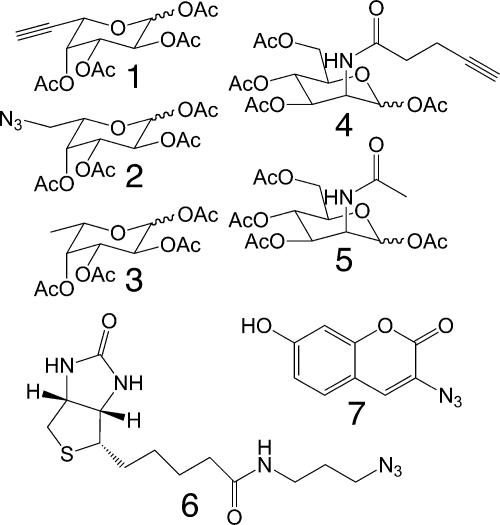
Abstract
Developing tools for investigating the cellular activity of glycans will help to delineate the molecular basis for aberrant glycosylation in pathological processes such as cancer. Metabolic oligosaccharide engineering, which inserts sugar-reporting groups into cellular glycoconjugates, represents a powerful method for imaging the localization, trafficking, and dynamics of glycans and isolating them for glyco-proteomic analysis. Herein, we show that the alkyne-reporting group can be incorporated into cellular glycans. The alkyne group is a small, inert, bio-orthogonal handle that can be chemoselectively labeled by using the Cu(I) catalyzed [3 + 2] azide-alkyne cycloaddition, or click chemistry. Alkynyl sugar monomers, based on fucose (Fuc) and N-acetylmannosamine (ManNAc), were incorporated into fucosylated and sialylated glycans in several cancer cell lines, allowing for cell surface and intracellular visualization of glycoconjugates, as well as, observation of alkyne-bearing glycoproteins. Similarly to our previous results with an azido Fuc/alkynyl probe system, we demonstrated that click-activated fluorogenic probes are practical tools for efficiently and selectively labeling alkynyl-modified glycans. Because Fuc and sialic acid are terminal glycan residues with a notably increased presence in many tumors, we hope that our method will provide useful information about their roles in cancer and ultimately can be used for diagnostic and therapeutic purposes.
Keywords: click chemistry, fluorescent imaging, fucose, sialic acid
Glycosylation is an important bioinformational process that occurs co- or posttranslationally on >50% of eukaryotic proteins. In living organisms, it affects protein bioactivity and metabolic turnover. Inside of cells, it mediates protein folding, stability, and trafficking. At the cell surface, glycans participate in molecular recognition events that are central to biological and pathological processes like cell-cell interactions involved in adhesion, migration, and metastasis; host-pathogen interactions critical for bacterial and viral infections; and, initiation of immune response (1). Aberrant glycosylation is often observed in pathological conditions such as inflammation and cancer metastasis (2–6). In particular, altered terminal fucosylation and sialylation, which are believed to result from changes in expression locations and levels of fucosyltransferases and sialyltransferases, are associated with tumor malignancy. For example, glycan determinants like Lewis y, Lewis x, sialyl Lewis x, sialyl Lewis a, sialyl Tn, Globo H, fucosyl GM1, and polysialic acid are expressed at elevated levels in neoplastic tissues (7–11). For this reason, these epitopes are promising and eagerly pursued targets for glycan-based vaccines. Further understanding of the molecular details and correlations between altered glycosylation and pathological status is of great interest and is likely to provide useful information for diagnosis and disease prognosis, in addition to unveiling new therapeutic targets. However, cellular glycans are complex, microheterogeneous populations, resulting from a non-template-driven process that cannot be manipulated genetically. This complexity makes the isolation and identification of glycans for structural analysis one of the most challenging and defining tasks in glycobiology.
Specific glycan-tagging systems provide a powerful method for probing the structure of heterogeneous glycans. The key to glycan tagging entails incorporating chemically modified-sugars into cellular glycans via normal biosynthetic pathways, and then detecting them by using a specific and chemically complementary probe. Many selective chemical probing techniques have been used for tagging glycoconjugates in cells. These methods include different bioorthogonal reactions such as ketone-aminooxy/hydrazide ligation (12, 13), Staudinger ligation (14), Michael addition (15), and the strain-promoted, and Cu(I)-catalyzed [3 + 2] azide-alkyne cycloaddition (16–19). Many sugar analogs with small functional handles, including ketones and azides, are tolerated and have been incorporated into glycoconjugates (14, 20, 21). These reporting sugars have been coupled with various tags such as FLAG peptides, biotin, and fluorescent or fluorogenic molecules. The strength of these systems is that the tagged glycan products have the potential to be enriched and isolated for further analysis by means of mass spectrometry, detected by flow cytometry for quantitative studies, or visualized through microscopy such that their localization, trafficking, and dynamics can be established.
