Abstract
Organ and tissue integrity is often maintained in animals by a specialized extracellular matrix structure called the basement membrane (BM). Accumulated evidence indicates that BM remodeling occurs during development and tumor invasion. Although the BM organizes and functions at the organ level, most past studies have explored its biochemical and in vitro properties. In this study, we monitor the BM in vivo during developmental tissue invasion for disc eversion and tumor invasion in Drosophila and modulate BM integrity with genetic alterations affecting either the whole organism or the targeted discs or tumors. We observe that the degradation of BM by the discs or the tumors is an early event during invasion processes and that preventing BM degradation completely blocks both tissue and tumor invasion, indicating that modulation of BM is essential for developmental and tumor invasion. Furthermore, elements of the invasion machinery, including JNK-induced matrix metalloproteinase (MMP) expression, are shared by both disc eversion and tumor invasion processes. Moreover, we show that although expression of MMP inhibitor, TIMP, is sufficient to halt developmental invasion, inhibition of proteases by both TIMP and RECK are required to block tumor invasion. These data suggest that tumor cells have a more robust invasion mechanism and could acquire metastatic behavior by co-opting developmental invasion programs. This type of co-option may be a general feature contributing to the progression of tumors. Finally, although past efforts using MMP inhibitors have not yielded much success, our results strongly argue that BM modulation could be a critical target for cancer therapy.
Keywords: developmental invasion, JNK signaling, tumor metastasis, matrix metalloproteinase
The basement membrane (BM) is a highly specialized form of extracellular matrix found on the basal side of polarized animal epithelia and is made up primarily of protein networks containing collagen and other glycoproteins (1). The importance of BM is inferred from the facts that this structure surrounds organs and tissues and is evolutionarily conserved from invertebrates to mammals. Proper regulation of BM is of critical importance for the functioning of many organs. Consistent with this notion, defects in BM are associated with several diseases, including thickened renal glomerular capillary and tubular BM in diabetes mellitus patients, ectopic expression of embryonic collagen in Alport's kidney syndrome, and loss or overproduction of BM components in muscular dystrophy and rheumatoid arthritis (2, 3). The observation that BM exhibits dynamic changes in developing tissues (4, 5) and in tumors (6) further suggests that modulation of BM could play a vital role in normal and pathologic processes and might be regulated by signaling events.
It has been suspected that modulation of BM could be an important step in tumor metastasis because the BM would be a barrier for tumor cells to leave their tissues of origin and invade different organs. The matrix metalloproteinases (MMPs) are zinc-dependent endopeptidases that have been shown to cleave components of the BM/extracellular matrix (7). The role of MMPs in tumor development and metastasis has been a focus of intense investigation over the past decade. However, it appears that MMPs play complex roles during tumor growth, and metastasis and several studies have shown that MMPs could have both pro- and antitumor growth and metastasis roles (8). Clinical trials targeting MMPs have yielded little success (8), which further clouds the role of MMP and BM in tumor growth and metastasis.
BM modulation could also play an important role during development. Much of the adult external structures in Drosophila are formed from sacs of larval epithelial cells called the imaginal discs. These discs initially hang inside the larval body cavity and must be brought to the outside during metamorphosis through an invasive process called disc eversion (9). Disc eversion requires cells to break out from imaginal tissues and break through the larval tissues, both engulfed by BM. However, other developmental invasion and migration processes do not appear to require cells to break through BM. Examples include the migration of the so-called epithelial border cells within the ovary during Drosophila oogenesis (10) and the migration of the germ cells in the embryo to reach their niche in the posterior region (11). Thus, it is not clear whether BM degradation is a critical step for initiating developmental invasion, in particular for disc eversion.
In addition to developmental invasion, Drosophila has also evolved into an important in vivo model to study tumor invasion. We have developed a genetic scheme for identifying and studying mutations that can cause otherwise noninvasive GFP-labeled tumors of the developing eye to exhibit metastatic behaviors (12). The fly tumors caused by oncogenic RasV12 and loss-of-cell polarity mutations exhibit strikingly similar metastatic hallmarks of malignant cells observed in human cancers. These behaviors include BM degradation, loss of E-cadherin expression, induction of cell migration, invasion of nearby tissues, and formation of distinct secondary foci.
Although the BM organizes and functions at the organ level, much of the past work had been focused on exploring biochemical and in vitro properties of BM (1). Taking the advantage that components of the BM as well as their regulators are conserved from flies to humans (13), we have explored the in vivo regulation of BM during disc eversion and tumor invasion. We show that BM degradation is essential for both developmental and tumor invasion, suggesting that modulation of BM degradation should still be a critical target for cancer therapy. We also find that disc eversion and tumor invasion share common invasive molecular machinery, further supporting the notion that tumor cells may hijack developmental programs for metastasis.
