Abstract
Pairs of Au nanoparticles have recently been proposed as “plasmon rulers” based on the dependence of their light scattering on the interparticle distance. Preliminary work has suggested that plasmon rulers can be used to measure and monitor dynamic distance changes over the 1- to 100-nm length scale in biology. Here, we substantiate that plasmon rulers can be used to measure dynamical biophysical processes by applying the ruler to a system that has been investigated extensively by using ensemble kinetic measurements: the cleavage of DNA by the restriction enzyme EcoRV. Temporal resolutions of up to 240 Hz were obtained, and the end-to-end extension of up to 1,000 individual dsDNA enzyme substrates could be simultaneously monitored for hours. The kinetic parameters extracted from our single-molecule cleavage trajectories agree well with values obtained in bulk through other methods and confirm well known features of the cleavage process, such as DNA bending before cleavage. Previously unreported dynamical information is revealed as well, for instance, the degree of softening of the DNA just before cleavage. The unlimited lifetime, high temporal resolution, and high signal/noise ratio make the plasmon ruler a unique tool for studying macromolecular assemblies and conformational changes at the single-molecule level.
The optical characterization of the function and dynamics of single biomolecules such as molecular motors (1–3), RNA ribozymes (4), and DNA helicases and binding proteins (5, 6) is an important tool for biomedical research (reviewed in refs. 7–9). The predominant technique for optical single molecule studies is FRET between individual organic donor and acceptor dye molecules (10, 11). FRET has proven to be an effective tool for revealing the dynamics of molecular machines and monitoring their composition. However, conventional organic dyes typically photobleach after absorbing ≈107 photons and exhibit complex photophysics including long-lived dark-states (12). Single-molecule FRET studies thus remain challenging because of low signal/noise ratio, limited continuous observation time, limited accessible distance range, and probe blinking.
We recently reported an alternative method for dynamic distance measurements on the nanometer scale by using pairs of 40-nm gold nanoparticles (13). Gold nanoparticles efficiently scatter visible light and do not blink or photobleach. Their optical properties are controlled by their plasmons, which are collective oscillations of their conduction electrons. The plasmon wavelength can be tuned from blue into infrared (14, 15) by varying their shape and structure (hollow/solid). The plasmon wavelength is also sensitive to the proximity of other particles, because plasmons couple (16–23) in a distance-dependent matter. With decreasing interparticle distance, the plasmon resonance wavelength red-shifts (24) and the scattering cross-section increases (25). Plasmon coupling can be used for colorimetric detection of analytes in bulk, as pioneered by Mirkin and coworkers (18).
The distance dependence of plasmon coupling can also be used to monitor the spacing between two nanoparticles linked by DNA (13). Individual pairs of biopolymer-linked noble metal nanoparticles therefore act as “plasmon rulers.” The inherent brightness of plasmon rulers makes them good candidates for highly parallel single-molecule assays able to reveal the dynamics of biological processes and biopolymers. A drawback of plasmon rulers compared with FRET methods is the relatively large size of the nanoparticles compared with organic fluorophores (≈30–40 nm vs. ≈1 nm).
One experimental geometry where the drawback of larger probe size is outweighed by the advantages of unlimited lifetime and larger dynamic range are studies of DNA bending by proteins. DNA bending plays a crucial role in determining the specificity in DNA-protein recognition (26), transcription regulation (27–31), and DNA packaging (32, 33). Typically, these DNA-bending processes are quite slow (millisecond timescales), and it is desirable to monitor a particular DNA for extended durations (milliseconds to days), so that the effects of enzyme concentration, ionic strength and pH changes, and presence of cofactors can be explored.
Using plasmon rulers, we investigated the dynamics of the EcoRV-catalyzed DNA cleavage reaction in a highly parallel, high-bandwidth (up to 240 Hz) single-molecule assay. We picked the EcoRV restriction enzyme because it is a member of the type II restriction endonucleases, which are paradigms for the study of protein–nucleic acid interactions (34–37). EcoRV transiently bends its DNA substrate, and the bend angle is known from crystal structures to be ≈52° (38, 39). Using plasmon rulers, we were able to follow certain steps in the catalytic cycle of EcoRV and directly observe DNA bending immediately preceding cleavage, confirming the standard model of how this enzyme works. By analysis of the interparticle potentials, we were also able to see the softening of the DNA resulting from its interactions with the enzyme before cleavage.
Results and Discussion
Three technical challenges needed to be overcome to be able to use plasmon rulers to monitor the dynamics of single enzyme–DNA complexes. First, nonspecific protein–particle interactions needed to be suppressed. Second, methods were needed to synthesize homogeneous samples of plasmon rulers. Third, the plasmon rulers' temporal resolution needed to be significantly improved from its previous value of ≈1 Hz (13).
To eliminate nonspecific interactions between the gold surface and the enzyme, we used a stepwise ligand exchange strategy in which we passivated the particle surfaces with biocompatible (40) low-molecular-mass PEGs. We then synthesized DNA-linked nanoparticle dimers (plasmon rulers) with dsDNA spacers between 30 and 60 bp by using a liquid phase DNA-programmed assembly strategy [see supporting information (SI) Fig. 6].
Homogeneous samples of plasmon rulers were obtained by gel-electrophoretic purification (SI Fig. 6). Sample homogeneity and quality was confirmed by TEM and small-angle x-ray scattering (SAXS). The SAXS measurements were needed because interparticle distances in aqueous solution cannot be inferred from TEM measurements. The pair distribution functions of isolated plasmon rulers obtained by SAXS (SI Fig. 6) contain strong dimer peaks of separated 40-nm gold particles and thus confirm that our synthetic strategy leads to DNA tethered dimers (SI Fig. 6).