The incorporation of exogenous natural or unnatural sugars into glycans is achieved by cellular biosynthetic pathways. These processes involve multistep enzymatic transformations that render free sugars in the cytosol into nucleotide-donor sugars, the substrates for glycosyltransferases. In the case of fucose (Fuc), a salvage pathway consisting of Fuc kinase and GDP-Fuc pyrophosphorylase contributes to the production of GDP-Fuc, which is then exploited by fucosyltransferases located in the Golgi apparatus to add Fuc onto glycoconjugates (22). Previous work by our laboratory and others has shown that modifications at the 6-position of Fuc are tolerated by the salvage pathway and fucosyltransferases (18, 19). In the sialic acid biosynthetic pathway, the precursor N-acetylmannosamine (ManNAc) is derived from GlcNAc or UDP-GlcNAc through specific epimerases, then sequentially converted to sialic acid by the cytosolic enzymes ManNAc 6-kinase, sialic acid-9-phosphate synthase, and sialic acid-9-phosphate phosphatase. CMP-sialic acid is subsequently formed in the nucleus, and transported to the Golgi apparatus for glycan elaboration by sialyltransferases (23). Studies on metabolic delivery of mannosamine or ManNAc analogs show that N-acyl chains up to five carbon atoms long are tolerated by the sialic acid biosynthetic pathway (23). This promiscuity led to our design of alkynyl ManNAc 4, N-4-pentynoylmannosamine, for recognition and incorporation into sialylated glycoconjugates in cells (see Scheme 1).
Scheme 1.
Modified sugar analogs and probes used in this study.
Previously, we reported a fluorescent labeling technique for probing metabolically labeled fucosylated glycans in cells (19). In this approach, azido Fuc analogs incorporated into glycans were labeled with a 1,8-naphthalimide fluorogenic probe, by using Cu(I)-catalyzed [3 + 2] cycloaddition, or azide-alkyne “click” reaction (24, 25). Our click-activated fluorescent derivatization demonstrated a method for specifically labeling fucosylated glycoconjugates. Here, we extend this system by including alkynyl Fuc and ManNAc analogs into the repertoire of reporting saccharides that can be used to label fucosylated and sialylated glycans in mammalian cells. Our studies show that alkynyl Fuc shows greatly reduced toxicity to cells when compared with its azido counterpart. Moreover, when these alkynyl sugars are coupled with biotin, click-activated fluorogenic coumarin, and other fluorescent probes, this technique allows for the isolation of fucosylated and sialylated glycoconjugates for further analysis, and fluorescent imaging, a capability that we hope to use for visualizing glycan dynamics inside of cells and to identify important glycan markers.
Results
Synthesis of Alkynyl Sugars and Biotinylated Azide Probe.
Peracetylated alkynyl derivatives of Fuc 1 and ManNAc 4, were synthesized in an effort to label fucosylated and sialylated glycoconjugates, respectively, in vivo. In our design, the sugar derivatives were synthesized in peracetylated form, as this modification is known to increase their cellular uptake efficiency (26, 27). The acetate esters are subsequently hydrolyzed in the cytosol. The synthesis of alkynyl Fuc [1, supporting information (SI) Scheme 2] proceeded from a known four-step transformation, beginning with l-(+)-galactonic acid γ-lactone and ending with the alkynyl diisopropylidene-Fuc intermediate. Subsequent protecting group removal followed by acetylation of the intermediate yielded the desired compound 1, as a mixture of pyranoside and furanoside forms. This mixture was then used directly for labeling fucosylated glycans in cells. For labeling sialylated glycoconjugates, compound 4 was synthesized. d-Mannosamine hydrochloride was reacted with N-succinimidyl 4-pentynoate in triethylamine to yield alkynyl ManNAc derivative (SI Scheme 3). The alkynyl ManNAc 4 was subsequently obtained by acetylation. The coupling partner, biotinylated azido probe 6, was synthesized by coupling of biotin to 1-azido-3-aminopropane (SI Scheme 4). Fluorogenic probe 7, 3-azido-7-hydoxycoumarin, was synthesized as reported (28). Modified sugar analogs and probes used in this study are illustrated in Scheme 1.
Fluorescent Labeling of Alkynyl Glycoconjugates at the Cell Surface.
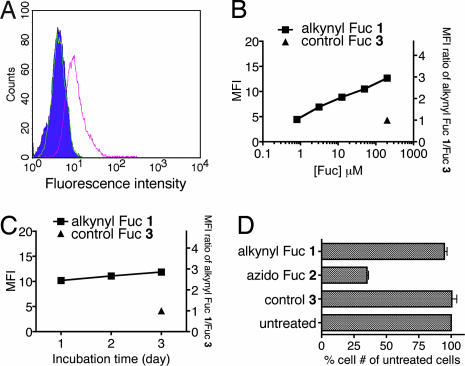
To demonstrate the labeling of fucosylated glycoconjugates at the cell surface, Jurkat cells were grown in the presence of alkynyl Fuc 1. After treatment, cells were subjected to Cu(I)-catalyzed [3 + 2] cycloaddition (click chemistry) to couple biotinylated azido probe 6 with any alkynyl Fuc-bearing glycans, and stained with fluorescein-conjugated streptavidin. As shown in Fig. 1A, the alkynyl Fuc 1-treated cells showed increased fluorescence intensity compared with control Fuc (3)-treated cells, as analyzed by flow cytometry. This result indicates that alkynyl-modified Fuc residues were incorporated into cell surface glycans and that these modified sugars can serve as chemical handles for selective labeling. Incorporation of the modified Fuc analogs into fucosylated glycoconjugates likely occurs via the Fuc salvage pathway. Alkynyl Fuc analog 1-treated cells showed a dose-dependent increase of fluorescence signal, with a 3-fold greater mean fluorescence intensity (MFI) compared with Fuc (3)-treated cells at 200 μM (Fig. 1B). The data also showed saturation of alkynyl Fuc incorporation within one-day of incubation, although there was a slight increase of labeling signal on cells treated for three days with alkynyl Fuc 1 (Fig. 1C).