Results
Reduction of MMP Activities Results in Disc Eversion Defects.
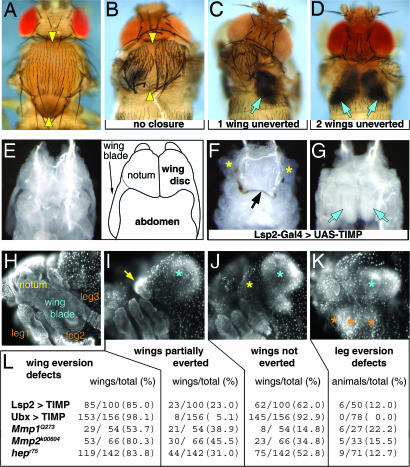
To investigate how BM remodeling contributes to tissue development, we chose to inhibit MMP function by targeted overexpression of the Drosophila tissue inhibitor of metalloproteases (TIMP) (14) using the UAS/GAL4 system (15). Overexpression of TIMP in the larval fat body, driven by Lsp2-GAL4 (Lsp2>TIMP) (16) and in the peripodial epithelium of the larval wing imaginal discs, by Ubx-GAL4 (Ubx>TIMP) (17), resulted in striking phenotypes that we focused on for further characterization [Fig. 1 and supporting information (SI) Fig. 6]. Despite predominant pupal lethality, dead pharates and adult escapers showed thoracic defects that ranged from clefts in the notum at the dorsal midline to thorax missing one or both heminota (Fig. 1 A–D). The missing heminota in these animals were found, upon dissection, to remain inside the animal together with the corresponding wing (Fig. 1 C and D). Similar phenotypes have been shown to result from defects in the eversion of imaginal wing discs, a process that takes place early in metamorphosis from 3.5 to 4 h after puparium formation (APF) (9).
Fig. 1.
Disc eversion requires MMP function. (A) Wild-type thorax. (B–D) A reduction of MMP function by overexpression of the MMP inhibitor TIMP in the fat body (Lsp2>TIMP) causes a range of thoracic phenotypes in pharate and adult escapers. (B) Failure in fusion of the discs at the dorsal midline (yellow arrowheads) results in a clefted thorax. (C and D) Lack of external structures derived from one or both wing discs, respectively, which remain inside the animal (blue arrows). (E–G) Disc eversion defects in the early pupa. (E) Dorsal view of a 6 h APF wild-type thorax with its schematic representation to the right. (F and G) TIMP overexpression can partially (F) or completely (G) halt disc eversion, as seen in pupae dissected 6 h APF. In F only the notum part of the wing disc has everted (yellow asterisks); failure of the heminota to fuse causes a dorsal hole encircled by a ribbon of necrotic tissue (black arrow). In G, the discs lie uneverted inside the animal (blue arrows). (H–K) Lateral views of 6 h APF pupae stained with DAPI to illustrate the range of disc eversion phenotypes observed in genotypes listed in L. (H) Wild-type pupa. (I) Partial eversion failure: the notum (yellow arrow), but not the rest of the wing disc (blue asterisk), is outside. (J) Complete failure of disc eversion: both the wing blade (blue asterisk) and the notum (yellow asterisk) are inside the animal. (K) Cases of leg eversion failure are also observed (orange asterisks). Complete genotypes in all figures are listed in SI Materials and Methods.
To confirm whether the phenotypes associated with overexpression of TIMP result from a failure in the invasion process of disc eversion and to assess their real penetrance, we looked at pupae 6 h APF, a time when disc eversion should be complete in wild-type animals and right before the onset of lethality for TIMP-overexpressing animals. Indeed, we discovered a range of disc eversion defects associated with the genotypes summarized in Fig. 1 I–L and SI Fig. 6. There are two MMP genes, Mmp1 and Mmp2, in Drosophila, and they have been shown to be required for the development of multiple tissues (13). Analysis of hypomorphic Mmp1 and Mmp2 mutant pupae also revealed similar disc eversion defects (Fig. 1L and SI Fig. 6), further supporting a requirement for MMP function in disc eversion. The thoracic phenotype obtained because of overexpression of TIMP is reminiscent of a defect in JNK signaling and JNK pathway mutants show a similar lack of disc eversion (9, 18).
Disc Eversion Defect Is Associated with Lack of BM Degradation.