To increase the temporal resolution of the plasmon ruler measurements, we investigated the dependence of the scattering intensity on the interparticle distance. In previous applications of plasmon rulers, we used the distance dependence of the resonance wavelength to measure distances and distance changes in DNA (13, 24). However, the spectral analysis of individual plasmon rulers relies on the dispersion of light, forcing us to integrate photon counts for several hundred milliseconds to obtain a detectable signal in our set-up. This rate is too slow to resolve the dynamics of biological processes such as enzymatic DNA bending and cleavage, which typically occur on timescales of micro- to milliseconds.
We reasoned that intensities can be collected with a much higher temporal resolution than spectral information, because no dispersion is required. It can be shown that, like the plasmon resonance wavelength, the scattering cross-section of a coupled pair of dimers depends on the interparticle distance. In a first approximation, each particle is treated as a simple dipole. Using this assumption, the cross-section of two coupled particles with identical diameters is proportional to the square of the pair polarizability Λ, for which the following expression can be derived by averaging over longitudinal and perpendicular plasmon modes (25):
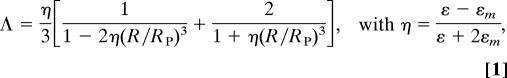 |
where ε is the wavelength-dependent dielectric function of gold (41), εm is the dielectric constant of the surrounding medium, R the interparticle distance, and RP is the particle radius. We set the dielectric constant of the medium to εm = 1.6, as before (24). Eq. 1 is a highly simplified description of the relationship between scattering cross-section and interparticle distance; among other things, we are using a dielectric function for an idealized bulk metal in an isotropic environment, and we are neglecting multipole contributions. Therefore, we expect qualitative rather than quantitative agreement of Eq. 1 with experiment.
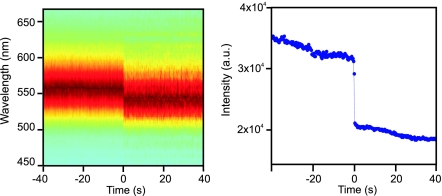
To validate this intensity-based approach, we recorded the scattering spectra of EcoRV-catalyzed DNA cleavage reactions by using an intensified CCD detector. The biological aspects of this experiment are described below; here it is only important to know that DNA cleavage by the EcoRV enzyme leads to plasmon ruler disassociation. As the DNA tethering the particles is cut, the interparticle distance suddenly increases, and consequently the scattering wavelength blue-shifts (Fig. 1) in a well understood manner (24). The particle plasmons cease to couple when the dimers dissociate, and consequently their resonance wavelengths and scattering cross-sections relax to the monomer values.
Fig. 1.
Correlation of plasmon resonance wavelength and scattering intensity. Scattering wavelength (Left) and integrated intensity (Right) during EcoRV-catalyzed DNA tether cleavage reaction recorded at 2 Hz. Dimer dissociation leads to a sudden spectral blueshift (Left) and a drop in total intensity (Right). Spectral shift and change in total intensity are highly correlated (Pearson correlation coefficient > 0.9).
To correlate the spectral shifts with changes in scattering intensity, we computed the total scattering intensities by integrating photon counts on the spectrometer over all wavelengths for six individual wavelength trajectories. The average Pearson correlation coefficient between integrated intensity and peak wavelength in individual trajectories is r = 0.92, confirming that color (peak wavelength) and scattering intensity are correlated and that the scattering intensity can be used to monitor interparticle distance. Ultimately, we achieved temporal resolutions of 240 Hz in a 20 × 20-μm field of view.
Highly Parallel Single EcoRV Restriction Digestion Assay.
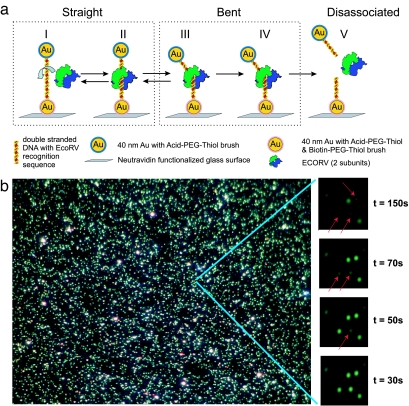
Having enriched samples of surface passivated plasmon rulers and a method for monitoring the ruler's interparticle separation with high temporal resolution, we set out to monitor the bending and cleavage dynamics of single-EcoRV enzymes. We immobilized plasmon rulers on the surface of a glass flow chamber via a biotin–Neutravidin bond between the biotin functionalized particle and surface (Fig. 2a). After equilibrating the chamber with 10 mM Mg2+ reaction buffer, we introduced EcoRV and monitored the plasmon rulers' color and intensity. Because of the brightness of the plasmon rulers (scattering cross-section of 40-nm particles, Csca = 130 nm2) (42), we could simultaneously monitor 500–1,500 plasmon rulers by using a low numerical aperture and low-magnification ×40 objective. The complete field of view in these experiments is limited to 150 × 100 μm by our camera and magnification.
Fig. 2.
Highly parallel single EcoRV restriction enzyme digestion assay. (a) The plasmon rulers are immobilized with one particle to a glass surface through biotin–Neutravidin chemistry. The homodimeric EcoRV enzyme binds nonspecifically to DNA bound between the particles (I), translocates and binds to the target site (II), bends the DNA at the target site by ≈50° (III), cuts the DNA in a blunt-ended fashion by phosphoryl transfer (54) (IV), and subsequently releases the products (V). (b) A 150 × 100-μm field of view with surface immobilized plasmon rulers. Individual dimers are visible as bright green dots. Dimer dissociation upon EcoRV-catalyzed DNA cleavage leads to a strong change in scattering intensities. The dimers are converted into monomers as shown for selected particles (red arrows, right side bar). EcoRV is added at t = 0 s. The restriction digestion in the complete field of view is provided in SI Movie 1.