Fig. 1.
Analysis of cells labeled with Fuc analogs. (A) Flow cytometry analysis of Jurkat cells treated with Fuc analogs and labeled with biotin/fluorescein-conjugated streptavidin (filled histogram, untreated cells; green, cells treated with Fuc 3; pink, cells treated with alkynyl Fuc 1). (B) Dose-dependency of fucosyl glycan labeling by alkynyl Fuc 1 over 3 days. (C) Time course of fucosyl glycan labeling by 200 μM alkynyl Fuc 1. (D) Cell growth analysis after treatment with different Fuc analogs. Jurkat cells were grown in the presence of 200 μM each Fuc analog for 3 days before cell numbers were counted. The data represent the percentage of treated cells vs. untreated cells (n = 4).
We next evaluated how treatment with exogenous Fuc derivative affects cell growth rate. As shown in Fig. 1D, the number of cells after 3 days was similar whether they were treated with 200 μM alkynyl Fuc 1, 200 μM Fuc 3, or grown without exogenous Fuc. In contrast, the addition of 200 μM azido Fuc 2 inhibited cell growth considerably, by 65% when compared with the untreated cells. These results indicate that azido Fuc 2, which has been used previously for probing fucosylation (18, 19), is more toxic to cells than alkynyl Fuc 1. Such toxicity may lead to global change in expression, therefore a nontoxic probe is preferred for accurate probing of glycoconjugate expression.
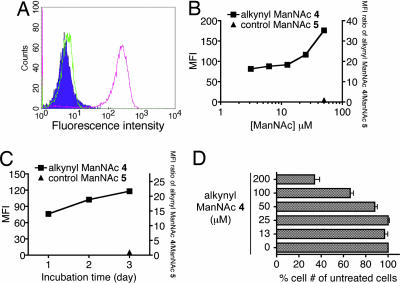
It has been demonstrated that the majority of an exogenous ManNAc analog, N-levulinoylmannosamine, acquired by cells is converted into sialic acid via biosynthetic pathways (29). Accordingly, we expected that treating cells with alkynyl ManNAc 4 would result in alkyne-bearing sialyl glycans. To test for such labeling, cells were treated with 4 at various concentrations for 1–3 days. Incorporation onto the cell surface glycans was analyzed by monitoring fluorescence intensity with flow cytometry after clicking on the biotin azide probe 6 and staining cells with fluorescein-conjugated streptavidin. Labeling with alkynyl ManNAc 4 yielded a specific signal on the cell surface compared with the control values obtained from cells treated with control ManNAc 5 (Fig. 2A). Dose-dependent labeling was observed in cells treated with alkynyl ManNAc 4 (Fig. 2B). Compared with the MFI of controls, there was significant labeling on cells treated with alkynyl ManNAc 4, even at concentrations as low as 3 μM (15-fold increase). Time-course experiments revealed that treatment with alkynyl ManNAc 4 from one to three days gave a 15- to 23-fold increase in labeling intensity over control levels (Fig. 2C). The optimal concentration of 4 for labeling sialyl glycans falls between 25 and 50 μM. In this concentration range, 4 showed little or no toxicity, although it is more toxic above 100 μM (Fig. 2D).
Fig. 2.
Analysis of cell surface labeling of sialyl glycoconjugates. (A) Flow cytometry analysis of Jurkat cells labeled with alkynyl ManNAc (filled histogram, untreated cells; green, cells treated with control 5; pink, cells treated with alkynyl ManNAc 4). (B) Dose dependency of sialyl glycan labeling by alkynyl ManNAc 4 for 3 days. (C) Time course for labeling sialyl glycans by treatment with 25 μM alkynyl ManNAc 4. (D) Growth rate of Jurkat cells treated with different doses of alkynyl ManNAc 4 after 3 days.