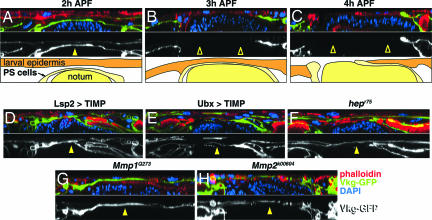
During disc eversion, the peripodial and stalk (PS) cells of the disc invade the larval epidermis. The first step in the invasive process is the basal-to-basal apposition of PS cells with the larval epidermis, which precedes the degradation of the BM in both epithelial layers (9). To monitor BM degradation we made use of a Viking-GFP protein fusion (GFP in the protein trap is fused with the Collagen IV protein from the viking gene) as a marker for the BM (19). Between 3 and 4 h APF, the BM is degraded in the wild-type animals (Fig. 2 A–C). However, the BM is not degraded in Mmp1Q273, Mmp2k00604, Lsp2>TIMP, and Ubx>TIMP pupae that fail to evert their wing discs, suggesting that the eversion defects are due to lack of BM degradation (Fig. 2 D, E, G, and H).
Fig. 2.
Disc eversion defects are associated with lack of BM degradation. (A–C) Confocal cross-sections of wild-type pupae. F-actin, nuclei, and BM are labeled with phalloidin (red), DAPI (blue), and Viking-GFP (green in Top and white pseudocolor in Middle), respectively. (A) During eversion, PS cells and the larval epidermis appose through their basal sides. (B) BM of both epithelial sheets degrades as eversion progresses. (C) This degradation precedes the invasion of the larval epidermis by the PS cells. The bottom panels schematically represent the tissues involved. (D–H) At 6 h APF, a time when disc eversion is complete in wild-type animals and BM is degraded, pupae that failed to evert their discs still present BM at the region where disc eversion takes place (yellow arrowheads). Arrowheads denote the presence (solid arrowheads) or absence (hollow arrowheads) of BM between the PS cells and the larval epidermis.
Because insufficient JNK signaling is also known to cause defective disc eversion (9, 18), we examined BM degradation in animals mutant for hemipterous (hep), which encodes the Drosophila JNK-kinase (20). In hepr75 pupae a highly penetrant failure of disc eversion (Fig. 1L and SI Fig. 6) is found, and this defect correlates with lack of BM degradation (Fig. 2F). This suggests a connection between MMP function and JNK signaling in BM degradation that we explored further.
MMPs Are Regulated by the JNK Signaling Pathway.
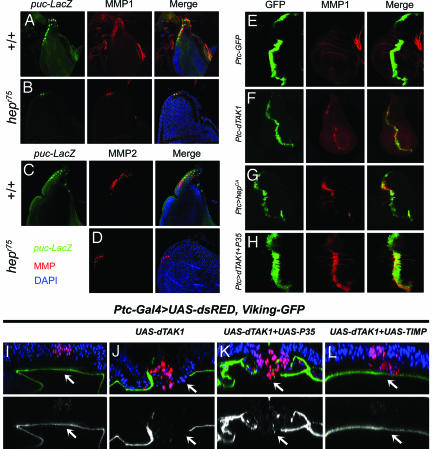
JNK signaling is known to drive, through the process of disc eversion, a partial epithelial-to-mesenchymal transition in PS cells. To check whether MMPs are expressed in PS cells we used an antibody for MMP1 (13) and a Gal4 enhancer trap in Mmp2 (SI Fig. 7). Both Mmp1 and Mmp2 are expressed in the stalk of wing imaginal discs (Fig. 3 A and C), coincident with the region of higher JNK activity among PS cells as assessed by the activation of an enhancer trap for the JNK target gene puckered (pucLacZ) (21). Furthermore, the expression of both MMPs is clearly reduced in the stalk of hepr75 mutant wing discs (Fig. 3 B and D). This indicates that JNK activity is necessary for normal MMP1 and MMP2 expression in PS cells.
Fig. 3.
JNK signaling pathway regulates MMP expression and BM degradation during development. (A–D) The stalk and the notum of the third instar larval wing disc are shown. (A) MMP1 is expressed (anti-MMP1 antibody, red) in the stalk of wing discs and colocalizes with puc-lacZ expression (green), a marker for JNK pathway activity. (B) In hepr75 mutant wing discs, JNK pathway activity is reduced, which results in substantially reduced MMP1 expression. (C) MMP2 is expressed in the stalk (MMP2>GFP, red) and partially colocalizes with puc-lacZ expression. (D) MMP2 expression is reduced in hepr75 mutant wing discs. (E–H) MMP1 expression is induced when JNK pathway is activated in a Ptc-Gal4 pattern (green) by overexpression of dTAK1 (F) and hepCA (G) but not in Ptc-Gal4 control (E). (H) The expression of MMP1 is more robust when cell death is blocked by p35. (I–L) JNK-mediated BM degradation can be suppressed by overexpressing TIMP. (I–L) Confocal cross-sections of wing discs. BM is marked by Viking-GFP (green in Upper and white pseudocolor in Lower). Ptc-Gal4-expressing cells are labeled with dsRED (red). Nuclei are stained with DAPI (blue). Activation of JNK pathway by overexpressing dTAK1 leads to degradation of the BM (J, arrow) in comparison to control (I). (K) Degradation of the BM is also seen when apoptosis associated with activated JNK pathway is suppressed by simultaneously expressing p35. (L) JNK signaling-induced BM degradation is suppressed by overexpressing TIMP.
