene (positions 338–536) yielded a single match in GenBank to KT71 with identity >97%. On the other hand, evidence for bacteriochlorophyll-based phototrophy has been observed in other members of the NOR5/OM60 clade (28), including strains HTCC 2080, HTCC 2148, and HTCC 2246 (see figure 1B in ref. 11), indicating that at least this trait might be widespread in the clade. HTCC 2080 has a puf operon identical to that of KT71, whereas puf operons in HTCC 2148 and HTCC 2246 appear to have been laterally transferred (28).
In conclusion, the remarkable tale of these Gammaproteobacteria AAnPs combined studies in microbiology and oceanography by using approaches from classical microbiology to environmental genomics and bacterial genomics. Like the discovery of proteorhodopsins (29) in P. ubique (SAR11) (25, 32) and marine Bacteroidetes (30), this study is a good example of the synergy between metagenomics and more traditional microbiology, where metagenomics and other cultivation-independent techniques uncover genes from environmentally significantly organisms and aid in the focus on cultivation and genomic sequencing efforts (31).
Footnotes
The authors declare no conflict of interest.
See companion article on page 2891.
References
- 1.Karl DM. Nature. 2002;415:590–591. doi: 10.1038/415590b. [DOI] [PubMed] [Google Scholar]
- 2.Kolber ZS, Van Dover CL, Niderman RA, Falkowski PG. Nature. 2000;407:177–179. doi: 10.1038/35025044. [DOI] [PubMed] [Google Scholar]
- 3.Kolber ZS, Plumley FG, Lang AS, Beatty JT, Blankenship RE, VanDover CL, Vetriani C, Koblízek M, Rathgeber C, Falkowski PG. Science. 2001;292:2492–2495. doi: 10.1126/science.1059707. [DOI] [PubMed] [Google Scholar]
- 4.Shiba T, Simidu U, Taga N. Appl Environ Microbiol. 1979;38:43–48. doi: 10.1128/aem.38.1.43-45.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shiba T, Shioi Y, Takamiya K, Sutton DC, Wilkinson CR. Appl Environ Microbiol. 1991;57:295–300. doi: 10.1128/aem.57.1.295-300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shiba T. J Gen Appl Microbiol. 1984;30:239–244. [Google Scholar]
- 7.Harashima K, Shiba T, Murata N. Aerobic Photosynthetic Bacteria. Berlin: Springer; 1989. [PubMed] [Google Scholar]
- 8.Yurkov VV, Beatty JT. Microbiol Mol Biol Rev. 1998;62:695–724. doi: 10.1128/mmbr.62.3.695-724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Béjà O. Curr Opin Biotechnol. 2004;15:187–190. doi: 10.1016/j.copbio.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Béjà O, Suzuki MT, Heidelberg JF, Nelson WC, Preston CM, Hamada T, Eisen JA, Fraser CM, DeLong EF. Nature. 2002;415:630–633. doi: 10.1038/415630a. [DOI] [PubMed] [Google Scholar]
- 11.Fuchs BM, Spring S, Teeling H, Quast C, Wulf J, Schattenhofer M, Yan S, Ferriera S, Johnson J, Glöckner FO, Amann R. Proc Natl Acad Sci USA. 2007;104:2891–2896. doi: 10.1073/pnas.0608046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLong EF. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuhrman JA, McCallum K, Davis AA. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giovannoni SJ, Britschgi TB, Moyer CL, Field KG. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt TM, DeLong EF, Pace NR. J Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glockner FO, Fuchs BM, Amann R. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pernthaler A, Pernthaler J, Schattenhofer M, Amann R. Appl Environ Microbiol. 2002;68:5728–5736. doi: 10.1128/AEM.68.11.5728-5736.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giovannoni SJ, Rappé M. In: Microbial Ecology of the Oceans, Kirchman DL, editor. New York: Wiley; 2000. pp. 47–84. [Google Scholar]
- 19.Suzuki MT, DeLong EF. In: Biodiversity of Microbial Life: Foundation of Earth's Biosphere, Staley JT, Reisenbach AL, editors. New York: Wiley; 2002. pp. 209–234. [Google Scholar]
- 20.Button DK, Robertson BR, Lepp PW, Schmidt TM. Appl Environ Microbiol. 1998;64:4467–4476. doi: 10.1128/aem.64.11.4467-4476.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho JC, Giovannoni SJ. Appl Environ Microbiol. 2004;70:432–440. doi: 10.1128/AEM.70.1.432-440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Connon SA, Giovannoni SJ. Appl Environ Microbiol. 2002;68:3878–3885. doi: 10.1128/AEM.68.8.3878-3885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zengler K, Toledo G, Rappé M, Elkins J, Mathur EJ, Short JM, Keller M. Proc Natl Acad Sci USA. 2002;99:15681–15686. doi: 10.1073/pnas.252630999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eilers H, Pernthaler J, Peplies J, Glockner FO, Gerdts G, Amann R. Appl Environ Microbiol. 2001;67:5134–5142. doi: 10.1128/AEM.67.11.5134-5142.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, Bibbs L, Eads J, Richardson TH, Noordewier M, et al. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 26.Buchan A, González JM, Moran MA. Appl Environ Microbiol. 2005;71:5665–5677. doi: 10.1128/AEM.71.10.5665-5677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alldredge AL, Cohen Y. Science. 1987;235:689–691. doi: 10.1126/science.235.4789.689. [DOI] [PubMed] [Google Scholar]
- 28.Cho J-C, Stapels MD, Morris RM, Vergin KL, Schwalbach MS, Givan SA, Barofsky DF, Giovannoni SJ. Environ Microbiol, 2007 doi: 10.1111/j.1462-2920.2007.01264.x. in press. [DOI] [PubMed] [Google Scholar]
- 29.Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL, et al. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- 30.Gómez-Consarnau L, González JM, Coll-Lladó M, Gourdon P, Pascher T, Neutze R, Pedrós-Alió C, Pinhassi J. Nature. 2007;445:210–213. doi: 10.1038/nature05381. [DOI] [PubMed] [Google Scholar]
- 31.Pedros-Alio C. Int Microbiol. 2006;9:191–197. [PubMed] [Google Scholar]
- 32.Giovannoni SJ, Bibbs L, Cho JC, Stapels MD, Desiderio R, Vergin KL, Rappe MS, Laney S, Wilhelm LJ, Tripp HJ, et al. Nature. 2005;438:82–85. doi: 10.1038/nature04032. [DOI] [PubMed] [Google Scholar]