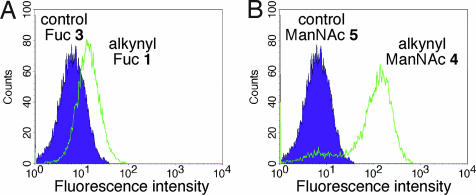
One of the advantages of Cu(I)-catalyzed [3 + 2] cycloaddition, or the click reaction, is the formation of a triazole unit, which can modulate the fluorescent emission of probes through electron-donating properties. We have previously shown that such click-activated fluorescence is useful in fluorescently labeling azido Fuc-bearing glycans using a 1,8-naphthalimide-alkyne probe (19). However, the azido version of the naphthalimide probe causes high background, making it less useful for labeling our alkynyl sugars. Recently, another click-activated fluorescent probe, based on coumarin, was reported in the literature (28). Thus, to demonstrate click-activated fluorescent cell labeling, we used this fluorogenic probe, 3-azido-7-hydroxycoumarin 7, as the coupling partner for alkynyl handles on labeled glycans. As shown in Fig. 3, cells treated with alkynyl Fuc 1 (Fig. 3A) or alkynyl ManNAc 4 (Fig. 3B) allowed significant fluorescent labeling after reacting with 3-azido-7-hydroxycoumarin probe, whereas cells treated with control sugars 3 and 5 gave very low background signals.
Fig. 3.
Labeling of cell surface glycans by alkynyl sugar analogs and click-activated probe 3-azido-7-hydoxycoumarin (7). Shown is flow cytometry analysis of Jurkat cells labeled with 200 μM alkynyl Fuc 1 (A) or 25 μM alkynyl ManNAc 4 (B) for 3 days. The fluorescence intensity was detected after clicking on coumarin probe 7. Filled histogram, cells treated with sugar analog 3 or 5; open histogram, cell treated with alkynyl sugar 1 or 4.
Visualization of Fluorescently Labeled Glycoconjugates in Cells.
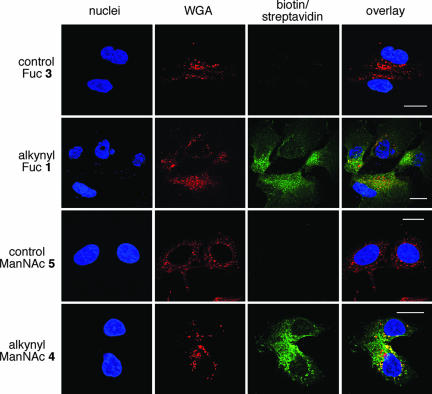
To visualize the localization of glycoconjugates with alkynyl sugar incorporated, adherent cells were grown on slides in the presence or absence of alkynyl sugars. After a 3-day-incubation, cells attached to the slides were fixed, permeabilized, and tagged with either biotin probe 6 or fluorogenic probe 7 for fluorescent signal analysis with confocal microscopy (Figs. 4 and 5 and SI). For comparison, samples were also stained with wheat germ agglutinin (WGA, a Golgi marker) and Hoechst 33342 (marker for cell nuclei). As demonstrated in Hep3B (hepatocellular carcinoma) and MCF-7 (breast adenocarcinoma) cells, treatment with alkynyl Fuc 1 resulted in a strong punctate-labeling signal after clicking on the biotin probe 6 and staining with fluorescein-conjugated streptavidin. This signal showed significant overlap with the WGA signal, indicating the labeled fucosyl glycans were localized in Golgi apparatus (Fig. 4 and SI Fig. 7). Similar results were obtained from cells treated with alkynyl ManNAc 4, which probes for sialic acid-containing glycans, when labeled by biotin probe 6 and fluorogenic probe 7 (Figs. 4 and 5 and SI Figs. 4 and 5). Consistent with the results from flow cytometry, confocal microscopic analysis of cells treated with control sugars 3 and 5 gave very low background after reacting with click probes, confirming the labeling of alkynyl containing glycans is specific and sensitive.
Fig. 4.
Fluorescent imaging of fucosyl and sialyl glycoconjugates in cells. Confocal microscopy of Hep3B cells treated with 200 μM Fuc analogs or 25 μM ManNAc analogs. Cells were biotin-tagged and stained with streptavidin (fluorescein, green), WGA lectin (Alexa Fluor 594, red), and Hoechst 33342 (blue). (Scale bars: 20 μm.)
Fig. 5.
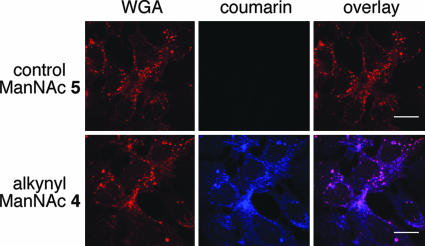
Visualization of sialyl glycoconjugates in cells using click-activated probe 7. Shown is confocal microscopy of coumarin-tagged Hep3B cells. Cells were treated with 25 μM ManNAc analogs 5 or 4 for 3 days, and then clicked with fluorogenic coumarin probe 7 (blue) and stained with WGA lectin (Alexa Fluor 594, red). (Scale bars: 20 μm.)
Labeling of Glycoproteins in Cell Extract.