To determine whether JNK signaling is sufficient to induce MMP expression, we activated the JNK pathway by overexpressing either the JNKK-kinase dTAK-1 (22) or a constitutively active form of the JNK-kinase Hemipterous (HepCA) (23) under the control of Ptc-Gal4 in a stripe of cells anterior to the A/P (anterior/posterior) boundary in the wing disc (Fig. 3E). To avoid cell death and lethality caused by constitutive activation of the JNK pathway, we used a temperature-sensitive Gal4 inhibitor, Gal80, to achieve transient expression (see SI Materials and Methods). Activation of the JNK pathway by overexpression of either dTAK1 (Fig. 3F) or hepCA (Fig. 3G) results in expression of MMP1. This expression is more robust when we coexpress the pan-caspase inhibitor p35 (24) (Fig. 3H), demonstrating that the expression of MMP1 in response to JNK pathway activation is not an indirect result of cell death. Similar activation is not seen for MMP2 (data not shown), suggesting that whereas JNK activity is sufficient to induce MMP1 expression, it is not sufficient for MMP2 expression in imaginal discs.
Activation of JNK Pathway Is Necessary and Sufficient for Inducing BM Degradation.
The lack of BM degradation during disc eversion in hepr75 animals (Fig. 2F) suggests that the JNK signaling may be able to trigger BM degradation in this process. To further test this possibility we overexpressed dTAK1 in discs that were also marked with Viking-GFP. Our results show that activation of the JNK pathway by overexpression of dTAK1 causes significant degradation of Viking-GFP (Fig. 3J compared with Fig. 3I) in a way that is again independent of cell death (Fig. 3K). To test whether this degradation was mediated by MMPs we coexpressed TIMP with dTAK1 and found that this resulted in inhibition of BM degradation (Fig. 3L), thus confirming that JNK-induced degradation of the BM is mediated by MMP activity.
Preventing BM Degradation Results in Suppression of Tumor Invasion.
Malignant transformation is a multistep process where various genetic alterations confer upon the affected cell, capabilities to breach the anticancer mechanisms of an organism. Hanahan and Weinberg (25) proposed six essential changes in cell physiology that lead to the breach of these mechanisms. One such critical change for cells to become malignant is the acquisition of the ability to degrade the BM.
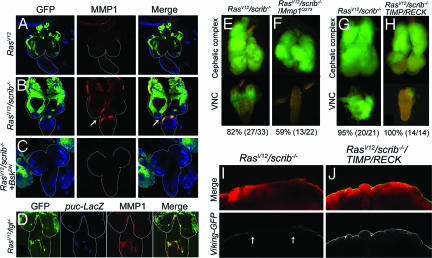
In Drosophila, cooperation between activated Ras (RasV12) and cell polarity mutations like scribbled (scrib) results in invasive behavior that is observed around day 8 after egg-laying in larval eye imaginal discs and is associated with BM degradation (12). It has also been shown that loss of cell polarity leads to activation of the JNK pathway (26). Staining of the tumors with an MMP1 antibody confirmed MMP1 up-regulation (Fig. 4B compared with Fig. 4A). This up-regulation is found across the thickness of the specimen as seen in several confocal sections in SI Fig. 8. This up-regulation was co-incident with the JNK pathway activation as assessed by puc-LacZ expression (Fig. 4D). For technical reasons the colocalization of the JNK pathway activation and MMP1 was looked at in lgl−/−/RasV12 clones (see SI Materials and Methods). Further, this up-regulation was suppressed by overexpression of a dominant negative form of the Jun-kinase basket (bskDN) (23) that reduces JNK pathway activity (Fig. 4C). To see whether MMP1 expression in the tumors was biologically relevant, we induced RasV12/scrib−/− clones in larvae mutant for Mmp1Q273, a hypomorphic allele of Mmp1 that survives past the third larval instar, and asked whether this would suppress the invasiveness of the tumors. This suppression of invasiveness was assessed by the inability of the tumors to invade the contiguous ventral nerve cord (VNC). Partial inhibition of invasive behavior was observed at day 8 (Fig. 4 E and F), suggesting that JNK-induced MMP1 expression contributes to invasive behavior in this Drosophila cancer model. A similar observation was also reported by Uhlirova and Bohmann (27). However, a complete suppression was not achieved as the tumors invaded the VNC beyond day 12 (data not shown). Consistent with this, overexpression of TIMP resulted in partial inhibition of the invasive phenotype, suggesting the possible involvement of other proteases in tumor invasion. A microarray study showed that another protease inhibitor, reversion-inducing-cysteine-rich protein with kazal motifs (RECK) (28), was down-regulated in invasive tumors (M. Wu, R.P., and T.X., data not shown). We thus tested whether inhibition of proteases by both TIMP and RECK could suppress tumor invasion. Indeed, co-overexpression of TIMP and RECK completely blocked VNC invasion (Fig. 4 G and H).