Upon addition of the enzyme, some plasmon rulers exhibited sudden intensity drops (Fig. 2b and SI Movie 1). Green dots are individual dimers, and red arrows denote locations of these intensity drops. Complete detachment of the dimer from the surface cannot account for the observed intensity drops, because a dim green spot, corresponding to a single surface-immobilized gold particle, remains after the intensity drop. Rather, the ruler dissociation events are due to enzymatic DNA cleavage (see below).
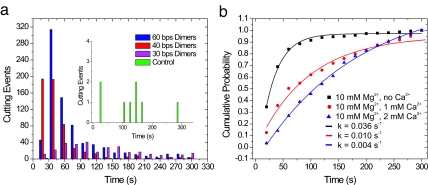
First, plasmon rulers constructed from DNA lacking the EcoRV restriction site do not dissociate (Fig. 3a): only 11 intensity drops are seen with the control DNA, whereas for dimers with the EcoRV recognition site, hundreds of dimer dissociations were observed. Second, the addition of Ca2+ [an inhibitor of DNA cleavage by EcoRV (43)] to the reaction buffer dramatically reduces the number of intensity drops (Fig. 3b). The first-order rate constants of the intensity drops decreased from 0.036 (no Ca2+) to 0.004 s−1 (2 mM Ca2+). Together, the observed sequence specificity and the inhibition of cleavage reaction by Ca2+ confirm that the observed dimer dissociation is the result of DNA cleavage by EcoRV.
Fig. 3.
Statistical analysis of EcoRV restriction digestion of plasmon rulers. (a) The cleavage reaction is highly specific. For all DNA spacers with an EcoRV restriction site, hundreds of cutting events are observed, and in the control experiments (60-bp dimer) without a restriction site, the cutting efficiency is almost zero. Each histogram contains the combined results of two independent cutting experiments per spacer length performed with similar surface coverage. (b) Percentage of plasmon rulers that have been cleaved as function of time for increasing Ca2+ concentrations. First-order kinetic fits are shown as continuous lines. EcoRV requires Mg2+ as a natural cofactor to catalyze DNA cleavage. Ca2+ can replace Mg2+ and facilitates formation of the enzyme–DNA complex, but the resulting complex does not catalyze the phosphodiester bond cleavage. Ca2+ inhibits the cleavage reaction in the presence of Mg2+. A Mg2+ concentration of 10 mM was retained throughout.
Plasmon rulers with 30-bp spacers exhibit a lower cutting rate than those with longer spacers (Fig. 3a), probably due to steric hindrance. According to the worm-like chain model for DNA (44) (contour length L = number of bases/10.5·3.4 nm, persistence length P = 53 nm; ref. 45), the interparticle distance for 30-bp DNA spacers is 9.4 nm compared with 12.4 and 18.3 nm for 40 and 60 bp, respectively. The PEG brush (L = 4.1 nm, P = 2 nm) adds ≈6 nm to the effective particle diameter, which leaves a gap of ≈3.4 nm for the enzyme in the case of the 30-bp spacer compared with 6.4 nm for the 40-bp spacer. It is known that the binding of one molecule of EcoRV covers ≈15 bp = 4.8 nm of DNA (38). Presumably, the binding step (Fig. 2a, step I) is sterically hindered by the PEG brush in the case of the 30-bp spacers, slightly reducing cleavage kinetics.
Magnitude of Intensity Changes Due to EcoRV-Catalyzed DNA Cleavage.
After assigning the sudden intensity drops as dimer disassociation resulting from DNA cleavage by EcoRV, we analyzed the intensity vs. time traces of individual cutting events at high temporal resolution. According to the standard model of DNA cleavage by EcoRV (Fig. 2a), DNA cutting occurs in a bent state (bend angle ≈ 52°). Based on an end-to-end distance from the worm-like chain model and a 52° bend, the interparticle distances in our geometries should decrease by 0.9 nm (Δ30bp), 1.0 nm (Δ40bp), and 1.3 nm (Δ60bp) due to DNA bending by EcoRV. Judging from the inverse pair polarizability–distance dependence given in Eq. 1, bending should slightly increase the scattering intensity in all three cases. Given that the predicted distance changes amount to only ≈10% of the DNA spacer length, this is a challenging test case for the sensitivity of the plasmon rulers.
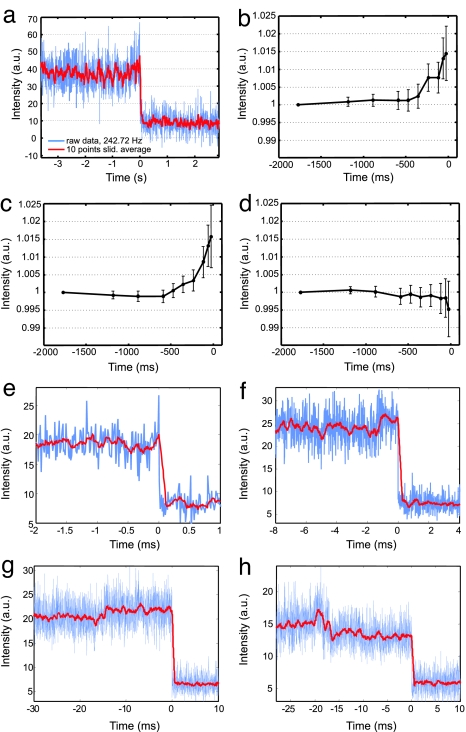
Our search for intensity changes due to bending was greatly facilitated by the large intensity drop upon dimer dissociation, because it allowed us to locate the disassociation time with high precision and thus also constrain the region that needed to be analyzed for evidence of DNA bending. Using the dissociation event as a fiduciary time-point, we calculated the average intensity preceding the dissociation from all recorded trajectories. Although temporal resolutions of 240 Hz could be achieved (see, e.g., Fig. 4a), we typically monitored the plasmon rulers' intensity at 85 Hz to increase the throughput with the larger field of view.