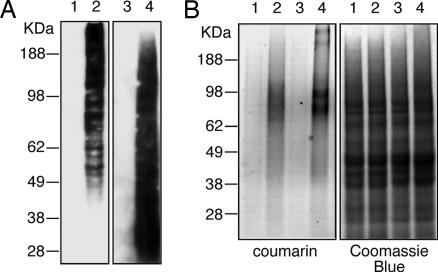
Because our probing system enables the identification of cellular glycoconjugates, it might also serve well in glyco-proteomic applications aimed at discovering unknown glycosylated targets for diagnostic and therapeutic purposes. To demonstrate the detection of individually labeled proteins, cell extracts were analyzed after growing cells with alkynyl sugars. Soluble lysate fractions were tagged with biotin probe 6, fluorogenic probe 7, or a standard rhodamine probe used in proteomics before separating proteins by SDS/PAGE (30). As shown in Fig. 6A, specific biotin-labeling signals were detected by Western blot in proteins from cells treated with alkynyl sugars 1 and 4. Fluorescent signal was also detected in alkynyl positive protein lysate when clicked with fluorogenic 3-azido-7-hydroxycoumarin 7 and rhodamine probes (Fig. 6B and SI Fig. 9). Proteins harvested from cells grown with control sugars 3 and 5 and processed under the same click condition, showed little to no signal by Western Blot or fluorescence. The labeling patterns for Fuc and ManNAc are notably different, indicating the detection of unique glycoproteins. The data here demonstrate the feasibility of labeling and identifying individual glycoproteins by using this probing system. Moreover, further processing, including an avidin enrichment or gel slice purification, will allow for comparative identification of unknown glycoproteins expressed at different cell status, for instance, un-differentiated verses differentiated cells, or normal verses cancer cells.
Fig. 6.
Detection of alkyne-labeled glycoproteins in cell extracts. Glycoproteins labeled with alkynyl sugars were derivatized and detected by immunoblotting of biotin tag (A) or fluorescent imaging of fluorogenic coumarin probe 7 (B). Protein extracts from cells grown with different sugars were analyzed (lane 1, control Fuc 3; lane 2, alkynyl Fuc 1; lane 3, control ManNAc 5; lane 4, alkynyl ManNAc 4). The protein gel (4–12%) was subsequently stained by Coomassie blue after fluorescent imaging, to verify equal protein loading.
Discussion
The ability to visualize and isolate cellular glycans will help to deconvolute the complexity and microheterogeneity that make it difficult to study their biological function (31). Toward this goal, several metabolic oligosaccharide engineering techniques have been developed, wherein the endogenous biosynthetic machinery for glycosylation is exploited to insert chemically modified sugar building blocks in place of their native counterparts. The remodeled glycans, which contain bioorthogonal chemical handles, can then be chemoselectively labeled with a complementary reactive probe for further manipulation, including visualization or isolation. Recently, we designed a system for incorporating and derivatizing 6-azido Fuc in cellular glycans using standard click chemistry, or the Cu(I)-catalyzed [3 + 2] cycloaddition. We also introduced the use of click-activated fluorogenic probes for selective and specific labeling of modified glycans at the cell surface as well as intracellularly in fixed-cell preparations. Here, we have expanded the scope of our specific glycan tagging system by establishing that another useful chemical reporter, the alkyne group, can also be incorporated into cellular glycans when appended on Fuc and ManNAc derivatives. Similar to its azide counterpart, the alkyne is a small, inert, bioorthogonal group that can be chemoselectively labeled by using click chemistry.
Our alkynyl Fuc and ManNAc sugars represent a robust platform for labeling fucosylated and sialylated glycans in vivo. Given the past success with incorporating azide sugar analogs into glycans, it is not surprising that our alkynyl sugars are also accepted by these relatively promiscuous biosynthetic pathways (18, 19). In our studies we found that the azido Fuc analog is quite toxic to cells at the levels required for efficient labeling, which might in turn lead to aberrant cellular glycan profiles. The alkynyl Fuc, on the other hand, is much less toxic, yielding higher signals and less background, when cellular incorporation was monitored by flow cytometry. Toxicity of alkynyl ManNAc relative to azido ManNAc was less of an issue, because very low levels of the modified sugar were required for efficient glycan labeling as observed by flow cytometry and microscopy. This observation likely reflects the higher relative abundance of sialic acid verses Fuc residues. The alkynyl sugars also proved to be efficient ligation partners for click-activated fluorogenic and standard click probes. Again, we found that tagging with click-activated probes is particularly useful because of the generation of an instant signal upon ligation with modified glycans that does not produce any significant background. As established by all of our methods (flow cytometry, confocal microscopy, and SDS/PAGE), the signal generated by the click-activated probe is equivalent to that of the biotin-secondary detection systems; however, it requires one less incubation step and no washing. Furthermore, the click-activated probes are small and hydrophobic, making them more amenable to intracellular penetration and tagging in cells.