Fig. 4.
JNK pathway-mediated expression of MMP1 is required for invasive behavior and BM degradation in Drosophila tumors. (A–D) GFP marked clones of tumor cells (green) in the eye antennal discs. The brain hemispheres and the VNC are outlined. (A) MMP1 expression (red) is not detected in RasV12 cells, which do not exhibit invasive behavior. (B) RasV12/scrib−/− tumor cells invade the VNC and also express MMP1 (arrow). (C) Inhibition of JNK signaling in RasV12/scrib−/− tumor cells by overexpressing BskDN suppresses invasive behavior and MMP1 expression. (D) MMP1 expression and JNK activation (puc-LacZ, blue) colocalizes in the invasive tumor cells. (E–H) Suppression of tumor invasion by inhibition of MMP function. Upper parts of panels show tumor growth in the cephalic complex, and lower parts of panels show tumor invasion in VNC. The invasion of VNC by RasV12/scrib−/− tumor cells in 8-day-old larvae (E) is suppressed in the Mmp1Q273 homozygous background (F). (G and H) Overexpressing TIMP and RECK suppresses the invasive behavior in 12-day-old larvae. The percentage of animals displaying the phenotype shown is given below E–H. The raw numbers used to calculate the percentages are shown in parenthesis. (I and J) Degradation of BM in RasV12/scrib−/− tumors (I, arrows) is suppressed by overexpressing TIMP and RECK (J). Shown are RFP-labeled tumors (red) and Viking-GFP-labeled BM (green in Upper and white pseudocolor in Lower).
To see whether preventing BM degradation is the cause of the blocking of invasive behavior, we examined BM in RasV12/scrib−/− tumors that also expressed TIMP and RECK. Indeed, we observed that BM degradation is suppressed in these animals (Fig. 4 I and J and SI Fig. 9). These results indicate that BM degradation is critical for initiating tumor invasion, and multiple proteases are likely to be involved in the process.
Discussion
Disc Eversion Requires MMP-Mediated BM Degradation.
Deposition, remodeling, and degradation of BM are observed in many processes of epithelial morphogenesis. Two well studied examples in mice are palate fusion (29) and the branching of salivary glands (30). The major proposed effectors of BM degradation are MMPs. The functions of many of the different MMP genes found to date in mammals have been analyzed in genetically modified mice (31). These studies confirmed the involvement of several MMP genes in tumor progression. However, regarding MMP functions in normal development, the phenotypes of these mutant mice are quite subtle, with the exception of defective bone development in MMP14−/− mice (32). The existence of more than 20 MMPs in mammals has constrained genetic dissection of in vivo function (8, 31). Mutations in the two fly MMPs do not affect embryonic development but do affect tracheal adhesion, morphogenesis of the adult epidermis and larval gut histolysis larvae, and metamorphosis (13). Similar to mammalian MMPs, however, the relation of these phenotypes with the proposed function of MMPs in BM degradation has not been addressed.
Here we have discovered that BM regulation by MMPs is involved in disc eversion during Drosophila metamorphosis and provided direct genetic evidence of the involvement of MMPs in BM degradation during normal development. Disc eversion involves two epithelial sheets: the imaginal disc and the larval epidermis (9). The PS cells in the imaginal disc first appose to the larval epidermis. Then, PS cells invade and ultimately replace the larval epidermis at the surface of the animal. Mmp1 and Mmp2 mutants, as well as animals overexpressing the MMP inhibitor TIMP, show partial or complete failure in the eversion of the imaginal wing discs (Fig. 1 and SI Fig. 6). The failure of disc eversion correlates with persistence of BM in between PS cells and the larval epidermis (Fig. 2). Furthermore, Mmp1 and Mmp2 are expressed in PS cells (Fig. 3 A and C) and are both required for disc eversion, suggesting that they may have different substrates. Our findings, thus, show that MMP expression in PS cells drives degradation of the BM of both PS cells and the apposed larval epidermis, which is essential for this invasive developmental process. Another developmental invasion process is the penetration of the wing disc by the trachea (33). It would be interesting to learn whether a similar mechanism is involved.
JNK Regulates BM Degradation During Development Through Activation of MMP Expression.