Fig. 4.
Single DNA cleavage events monitored by plasmon coupling. (a) Cleavage trajectory recorded at 240 Hz by using an intensified CCD detector. (b–d) Average scattering intensities at defined intervals preceding the dimer dissociation (0 ms) for 30 (b), 40 (c), and 60 (d) bp plasmon rulers (raw data recorded at 85 Hz). We set the time of the plasmon ruler dissociation equal to 0 ms and calculated average intensities for shortening time intervals between [−1,770 to 0] and [−24 to 0] ms. To account for different starting intensities, we normalized the trajectories and set the average intensities of the longest time interval equal to one for each trajectory [total number of trajectories: 86 (30 bp), 96 (40 bp), and 114 (60 bp)]. Error bars indicate standard errors of the mean. Increasing scattering intensities for 30- and 40-bp spacers are consistent with a decreasing interparticle distance due to bending in the enzyme–DNA complex. For the 60-bp spacers, the plasmon coupling is too weak to observe the bending (see text). The slight drop in the last data point of d is due to some uncertainty in defining the moment of dissociation. In some trajectories, points during or shortly after the dissociation were picked, leading to an overall lower scattering intensity. (e–h) Individual scattering trajectories for 40-bp plasmon rulers recorded at 85 Hz. The raw data are plotted in blue; sliding averages [over 10 (e), 25 (f), and 50 (g and h) frames] are included in red to guide the eye. A common feature observed in many trajectories is a slight increase in scattering intensity preceding dimer dissociation (0 s). The dwell times in the high-intensity state range from a few (e) to hundreds (f) of milliseconds to tens of seconds (g). (h) In ≈4% of the recorded trajectories, we first observed a slight drop in intensity before dimer dissociation. This result is consistent with subsequent cleavage event in plasmon rulers with multiple tethers.
The average intensities as a function of time are shown in Fig. 4 b–d. For both the 30- and 40-bp DNA spacers, there is a significant increase in average scattering intensity of up to 1.5% immediately before dimer dissociation. The measured average intensity changes [1.5 ± 0.8% (30 bp), 1.6 ± 0.9% (40 bp)] are in reasonable agreement with the order-of-magnitude calculation [Eq. 1, 2.4% (30 bp), 1.6% (40 bp)]. For the 30-bp spacer, the predicted value is slightly above the experimental error margin. For the rulers with largest initial interparticle separation (60 bp), no statistically significant intensity change was observed (predicted: 0.9%, measured: −0.5 ± 0.8%), consistent with the known inverse relationship between sensitivity and spacer length (24) and suggesting that the rulers' sensitivity was decreasing faster as predicted by Eq. 1. Given assumptions that went into the estimates obtained from Eq. 1, including the magnitude of the bend caused by the EcoRV enzyme, the end-to-end extension of the DNA, the thickness of the PEG brush, the uncertainty in the dielectric function, and the omission of multipole terms, the agreement between theory and experiment is considerably better than expected.
Is DNA bending by the EcoRV restriction enzyme the most probable explanation for observed intensity increases before the cut? The scattering intensity also depends, next to the interparticle distance, on the dielectric constant of the surrounding medium. It is therefore conceivable that enzyme binding (Fig. 2a, step II) and not DNA bending (step III) causes the observed intensity change by a local change of the refractive index. Therefore, we decoupled binding from bending by letting EcoRV react with 40-bp plasmon rulers in the absence of divalent metal ions (KD in the absence of Mg2+: ≈1 × 10−8 M) (46), preventing cleavage but not binding.‖ We then flushed the unbound enzyme out of the chamber with divalent-free buffer. Finally, we introduced Mg2+, allowing bound enzymes to cleave their DNA substrate. As shown in SI, these trajectories exhibit intensity jumps preceding dimer dissociation, confirming our interpretation of the intensity jumps as DNA bending and not protein–DNA binding or sticking of proteins to the gold particles. Thus, the high-intensity state most likely corresponds to steps III–IV in Fig. 2a in which the DNA is bent by 52°.
Individual traces show large variations in bending times. The trajectory shown in Fig. 4e exhibits a spike in intensity before the dimers dissociate. In this trajectory, only the very last frame before dissociation has an increased intensity, whereas in Fig. 4 f and g the dwell times are on the order of a few seconds (f) and a few tens of seconds (g) respectively. Student's t tests show that in 76% of all 40-bp dimer trajectories with “high intensity states” of >100 frames, high- and low-intensity states differ significantly (P < 0.05). In 4% of all collected trajectories, the large drop in intensity due to the DNA cleavage is preceded by a smaller intensity drop as shown in Fig. 4h. This behavior suggests two subsequent DNA cutting events in dimers that have two instead of one DNA tether. The cleavage of the first tether does not lead to the dissociation of the particles but only to a slight increase in equilibrium distance. The low probability of this behavior suggests that most of the dimers are indeed tethered by only one DNA molecule.