The utility of our approach for probing interesting glycans was demonstrated by treating several human cancer cell lines with the alkynyl sugar substrates and subjecting them to several methods of analysis. In all cases, fluorescent-labeling of cell surface glycoconjugates was witnessed by flow cytometry. Information about intracellular glycan labeling and localization was determined by using confocal microscopy. Here, we show that both alkynyl Fuc and ManNAc modified glycans are localized in the Golgi, consistent with their proposed site of transfer. Notably, detailed analysis of microscopy images can supply quantitative data within regions of colocalization, which may provide a useful tool for monitoring glycan levels and trafficking. Finally, we demonstrate that individual modified glycoproteins can be separated and visualized by SDS/PAGE analysis, setting the stage for further proteomic analysis. In future studies, we plan to extend and combine these methodologies to obtain information about cellular glycans under different physiological disease states and cellular statuses, such as stress, apoptosis, or inflammation. Comparative studies between various stages of cancer progression, in addition to pulse–chased techniques to follow the dynamics of newly synthesized proteins within individual cellular systems should provide much needed snapshots of critical glycan behavior. Indeed, in preliminary studies with prostate cancer cells, we observed an increase in the fucosylated glycan signal when compared with noncancerous prostate controls (SI Fig. 10). This enhanced signal indicates that there might be some interesting correlations between increased Fuc expression and prostate cancer, a fact that is already well known for numerous cancers (3).
Notably, it is important to consider that the introduction of modified sugars might change the cellular activity of certain glycans. Perturbations in glycan-mediated binding have been noted with viral receptors and lectin interactions in metabolic oligosaccharide engineering studies where sialic acid derivatives were introduced into cellular glycans (23, 32). Accordingly, our studies also found that some Fuc lectins, including aleuria aurantia lectin (AAL; specific for α-1,6- or α-1,3-linked Fuc) and Ulex Europaeus Agglutinin I (UEA-I; specific for α-1,2-linked Fuc), bound with significantly lower avidity among cells treated with alkyne Fuc verses control (SI Fig. 11). These results are not surprising, considering that a change from a methyl to a more bulky alkynyl group may interfere with the recognition in the small conserved hydrophobic pocket found in many Fuc lectins (33, 34). Indeed, in some cases, altered biological responses may prove useful for perturbing and profiling the function of unknown carbohydrate binding proteins (32). On the other hand, some glycan modifications do not seem to greatly affect binding interactions, past studies analyzing the binding of selectins to synthetic analogues of sialyl Lewis x showed a significant tolerance for N-acyl modification on sialic acid (35). Thus, the analysis of cells treated with modified sugars over an extended period must be evaluated carefully. To circumvent any artifacts from altered activity or differential cellular uptake, pulse–chase experiments may be useful. These experiments would result in lower levels of modified glycans, while providing a comparable cell-to-cell snapshot of glycan behavior.
In our experimental system, cells are fixed and permeabilized to enable intracellular labeling, and addition of toxic Cu(I) is required. However, the presence of an intracellular copper pool localized in the Golgi apparatus and mitochondria (36), may allow for intracellular labeling without addition of exogenous Cu(I). Along with the development of new cell-permeable probes, labeling may be achieved in living cells without toxicity.
In conclusion, we have developed a method for metabolic oligosaccharide engineering that can incorporate alkyne-bearing sugar analogs in cellular glycans. The utility of the alkynyl system has been demonstrated by incorporating Fuc and ManNAc derivative sugars into cancer cell lines, where they were visualized at the cell surface, intracellularly, and as individual glycoproteins. We purposefully choose to use sugars that report on Fuc (alkynyl Fuc) and sialic acid (alkynyl ManNAc) because these residues, in particular, have been linked to many aberrant glycans in cancer. Although several epitopes are known, there are likely many other as yet unidentified glycans and activities that contribute. Identification of these glycan-related biomarkers and targets for therapeutic intervention is one of the key objectives in our strategy.
Materials and Methods
Details describing the synthesis of the compounds can be found in http://www.pnas.org/cgi/content/full/0611307104/DC1SI Materials and Methods.
Flow Cytometric Analysis of Fluorescent Labeling on Cell Surface.
Jurkat cells were cultivated (2 × 106/10 ml) in RPMI medium 1640 supplemented with 10% FCS and various concentrations of peracetylated alkynyl sugars 1, 2, or 4 or native sugars 3 or 5, for 1–3 days at 37°C. Cells were then harvested, washed with 0.1% FCS/PBS, and resuspended (106 cells) in 100 μl of click reaction solution (0.1 mM biotin probe 6 or fluorogenic probe 7/0.1 mM Tris-triazoleamine catalyst/0.1 mM CuSO4/0.5 mM sodium ascorbate, in PBS). The reaction was incubated at room temperature for 30 min, and then the cells were washed twice with 0.1% FCS/PBS. Cells treated with biotin probe 6 were subsequently stained with fluorescein-conjugated streptavidin (0.5 μg per sample in 50 μl of 1% FCS/PBS) for 30 min at 4°C, and washed three times with 1% FCS/PBS. Data were acquired by BD LSR II with FACSDiva software, and were analyzed by CellQuestPro software (BD Biosciences, San Jose, CA). Detection of fluorescent adduct with probe 7 was monitored with a 408 nm laser and a 440/40 bandpass filter for excitation and emission, respectively.
Microscopic Analysis of Fluorescent Labeling in Cells.