The dynamic change of BM during development suggests that it is regulated by signaling events. JNK signaling is a leading candidate regulating the process during thoracic development. JNK activity in PS cells is required for disc eversion and their subsequent closure to form the adult epidermis (18). Similar to Mmp1 and Mmp2 mutants, animals mutant for dJNK-kinase hep also show partial or complete failure of wing disc eversion, along with persistence of the BM between PS cells and the larval epidermis (Fig. 2). Furthermore, expression of Mmp genes in the wing disc is reduced in hep mutants (Fig. 3 B and D). Moreover, JNK signaling can activate expression of MMP1 (but not MMP2) and effectively cause BM degradation (Fig. 3 E–K). These findings show that JNK signaling regulates BM degradation by controlling MMP expression, providing a molecular explanation for how JNK signal regulates disc eversion.
Co-Option of a Developmental Invasion Program by Tumor Cells.
It is starting to become increasingly clear that whole modules of genes, specifying developmental actions, are co-opted in tumor cells to reenact complete developmental programs. One example is the stem cell program (34). Tumor cells with stem cell characteristics display undifferentiation and unlimited proliferation potential. Another example is epithelial-to-mesenchymal transition (EMT) (35). The invasive behavior of epithelial cancers resembles EMT during development. The networks of genes controlled by the transcription factors Snail (36, 37) and Twist (38), which drive EMT during development both in Drosophila and mammals, are expressed in tumor cells.
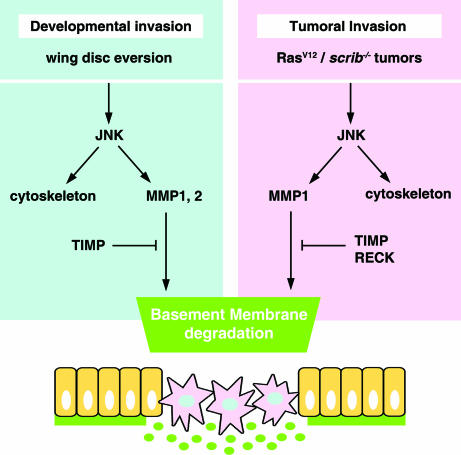
Our observation that JNK signal controls a developmental invasion process by inducing MMP expression and BM degradation suggests that the JNK/MMP invasion module could be co-opted by tumor cells (Fig. 5). Consistent with this hypothesis, it has been recently shown that JNK activity is required for the invasive and metastatic behavior of tumors in Drosophila (12, 26, 39). We observed that JNK activation induces MMP1 expression and MMP1 in turn promotes the invasive behavior of the tumor (Fig. 4 A–F). Thus, it seems that JNK signaling, once activated by loss of cell polarity (26), is driving the invasive behavior of the tumor cells in a way similar to how it does during disc eversion in PS cells.
Fig. 5.
Co-option of a developmental pathway by invasive tumors. A model summarizing the relationships between JNK pathway, MMPs, TIMP, and RECK and their role in BM degradation during the developmental invasive process of disc eversion and tumor invasion. During the disc eversion process, both MMP1 and MMP2 are under the control of the JNK pathway and result in BM degradation. In tumor invasion, part of the developmental pathway is hijacked to affect BM degradation and invasiveness. MMP1 and MMP2 function is inhibited by TIMP and/or RECK. It is possible that RECK could also block other proteases activated by RasV12 and/or scrib−/−, which are independent from JNK signaling.
JNK signaling in Drosophila is known to be involved, besides disc eversion and closure, in embryonic dorsal closure (40) and wound healing (41). In these processes JNK is active in epithelial cells regulating their adhesive and migratory properties. There is also evidence in vertebrates for a role of JNK in several processes of epithelial morphogenesis and in wounds (42–44), which raises the possibility of a conserved module of genes exerting epithelial remodeling under the control of JNK as a “master signal.” The contribution of JNK to tumor progression could include the activation of MMPs and other genes driving migration and invasion in a way similar to disc eversion and closure (Fig. 5).
Preventing BM Degradation Blocks Tumor Invasion.
In contrast to the effect on disc eversion, TIMP overexpression or Mmp1 mutant could only partially suppress tumor invasion. A total block of invasiveness was achieved, however, when we coexpressed more than one protease inhibitor (TIMP and RECK) (Fig. 4 G and H). We found that the size of these TIMP and RECK overexpressing tumors was the same as controls. In addition, same as controls, the cells in these tumors still lost epithelial characteristics. Yet contrary to controls, BM degradation was suppressed (Fig. 4 I and J). This supports previous observations in this cancer model that tumor growth and tumor invasiveness can be separated (26). Our data also show that the degradation of the BM surrounding the tumors or the wing disc is a prerequisite for invading other tissues. Very importantly, these results also indicate that, apart from MMPs, additional proteases contribute to the degradation of BM in tumors. Although clinical trials for MMP inhibitors in cancer treatment have not yielded much success so far (8), our results argue that modulation of BM is indeed a potent target for tumor progression, and perhaps new protease or JNK inhibitors for cancer should still be considered.