Dynamics of EcoRV-Catalyzed DNA Cleavage as Measured with Plasmon Rulers.
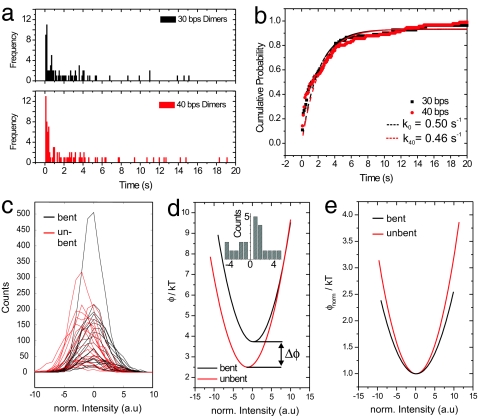
Having assigned the intensity jumps to DNA bending by the EcoRV enzyme and determined the average intensity changes before DNA cleavage/particle pair disassociation, we built up distributions of the duration of the high-intensity state for the 30- and 40-bp dimer trajectories. The results of the dwell time analyses are shown in Fig. 5a. The cumulative dissociation probability is plotted in Fig. 5b. In these plots the percentage of dimers that have been cleaved is plotted against the time they spend in the high-intensity state. Exponential fits to the decay curves in Fig. 5b give rate constants of k30 = 0.50 s−1 (30 bp, R2 = 0.94) and k40 = 0.46 s−1 (40 bp, R2 = 0.91). The similarity of the two rate constants is in agreement with the assertion that the trajectories comprise molecular events like DNA bending, DNA hydrolysis, and product release, but not DNA binding, which is expected to be much slower in case of the 30-bp spacer because of steric hindrance.
Fig. 5.
Kinetic and thermodynamic analysis of single-molecule data. (a and b) Dwell-time analysis of plasmon rulers in high-intensity state. (a) Histograms with a bin size of 100 ms for 30 (Upper) and 40 (Lower) bp plasmon rulers. (b) Cumulative probability of plasmon ruler dissociation. Plots show the percentage of dimers with dwell times less than the indicated time. First-order kinetic fits are included as dashed lines (k30 = 0.50 s−1, k40 = 0.46 s−1). (c) Histogram of normalized intensities for 21 individual cleavage trajectories of plasmon rulers with 40-bp DNA (selection criterion: system remains in bent state for >100 frames). The intensities are normalized to the maximum of the intensity distributions in the bent states (arbitrarily set to 0). Distributions of the bent state (black) are shifted to higher-intensity values compared with the straight state (red). (d) Average interaction potentials of the particles in plasmon rulers with bent (black) and unbent (red) DNA spacer obtained from c. The potential of the bent state is shifted to higher intensity (= shorter interparticle distance) and is offset in energy by Δφ = 1.2 kT. This energetic offset corresponds to the energy required for bending the DNA. The histogram shows the intensity distribution of the penultimate frames before dimer dissociation for all 21 trajectories. (e) Superposition of the interparticle potentials of the bent and unbent states show that the width of the bent potential is larger indicating a decreased DNA spring constant in the bent state.
The catalytic cycle of EcoRV with short (7–14 bp) substrate DNAs has been investigated in bulk by using FRET, stopped-flow fluorescence, fluorescence anisotropy, and quench-flow methods (46, 48, 49). These studies found that the turnover rate kcat is limited by rates of DNA hydrolysis and product release (Fig. 2a, steps IV and V), which are approximately equivalent in magnitude (46), whereas DNA bending is nearly simultaneous with binding. It is thus possible to compare k30 and k40 with kcat. The determined k30 and k40 are lower than the bulk steady state kcat = 0.7 s−1 for 10 mM Mg2+ and are closer to the bulk kcat for 4 mM Mg2+ (49). Our rate constants are very close to the first-order rate constant of 0.4 s−1 as obtained by FRET for the later stages of the reaction cycle (46). Thus, although deviations from the bulk rates are to be expected in our experiments (in our case, the DNA substrate is tethered on the surface and has less configurational freedom; the gold probes may slow the dynamics of the enzymatic reaction, and there may be a small nonzero force between the two particles**), such deviations are therefore small (not more than 0.2 s−1).
Besides elucidating the dynamics of DNA bending and cleavage by EcoRV, the scattering trajectories can also be used to estimate the interaction potential φ between the two DNA-linked nanoparticles. Just as the probability P of a DNA-tethered microsphere to be at a certain height h above a surface depends on the potential between the surface and the microsphere (51, 52), the probability of an individual plasmon ruler to be in a certain intensity state I is related to the interparticle potential φ. The interparticle potential φ determines the equilibrium interparticle distance. We used the relationship P(I) ∼ exp(−φ(I)/kT) to construct the potential φ in the bent and unbent states for the 40-bp plasmon rulers. P(I) is obtained from scattering intensity histograms of the high- and low-intensity states from single scattering trajectories with >100 frames in the high-intensity state (Fig. 5c). The resulting average potentials for the bent and unbent states are shown in Fig. 5d. The repulsive part of the interparticle potential is mainly determined by the surface coating of the particles and thus is identical for the bent and unbent states. Consequently, the arbitrary additive constant of the potentials has been chosen so that the repulsive parts of the potentials coincide. Comparing the two potentials reveals that the bent state is energetically disfavored by Δφ = 1.2kT.