Human hepatocellular carcinoma cells (Hep3B) or breast adenocarcinoma cells (MCF-7) were seeded onto six-well plates (3 × 105/2 ml per well) containing glass coverslips, and were cultivated in 10% FCS/DMEM or 10% FCS/RPMI medium 1640. Growth medium was supplemented with a control sugar (200 μM Fuc 3 or 25 μM ManNAc 5) and an alkynyl-modified sugar (alkynyl Fuc 1 or alkynyl ManNAc 2 at the same concentration as control sugars). After growing for 3 days, cells on coverslips were fixed and permeabilized with acetone for 10 min, then subjected to the probe labeling reaction: 0.1 mM biotin probe 6 or fluorogenic probe 7/0.1 mM Tris-triazoleamine catalyst/1 mM CuSO4/2 mM sodium ascorbate, in PBS, at room temperature for 30 min. Subsequently, the fixed and labeled cells were rinsed with PBS and stained with Alexa Fluor 594-conjugated WGA lectin (2 μg/ml in 5% BSA/PBS) and/or fluorescein-conjugated streptavidin (2 μg/ml in 5% BSA/PBS) at room temperature for 30 min. Hoechst 33342 (10 μg/ml in PBS) was used to stain nuclei. Fluorescent images were captured by Bio-Rad (Carl Zeiss, Thornwood, NY) Radiance 2100 Rainbow laser scanning confocal microscopy system.
Labeling and Detection of Glycoproteins in Cell Extracts.
Cells were seeded at 3 × 106/8 ml per 10-cm dish and treated with control and test sugars (200 μM Fuc 3 vs. alkynyl Fuc 1, or 25 μM ManNAc 5 vs. alkynyl ManNAc 2) in growth medium at 37°C. After 3 days, cell extracts were prepared by resuspending the cells in 1 ml of lysis buffer (1% Nonidet P-40/150 mM NaCl/protease inhibitor/100 mM sodium phosphate, pH 7.5). Protein extract (1 mg/ml) was labeled for 1 h at room temperature (conditions as outlined in microscopic analysis; the azido rhodamine probe was a gift from Benjamin F. Cravatt, The Scripps Research Institute). Labeled protein lysate was resolved by SDS/PAGE. For immunoblotting of biotin-labeled glycoproteins, electrophoresed proteins were transferred onto PVDF membranes, blocked for 20 min with SuperBlock Blocking Buffer, probed for 1 h with anti-biotin MAb (1 μg/ml), and incubated with peroxidase-conjugated goat anti-mouse IgG (1:7,500 dilution) for 30 min. Each step was followed by a wash with 0.02% Tween 20/PBS (PBST). Signal was developed with SuperSignal Chemiluminescent Substrate and detected by exposure to x-ray film. For detecting the coumarin-labeled glycoproteins, gels were examined under 365 nm UV light with a 535 ± 50 nm filter. Images were taken by using a BioDoc-It imaging system (UVP, Upland, CA). Rhodamine gels were analyzed as described (30).
Supplementary Material
Acknowledgments
We thank Dr. William B. Kiosses for help with microscopy and Drs. William A. Greenberg and Clay S. Bennett for helpful discussion. T.-L.H. is grateful for postdoctoral support from Taiwan Merit Scholarships TMS-094-1-B-033. S.R.H. acknowledges a predoctoral fellowship from the Achievement Rewards for College Scientists (ARCS) Foundation. This research was supported by the National Institutes of Health and The Skaggs Institute for Chemical Biology.
Abbreviations:
- Fuc
fucose
- ManNAc
N-acetylmannosamine
- WGA
wheat germ agglutinin.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611307104/DC1.
References
- 1.Varki A, Cummings R, Esko JD, Freeze H, Hart GW, Marth J. Essentials of Glycobiology. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1999. pp. 1–635. [PubMed] [Google Scholar]
- 2.Axford JS. Biochim Biophys Acta. 1999;1455:219–229. doi: 10.1016/s0925-4439(99)00057-5. [DOI] [PubMed] [Google Scholar]
- 3.Dube DH, Bertozzi CR. Nat Rev Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 4.Mackiewicz A, Mackiewicz K. Glycoconj J. 1995;12:241–247. doi: 10.1007/BF00731326. [DOI] [PubMed] [Google Scholar]
- 5.Meezan E, Wu HC, Black PH, Robbins PW. Biochemistry. 1969;8:2518–2524. doi: 10.1021/bi00834a039. [DOI] [PubMed] [Google Scholar]
- 6.Turner GA. Clin Chim Acta. 1992;208:149–171. doi: 10.1016/0009-8981(92)90073-y. [DOI] [PubMed] [Google Scholar]
- 7.Orntoft TF, Vestergaard EM. Electrophoresis. 1999;20:362–371. doi: 10.1002/(SICI)1522-2683(19990201)20:2<362::AID-ELPS362>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 8.Sell S. Hum Pathol. 1990;21:1003–1019. doi: 10.1016/0046-8177(90)90250-9. [DOI] [PubMed] [Google Scholar]