Materials and Methods
Strains and Crosses.
All crosses were carried out at 25°C on standard medium unless otherwise specified. The genotypes of the animals are provided in SI Materials and Methods. In Fig. 3 E–L, Tubulin Gal80ts was used to either inhibit or allow Gal4 activity at 18°C and 29°C, respectively. Transgenes for JNK pathway components were activated for 20–24 h before wandering third instar larvae were dissected and fixed. Invasive tumors were analyzed according to ref. 12.
Drosophila RECK cDNA (CG5392) was cloned into EcoRI of pUAST to generate UAS-RECK.
Immunohistochemistry.
Tissues were fixed and stained according to ref. 12 with the following antibodies: rabbit anti-β-galactosidase (Cappel, Durham, NC; 1:200 dilution, overnight), mouse anti-MMP1 (gift of A. Page-McCaw, Rensselaer Polytechnic Institute; 1:40 dilution, overnight), and secondary Cy3-FITC antibodies (Jackson Immuno Research, West Grove, PA; 1:250 dilution for 90 min). Tissues were mounted in a drop of Vectashield-DAPI (Vector Laboratories, Burlingame, CA). A Zeiss LSM510 Meta Confocal Microscope (Zeiss, Jena, Germany) was used for analysis.
Supplementary Material
Acknowledgments
We thank A. Page-McCaw, K. Irvine, L. S. Shashidhara, and W. Wang for reagents and strains and L. Xue, D. Nguyen, and T.X. laboratory members for comments. A.S. and J.C.P.-P. were supported by the Anna Fuller Fund Postdoctoral Fellowship in Molecular Oncology and the Spanish Ministry of Education postdoctoral fellowship, respectively. T.I. was supported in part by a fellowship from the Yamanouchi Foundation for Research on Metabolic Disorders and by the long-term fellowship from the Human Frontier Science Program. This work was supported by a grant from the National Institutes of Health, National Cancer Institute (to T.X.). T.X. is a Howard Hughes Medical Institute Investigator.
Abbreviations
- BM
basement membrane
- MMP
matrix metalloproteinase
- PS
peripodial and stalk
- APF
after puparium formation
- VNC
ventral nerve cord.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611666104/DC1.
References
- 1.Quondamatteo F. Histochem J. 2002;34:369–381. doi: 10.1023/a:1023675619251. [DOI] [PubMed] [Google Scholar]
- 2.Bedossa P, Paradis V. J Pathol. 2003;200:504–515. doi: 10.1002/path.1397. [DOI] [PubMed] [Google Scholar]
- 3.Tsilibary EC. J Pathol. 2003;200:537–546. doi: 10.1002/path.1439. [DOI] [PubMed] [Google Scholar]
- 4.Medioni C, Noselli S. Development (Cambridge, UK) 2005;132:3069–3077. doi: 10.1242/dev.01886. [DOI] [PubMed] [Google Scholar]
- 5.Werb Z, Chin JR. Ann NY Acad Sci. 1998;857:110–118. doi: 10.1111/j.1749-6632.1998.tb10111.x. [DOI] [PubMed] [Google Scholar]
- 6.Egeblad M, Werb Z. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 7.Mott JD, Werb Z. Curr Opin Cell Biol. 2004;16:558–564. doi: 10.1016/j.ceb.2004.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Overall CM, Kleifeld O. Nat Rev Cancer. 2006;6:227–239. doi: 10.1038/nrc1821. [DOI] [PubMed] [Google Scholar]
- 9.Pastor-Pareja JC, Grawe F, Martin-Blanco E, Garcia-Bellido A. Dev Cell. 2004;7:387–399. doi: 10.1016/j.devcel.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 10.Montell DJ. Nat Rev Mol Cell Biol. 2003;4:13–24. doi: 10.1038/nrm1006. [DOI] [PubMed] [Google Scholar]
- 11.Santos AC, Lehmann R. Curr Biol. 2004;14:R578–R589. doi: 10.1016/j.cub.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 12.Pagliarini RA, Xu T. Science. 2003;302:1227–1231. doi: 10.1126/science.1088474. [DOI] [PubMed] [Google Scholar]
- 13.Page-McCaw A, Serano J, Sante JM, Rubin GM. Dev Cell. 2003;4:95–106. doi: 10.1016/s1534-5807(02)00400-8. [DOI] [PubMed] [Google Scholar]
- 14.Godenschwege TA, Pohar N, Buchner S, Buchner E. Eur J Cell Biol. 2000;79:495–501. doi: 10.1078/0171-9335-00072. [DOI] [PubMed] [Google Scholar]