This interpretation of the intensity distributions (Fig. 5c) must take various error sources into account. Specifically, the measured intensity distributions are convolutions of longitudinal and transverse fluctuations of the top particle with respect to the surface-bound particle and experimental noise. We are interested only in the variations of the interparticle separation, corresponding to the end-to-end distance of the tethering DNA. Assuming that the transverse fluctuations and the experimental noise are free from systematic errors, then their combined effect will be to broaden the intensity distributions. In consequence, the intensity distributions and all derived quantities represent upper bounds on the magnitude of the longitudinal fluctuations, and the potentials shown in Fig. 5d are lower bounds constraining the shape of the unknown and not directly measurable true potentials. Our Δφ of 1.2kT is therefore a lower bound on the true energy difference between the bent and straight states. Indeed, the worm-like chain predicts an energy difference of 1.6kT for a 52° kink in a 40-bp DNA molecule (53), exceeding our experimental value by 25%.
A second quantity, the relative change of the width of the two potentials, is less subject to various experimental errors because they should equally affect both intensity distributions and the assumption of coincidence of the repulsive parts of the potentials is not needed. Parabolic fits φ = 1 + bI2 to the superimposed potentials in Fig. 5e reveal that bbent is 27% smaller than bunbent (bbent = 0.016, bunbent = 0.022). The wider potential for the bent DNA state indicates that the interaction of the DNA with the enzyme decreases the spring constant of the DNA. This effect could account for the difference between the experimental and predicted Δφ and has mechanistic implications. By reducing the DNA stiffness, the enzyme decreases the energetic bias of the bent state and reduces the activation energies of the rate-determining steps, phosphate backbone hydrolysis and DNA dissociation, both of which occur in the bent state (Fig. 2a).
The plasmon ruler is a previously undescribed method for measuring dynamical distance changes in single-molecule biophysics experiments. In this work, we have taken an important step in validating the use of this type of ruler, by performing an in-depth study of DNA cleavage by the EcoRV restriction enzyme, a process that has been investigated by numerous established techniques. The plasmon ruler experiments recapitulate what is known from prior ensemble kinetic studies and single-molecule FRET experiments, indicating that the Au particles do not significantly perturb the system. The plasmon ruler also experiments afford the possibility of studying fluctuations in bending of DNA, which can be used to determine the potential energy change between the straight and bent DNA states. The plasmon ruler is now sufficiently well developed to be used as a tool to study a wide range of biological processes. The ability of the ruler to operate over extended time scales and to be effective in measuring distances over the 1- to 100-nm range make it a unique tool for studying macromolecular assemblies and conformational changes at the single-molecule level.
Methods
General.
Aqueous solutions of 40-nm gold particles with a concentration of 9.0 × 1010 particles per ml were from Ted Pella (Redding, CA). Freshly deionized water from a Millipore (Billerica, MA) water system was used for all experiments. All buffers were filtered with a 0.2-μm Millipore filter directly before use. Trithiolated oligonucleotides were from Fidelity Inc. (Gaithersburg, MD).
Passivation and Bulk Assembly of Plasmon Rulers.
We applied a scheme of sequential ligand exchanges to functionalize and passivate 40-nm gold particles (see SI Methods for details).
Optical Setup.
Single-molecule measurements were performed in an Axioplan2 upright microscope (Zeiss, Jena, Germany) with a darkfield condenser. The darkfield microscope as described (13, 24) was modified by the addition of an iXon EM+ CCD detector (512 × 512 pixel chip; Andor, South Windsor, CT) on the imaging port of the SpectraPro 2300i spectrometer (Acton, MA).
Surface Immobilization of Plasmon Rulers.
The flow chambers were prepared by first incubating for 15 min with a solution of 1 mg/ml BSA-biotin (Roche, Nutley, NJ) and then flushed with 300 μl T50 (50 mM NaCl/10 mM Tris, pH 8). Next, the chambers were reacted with a solution of 1 mg/ml Neutravidin (31000; Pierce, Rockford, IL) for 15 min and flushed with 300 μl of T50. The chambers then were incubated with SuperBlock (37515; Pierce) for 45 min and flushed again with 300 μl of T50. Next, a dilute solution of plasmon rulers was flushed into the chamber. The plasmon rulers bind only with biotinylated particle to the Neutravidin-coated glass surface, while the nonbiotinylated particle is free to move in solution. Control experiments verified that nonbiotinylated, pegylated particles did not stick to the passivated glass surfaces. Finally, the chamber was equilibrated with EcoRV reaction buffer (100 mM Tris·HCl/100 mM NaCl/10 mM MgCl2/1 mM DTT, pH 7.9).
Single-Molecule Experiments.
After equilibration of chambers with reaction buffer, 40 units of EcoRV restriction endonuclease (New England Biolabs, Beverly, MA) in 200 μl of reaction buffer was flushed into the chamber (≈1.75 nM EcoRV). The scattering intensities of the plasmon rulers in the field of view were continuously monitored with a color camera (Coolsnap cf.; Roper Scientific, Trenton, NJ) or for faster temporal resolution with the Andor CCD detector. To correlate intensity and wavelength information, we recorded the scattering spectra of individual plasmon rulers during the EcoRV-catalyzed cleavage reaction. To prebind EcoRV enzyme to plasmon rulers, we incubated plasmon rulers with EcoRV (4.4 nM) for ≈1 min in Mg2+ free buffer (50 mM Tris·Hcl/100 mM NaCl). Then we flushed the chamber with the buffer to remove excess enzyme. Reaction buffer containing Mg2+ then was introduced, and we started recording.
Data Analysis.