- 9.Taylor-Papadimitriou J, Epenetos A. Trends Biotechnol. 1994;12:227–233. doi: 10.1016/0167-7799(94)90121-X. [DOI] [PubMed] [Google Scholar]
- 10.Zhang S, Cordon-Cardo C, Zhang HS, Reuter VE, Adluri S, Hamilton WB, Lloyd KO, Livingston PO. Int J Cancer. 1997;73:42–49. doi: 10.1002/(sici)1097-0215(19970926)73:1<42::aid-ijc8>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 11.Zhang S, Zhang HS, Cordon-Cardo C, Reuter VE, Singhal AK, Lloyd KO, Livingston PO. Int J Cancer. 1997;73:50–56. doi: 10.1002/(sici)1097-0215(19970926)73:1<50::aid-ijc9>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 12.Mahal LK, Yarema KJ, Bertozzi CR. Science. 1997;276:1125–1128. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
- 13.Tai HC, Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. J Am Chem Soc. 2004;126:10500–10501. doi: 10.1021/ja047872b. [DOI] [PubMed] [Google Scholar]
- 14.Saxon E, Bertozzi CR. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 15.Sampathkumar SG, Li AV, Jones MB, Sun Z, Yarema KJ. Nat Chem Biol. 2006;2:149–152. doi: 10.1038/nchembio770. [DOI] [PubMed] [Google Scholar]
- 16.Agard NJ, Baskin JM, Prescher JA, Lo A, Bertozzi CR. ACS Chem Biol. 2006;1:644–648. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- 17.Agard NJ, Prescher JA, Bertozzi CR. J Am Chem Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 18.Rabuka D, Hubbard SC, Laughlin ST, Argade SP, Bertozzi CR. J Am Chem Soc. 2006;128:12078–12079. doi: 10.1021/ja064619y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawa M, Hsu TL, Itoh T, Sugiyama M, Hanson SR, Vogt PK, Wong CH. Proc Natl Acad Sci USA. 2006;103:12371–12376. doi: 10.1073/pnas.0605418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dube DH, Prescher JA, Quang CN, Bertozzi CR. Proc Natl Acad Sci USA. 2006;103:4819–4824. doi: 10.1073/pnas.0506855103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hang HC, Yu C, Kato DL, Bertozzi CR. Proc Natl Acad Sci USA. 2003;100:14846–14851. doi: 10.1073/pnas.2335201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becker DJ, Lowe JB. Glycobiology. 2003;13:41R–53R. doi: 10.1093/glycob/cwg054. [DOI] [PubMed] [Google Scholar]
- 23.Keppler OT, Horstkorte R, Pawlita M, Schmidt C, Reutter W. Glycobiology. 2001;11:11R–18R. doi: 10.1093/glycob/11.2.11r. [DOI] [PubMed] [Google Scholar]
- 24.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew Chem Int Ed Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 25.Wang Q, Chan TR, Hilgraf R, Fokin VV, Sharpless KB, Finn MG. J Am Chem Soc. 2003;125:3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 26.Jacobs CL, Yarema KJ, Mahal LK, Nauman DA, Charters NW, Bertozzi CR. Methods Enzymol. 2000;327:260–275. doi: 10.1016/s0076-6879(00)27282-0. [DOI] [PubMed] [Google Scholar]
- 27.Sarkar AK, Fritz TA, Taylor WH, Esko JD. Proc Natl Acad Sci USA. 1995;92:3323–3327. doi: 10.1073/pnas.92.8.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sivakumar K, Xie F, Cash BM, Long S, Barnhill HN, Wang Q. Org Lett. 2004;6:4603–4606. doi: 10.1021/ol047955x. [DOI] [PubMed] [Google Scholar]
- 29.Yarema KJ, Mahal LK, Bruehl RE, Rodriguez EC, Bertozzi CR. J Biol Chem. 1998;273:31168–31179. doi: 10.1074/jbc.273.47.31168. [DOI] [PubMed] [Google Scholar]
- 30.Speers AE, Cravatt BF. Chem Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 31.Hanson S, Best M, Bryan MC, Wong CH. Trends Biochem Sci. 2004;29:656–663. doi: 10.1016/j.tibs.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Luchansky SJ, Bertozzi CR. Chembiochem. 2004;5:1706–1709. doi: 10.1002/cbic.200400148. [DOI] [PubMed] [Google Scholar]
- 33.Fujihashi M, Peapus DH, Kamiya N, Nagata Y, Miki K. Biochemistry. 2003;42:11093–11099. doi: 10.1021/bi034983z. [DOI] [PubMed] [Google Scholar]
- 34.Wimmerova M, Mitchell E, Sanchez JF, Gautier C, Imberty A. J Biol Chem. 2003;278:27059–27067. doi: 10.1074/jbc.M302642200. [DOI] [PubMed] [Google Scholar]
- 35.Simanek EE, McGarvey GJ, Jablonowski JA, Wong CH. Chem Rev. 1998;98:833–862. doi: 10.1021/cr940226i. [DOI] [PubMed] [Google Scholar]
- 36.Yang L, McRae R, Henary MM, Patel R, Lai B, Vogt S, Fahrni CJ. Proc Natl Acad Sci USA. 2005;102:11179–11184. doi: 10.1073/pnas.0406547102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.