- 15.Brand AH, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 16.Cherbas L, Hu X, Zhimulev I, Belyaeva E, Cherbas P. Development (Cambridge, UK) 2003;130:271–284. doi: 10.1242/dev.00205. [DOI] [PubMed] [Google Scholar]
- 17.Pallavi SK, Shashidhara LS. Development (Cambridge, UK) 2003;130:4931–4941. doi: 10.1242/dev.00719. [DOI] [PubMed] [Google Scholar]
- 18.Agnes F, Suzanne M, Noselli S. Development (Cambridge, UK) 1999;126:5453–5462. doi: 10.1242/dev.126.23.5453. [DOI] [PubMed] [Google Scholar]
- 19.Morin X, Daneman R, Zavortink M, Chia W. Proc Natl Acad Sci USA. 2001;98:15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glise B, Bourbon H, Noselli S. Cell. 1995;83:451–461. doi: 10.1016/0092-8674(95)90123-x. [DOI] [PubMed] [Google Scholar]
- 21.Martin-Blanco E, Pastor-Pareja JC, Garcia-Bellido A. Proc Natl Acad Sci USA. 2000;97:7888–7893. doi: 10.1073/pnas.97.14.7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mihaly J, Kockel L, Gaengel K, Weber U, Bohmann D, Mlodzik M. Mech Dev. 2001;102:67–79. doi: 10.1016/s0925-4773(01)00285-4. [DOI] [PubMed] [Google Scholar]
- 23.Weber U, Paricio N, Mlodzik M. Development (Cambridge, UK) 2000;127:3619–3629. doi: 10.1242/dev.127.16.3619. [DOI] [PubMed] [Google Scholar]
- 24.Hay BA, Wolff T, Rubin GM. Development (Cambridge, UK) 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- 25.Hanahan D, Weinberg RA. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 26.Igaki T, Pagliarini RA, Xu T. Curr Biol. 2006;16:1139–1146. doi: 10.1016/j.cub.2006.04.042. [DOI] [PubMed] [Google Scholar]
- 27.Uhlirova M, Bohmann D. EMBO J. 2006;25:5294–5304. doi: 10.1038/sj.emboj.7601401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh J, Takahashi R, Kondo S, Mizoguchi A, Adachi E, Sasahara RM, Nishimura S, Imamura Y, Kitayama H, Alexander DB, et al. Cell. 2001;107:789–800. doi: 10.1016/s0092-8674(01)00597-9. [DOI] [PubMed] [Google Scholar]
- 29.Kaartinen V, Cui XM, Heisterkamp N, Groffen J, Shuler CF. Dev Dyn. 1997;209:255–260. doi: 10.1002/(SICI)1097-0177(199707)209:3<255::AID-AJA1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 30.Sakai T, Larsen M, Yamada KM. Nature. 2003;423:876–881. doi: 10.1038/nature01712. [DOI] [PubMed] [Google Scholar]
- 31.Sternlicht MD, Werb Z. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holmbeck K, Bianco P, Caterina J, Yamada S, Kromer M, Kuznetsov SA, Mankani M, Robey PG, Poole AR, Pidoux I, et al. Cell. 1999;99:81–92. doi: 10.1016/s0092-8674(00)80064-1. [DOI] [PubMed] [Google Scholar]
- 33.Sato M, Kornberg TB. Dev Cell. 2002;3:195–207. doi: 10.1016/s1534-5807(02)00202-2. [DOI] [PubMed] [Google Scholar]
- 34.Reya T, Morrison SJ, Clarke MF, Weissman IL. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 35.Thiery JP. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 36.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 37.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 38.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 39.Uhlirova M, Jasper H, Bohmann D. Proc Natl Acad Sci USA. 2005;102:13123–13128. doi: 10.1073/pnas.0504170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noselli S. Trends Genet. 1998;14:33–38. doi: 10.1016/S0168-9525(97)01320-6. [DOI] [PubMed] [Google Scholar]
- 41.Ramet M, Lanot R, Zachary D, Manfruelli P. Dev Biol. 2002;241:145–156. doi: 10.1006/dbio.2001.0502. [DOI] [PubMed] [Google Scholar]
- 42.Zenz R, Scheuch H, Martin P, Frank C, Eferl R, Kenner L, Sibilia M, Wagner EF. Dev Cell. 2003;4:879–889. doi: 10.1016/s1534-5807(03)00161-8. [DOI] [PubMed] [Google Scholar]
- 43.Weston CR, Wong A, Hall JP, Goad ME, Flavell RA, Davis RJ. Genes Dev. 2003;17:1271–1280. doi: 10.1101/gad.1087303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li G, Gustafson-Brown C, Hanks SK, Nason K, Arbeit JM, Pogliano K, Wisdom RM, Johnson RS. Dev Cell. 2003;4:865–877. doi: 10.1016/s1534-5807(03)00159-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.