By inspecting the movies recorded with the Andor CCD detector using software from the manufacturer, we detected all dimers in the field of view that showed a sudden intensity drop. The movies were then imported into Matlab (Mathworks, Natick, MA), where the exposed pixels were manually marked with a box. The CCD detector has a pixel size of 16 × 16 μm, and most of the light scattered from individual plasmon rulers were collected in one pixel. The intensity trajectories of individual plasmon rulers were calculated as sequence of the most intense pixels in the defined boxes. All recorded trajectories contained a clear drop in intensity that was set to 0 ms. For analysis of average scattering intensity before dimer dissociation (recorded at 85 Hz), we superimposed all trajectories for a given spacer length. First, we calculated the average scattering intensities over defined intervals preceding dimer dissociation for each trajectory (interval lengths: 1770.0, 1180.0, 885.0, 559.0, 472.0, 354.0, 236.0, 118.0, 59.0, and 23.6 ms). Within each trajectory, the average intensity of the longest interval was set to 1, and the normalized intensities for each interval were averaged over all trajectories.
Supplementary Material
Acknowledgments
We thank Merek Siu, Stephanie Hsiao, and Harish Agarwal. This work was supported in part by the University of California, Berkeley (J.L.), the Hellman Faculty Fund (J.L.), the Sloan and Searle foundations (J.L.), and the U.S. Department of Energy, Energy Biosciences Program and the Director of Basic Energy Biosciences, Materials Science, and Engineering Division Contract DE-AC02-05CH11231 (to A.P.A.).
Abbreviation
- SAXS
small-angle x-ray scattering.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607826104/DC1.
In the absence of divalent metal ions, EcoRV endonuclease binds nonspecifically to DNA, with no preference for its recognition site over any other (47). It is known from crystal structures that in noncognate DNA–EcoRV restriction enzyme complexes the DNA retains a B configuration-like structure and is not bent (38). This was confirmed by bulk FRET measurements that did not show an intensity change upon addition of EcoRV without divalent ions (46).
The gold probes might also slow the dynamics of the enzymatic reaction by perturbation of the two rate-determining steps, DNA hydrolysis and product release. Based on the Stokes-Einstein approximation, the diffusion coefficient of a free 40-nm particle in water at 298 K is ≈1.1 × 10−11 m2·s, and it has an rms displacement of 4.7 nm/μs. For estimation of the force exerted on the DNA by the nonimmobilized particles, we assume that displacement occurs completely along the interparticle axis. In that case a pertubating force of ≈1.6 pN could act on the enzyme–DNA complex. Wuite and coworkers (50) investigated the efficiency of DNA cleavage by EcoRV under tension and showed that cleavage efficiency is not affected by forces significantly below 30 pN.
References
- 1.Itoh H, Takahashi A, Adachi K, Noji H, Yasuda R, Yoshida M, Kinosita K. Nature. 2004;427:465–468. doi: 10.1038/nature02212. [DOI] [PubMed] [Google Scholar]
- 2.Kural C, Kim H, Syed S, Goshima G, Gelfand VI, Selvin PR. Science. 2005;308:1469–1472. doi: 10.1126/science.1108408. [DOI] [PubMed] [Google Scholar]
- 3.Yildiz A, Tomishige M, Vale RD, Selvin PR. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 4.Zhuang XW, Kim H, Pereira MJB, Babcock HP, Walter NG, Chu S. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 5.Bianco PR, Brewer LR, Corzett M, Balhorn R, Yeh Y, Kowalczykowski SC, Baskin RJ. Nature. 2001;409:374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 6.Shivashankar GV, Libchaber A. Biophys J. 1998;74:A242–A242. [Google Scholar]
- 7.Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 8.Weiss S. Nat Struct Biol. 2000;7:724–729. doi: 10.1038/78941. [DOI] [PubMed] [Google Scholar]
- 9.Xie XS, Lu HP. J Biol Chem. 1999;274:15967–15970. doi: 10.1074/jbc.274.23.15967. [DOI] [PubMed] [Google Scholar]
- 10.Weiss S. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 11.Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Proc Natl Acad Sci USA. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubois A, Canva M, Brun A, Chaput F, Boilot JP. Appl Opt. 1996;35:3193–3199. doi: 10.1364/AO.35.003193. [DOI] [PubMed] [Google Scholar]
- 13.Sonnichsen C, Reinhard BM, Liphardt J, Alivisatos AP. Nat Biotechnol. 2005;23:741–745. doi: 10.1038/nbt1100. [DOI] [PubMed] [Google Scholar]
- 14.Halas N. MRS Bull. 2005;30:362–367. [Google Scholar]
- 15.Hirsch LR, Gobin AM, Lowery AR, Tam F, Drezek RA, Halas NJ, West JL. Ann Biomed Eng. 2006;34:15–22. doi: 10.1007/s10439-005-9001-8. [DOI] [PubMed] [Google Scholar]
- 16.Kelly KL, Coronado E, Zhao LL, Schatz GC. J Phys Chem B. 2003;107:668–677. [Google Scholar]
- 17.Khlebtsov B, Melnikov A, Zharov V, Khlebtsov N. Nanotechnology. 2006;17:1437–1445. [Google Scholar]
- 18.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 19.Nordlander P, Prodan E. Nano Lett. 2004;4:2209–2213. [Google Scholar]
- 20.Maier SA, Brongersma ML, Kik PG, Atwater HA. Phys Rev B. 2002;65:193408. [Google Scholar]
- 21.Rechberger W, Hohenau A, Leitner A, Krenn JR, Lamprecht B, Aussenegg FR. Opt Commun. 2003;220:137–141. [Google Scholar]
- 22.Prodan E, Radloff C, Halas NJ, Nordlander P. Science. 2003;302:419–422. doi: 10.1126/science.1089171. [DOI] [PubMed] [Google Scholar]
- 23.Haynes CL, McFarland AD, Zhao LL, Van Duyne RP, Schatz GC, Gunnarsson L, Prikulis J, Kasemo B, Kall M. J Phys Chem B. 2003;107:7337–7342. [Google Scholar]
- 24.Reinhard BM, Siu M, Agarwal H, Alivisatos AP, Liphardt J. Nano Lett. 2005;5:2246–2252. doi: 10.1021/nl051592s. [DOI] [PubMed] [Google Scholar]
- 25.Kreibig U, Vollmer M. Optical Properties of Metal Clusters. Berlin: Springer; 1995. [Google Scholar]
- 26.Bloomfield VA, Crothers DM, Tinoco J, Ignacio . Nucleic Acids, Structures, Properties, and Functions. Sausalito, CA: University Science Books; 2000. [Google Scholar]
- 27.Chen L, Glover JNM, Hogan PG, Rao A, Harrison SC. Nature. 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- 28.Love JJ, Li XA, Case DA, Giese K, Grosschedl R, Wright PE. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 29.Gartenberg MR, Crothers DM. Nature. 1988;333:824–829. doi: 10.1038/333824a0. [DOI] [PubMed] [Google Scholar]
- 30.Ansari AZ, Bradner JE, Ohalloran TV. Nature. 1995;374:371–375. doi: 10.1038/374370a0. [DOI] [PubMed] [Google Scholar]
- 31.Rappas M, Schumacher J, Beuron F, Niwa H, Bordes P, Wigneshweraraj S, Keetch CA, Robinson CV, Buck M, Zhang XD. Science. 2005;307:1972–1975. doi: 10.1126/science.1105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Travers A, Drew H. Biopolymers. 1997;44:423–433. doi: 10.1002/(SICI)1097-0282(1997)44:4<423::AID-BIP6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 33.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 34.Perona JJ. Methods. 2002;28:353–364. doi: 10.1016/s1046-2023(02)00242-6. [DOI] [PubMed] [Google Scholar]
- 35.Pingoud A, Jeltsch A. Eur J Biochem. 1997;246:1–22. doi: 10.1111/j.1432-1033.1997.t01-6-00001.x. [DOI] [PubMed] [Google Scholar]
- 36.Wilcox DE. Chem Rev. 1996;96:2435–2458. doi: 10.1021/cr950043b. [DOI] [PubMed] [Google Scholar]
- 37.Vipond IB, Halford SE. Mol Microbiol. 1993;9:225–231. doi: 10.1111/j.1365-2958.1993.tb01685.x. [DOI] [PubMed] [Google Scholar]
- 38.Winkler FK, Banner DW, Oefner C, Tsernoglou D, Brown RS, Heathman SP, Bryan RK, Martin PD, Petratos K, Wilson KS. EMBO J. 1993;12:1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perona JJ, Martin AM. J Mol Biol. 1997;273:207–225. doi: 10.1006/jmbi.1997.1315. [DOI] [PubMed] [Google Scholar]
- 40.Zheng M, Davidson F, Huang XY. J Am Chem Soc. 2003;125:7790–7791. doi: 10.1021/ja0350278. [DOI] [PubMed] [Google Scholar]
- 41.Johnson PB, Christy RW. Phys Rev B. 1972;6:4370–4379. [Google Scholar]
- 42.Yguerabide J, Yguerabide EE. Anal Biochem. 1998;262:157–176. doi: 10.1006/abio.1998.2760. [DOI] [PubMed] [Google Scholar]
- 43.Vipond IB, Baldwin GS, Halford SE. Biochemistry. 1995;34:697–704. doi: 10.1021/bi00002a037. [DOI] [PubMed] [Google Scholar]
- 44.Kratky O, Porod G. Recueil Des Travaux Chimiques Des Pays-Bas-Journal of the Royal Netherlands Chemical Society. 1949;68:1106–1122. [Google Scholar]
- 45.Smith SB, Cui YJ, Bustamante C. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 46.Hiller DA, Fogg JM, Martin AM, Beechem JM, Reich NO, Perona JJ. Biochemistry. 2003;42:14375–14385. doi: 10.1021/bi035520w. [DOI] [PubMed] [Google Scholar]
- 47.Taylor JD, Badcoe IG, Clarke AR, Halford SE. Biochemistry. 1991;30:8743–8753. doi: 10.1021/bi00100a005. [DOI] [PubMed] [Google Scholar]
- 48.Waters TR, Connolly BA. Biochemistry. 1994;33:1812–1819. doi: 10.1021/bi00173a026. [DOI] [PubMed] [Google Scholar]
- 49.Baldwin GS, Vipond IB, Halford SE. Biochemistry. 1995;34:705–714. doi: 10.1021/bi00002a038. [DOI] [PubMed] [Google Scholar]
- 50.van den Broek B, Noom MC, Wuite GJ. Nucleic Acids Res. 2005;33:2676–2684. doi: 10.1093/nar/gki565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh-Zocchi M, Dixit S, Ivanov V, Zocchi G. Proc Natl Acad Sci USA. 2003;100:7605–7610. doi: 10.1073/pnas.1337215100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zocchi G. Biophys J. 2001;81:2946–2953. doi: 10.1016/S0006-3495(01)75934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bustamante C, Bryant Z, Smith SB. Nature. 2003;421:423–427. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- 54.Sam MD, Perona JJ. Biochemistry. 1999;38:6576–6586. doi: 10.1021/bi9901580. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.