Abstract
Wild barley (Hordeum spontaneum) represents a significant genetic resource for crop improvement in barley (Hordeum vulgare) and for the study of the evolution and domestication of plant populations. The Isa gene from barley has a putative role in plant defense. This gene encodes a bifunctional α-amylase/subtilisin inhibitor that inhibits the bacterial serine protease subtilisin, fungal xylanase, and the plant's own α-amylase. The inhibition of plant α-amylases suggests this protein may also be important for grain quality from a human perspective. We identified 16 SNPs in the coding region of the Isa locus of 178 wild barley accessions from eight climatically divergent sites across Israel. The pattern of SNPs suggested a large number of recombination events within this gene, indicating that the low-outcrossing rate of wild barley is not a barrier to recombinant haplotypes becoming established in the population. Seven amino acid substitutions were present in the coding region. Genetic diversity for each population was calculated by using Nei's diversity index, and a Spearman rank correlation was carried out to test the association between gene diversity and 16 ecogeographical factors. Highly significant correlations were found between diversity at the Isa locus and key water variables, evaporation, rainfall, humidity, and latitude. The pattern of association suggests selective sweeps in the wetter climates, with resulting low diversity and weaker selection or diversifying selection in the dryer climates resulting in much higher diversity.
Keywords: climatic selection, crop improvement, Hordeum spontaneum, Isa molecular polymorphism
Barley (Hordeum vulgare L.) has been cultivated for ≈10,000 years (1) and may have been the first plant domesticated by humans. It is a diploid (2n = 14) species and, along with the other important food plants, wheat and rye, belongs to the Triticeae tribe. Hordeum spontaneum is the wild progenitor of barley (2). Cultivated varieties are thought to have occurred through a single selection event (1), which has resulted in a depleted gene pool in cultivated barley (3, 4). Wild barley, therefore, is an important genetic resource for domesticated barley (5).
The Isa gene (6) codes for the protein bifunctional α-amylase/subtilisin inhibitor (BASI) (7), which inhibits a barley α-amylase AMY2 (found specifically in the Triticeae), the bacterial serine protease subtilisin (8), and fungal xylanase (9). This functional evidence combined with knowledge of BASI expression patterns in seed tissue (10), suggests the Isa gene has a role in pathogen resistance. The amount of genetic diversity at the Isa gene locus is significantly lower in cultivated barley than in wild populations (11), and recent studies involving DNA sequence polymorphism in functional genes from wild barley have revealed patterns of genetic variation consistent with processes of selection (11, 12).
Differences in genetic diversity among populations are derived from a complex evolutionary history involving multiple factors. Population bottlenecks, selection, genetic drift, mating system, and migration all have an effect on the genetic content of individuals and populations. Different loci in the same species also show different patterns of genetic diversity. Varied patterns of polymorphism and geographic structure are reported at different wild barley loci (12, 13). Selective association between genetic diversity in wild barley and ecogeographical factors has been shown by using applied fragment length polymorphism (14, 15) and microsatellite (16, 17) markers. We report here a study of diversity in the coding region of the Isa gene at the DNA sequence level.
Results
Genetic Diversity and Recombination.
Sixteen SNP sites defining 19 haplotypes were observed in the 584 bp of Isa coding sequence derived from the eight wild barley populations [supporting information (SI) Tables 5–12]. However, a sequence of sufficient quality was obtained for all of the 178 samples across a region covering only 14 SNP sites and defining 16 haplotypes spanning ≈500 bp of sequence (Table 1). These haplotypes were then used as the basis for frequency and genetic diversity calculations. Occurrence and frequency of haplotypes varied considerably among populations, with considerable variation also in genetic diversity index (He) at this locus (Table 2). Genetic diversity was found to be highest in the xeric populations of Sede Boqer and Wada Qilt, whereas lowest genetic diversity was found in Maalot and Mt. Meron. Based on Fisher's exact test on haplotype counts, each population was found to be significantly different from every other population (data not shown); from the 28 tests performed, 25 were significant at the 0.001 level, Mt. Meron/Tabigha Terra Rosa (TR) and Tabigha TR/Tabigha Basalt (B) (P = 0.003) were significant at the 0.01 level, and the subpopulations from the opposing slopes of “Evolution Canyon” were significant at the 0.05 level (P = 0.017).
Table 1.
SNP-based haplotypes of the coding region of the Isa gene found in populations of wild barley from Israel
| Base position* | 64 | 120 | 151 | 194 | 249 | 269 | 277 | 286 | 336 | 339 | 351 | 462 | 546 | 549 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hap 1 | C | C | G | A | G | T | T | A | G | C | T | C | T | C |
| Hap 2 | C | |||||||||||||
| Hap 3 | C | C | ||||||||||||
| Hap 4 | C | |||||||||||||
| Hap 5 | T | C | ||||||||||||
| Hap 6 | T | G | ||||||||||||
| Hap 7 | T | C | ||||||||||||
| Hap 8 | T | |||||||||||||
| Hap 9 | A | |||||||||||||
| Hap 10 | T | |||||||||||||
| Hap 11 | T | C | ||||||||||||
| Hap 12 | T | T | C | T | ||||||||||
| Hap 13 | T | C | C | G | ||||||||||
| Hap 14 | T | T | G | |||||||||||
| Hap 15 | T | T | C | G | C | |||||||||
| Hap 16 | C | G |
*Base positions are relative to the start codon of the Isa gene.
Table 2.
Frequency of haplotypes of the Isa gene in eight wild barley populations from Israel
| Haplotypes | Maalot | Mt. Meron | T (TR) | T (B) | EC (SF) | EC (NF) | W.Q. | S.B. |
|---|---|---|---|---|---|---|---|---|
| Sample size | 27 | 30 | 23 | 19 | 27 | 16 | 16 | 20 |
| Hap 1 (consensus) | 0.800 | 0.630 | 0.290 | 0.111 | 0.438 | 0.150 | ||
| Hap 2 | 0.889 | 0.033 | 0.050 | |||||
| Hap 3 | 0.111 | |||||||
| Hap 4 | 0.133 | 0.217 | 0.263 | 0.259 | 0.125 | 0.125 | 0.100 | |
| Hap 5 | 0.611 | 0.781 | 0.125 | |||||
| Hap 6 | 0.550 | |||||||
| Hap 7 | 0.152 | 0.447 | 0.050 | |||||
| Hap 8 | 0.063 | |||||||
| Hap 9 | 0.063 | |||||||
| Hap 10 | 0.063 | 0.125 | ||||||
| Hap 11 | 0.063 | |||||||
| Hap 12 | 0.033 | |||||||
| Hap 13 | 0.050 | |||||||
| Hap 14 | 0.031 | |||||||
| Hap 15 | 0.019 | |||||||
| Hap 16 | 0.050 | |||||||
| Number of haplotypes | 2 | 4 | 3 | 3 | 4 | 4 | 7 | 7 |
| He | 0.198 | 0.340 | 0.532 | 0.647 | 0.547 | 0.369 | 0.750 | 0.655 |
| Heterozygotes | 1 | 1 | 2 | 4 | 2 |
EC, “Evolution Canyon” (Nahal Oren); SF, south-facing slope; African slope, (AS); NF, north-facing slope; European slope (ES). T, Tabigha (Terra Rosa/Basalt), W.Q., Wadi Qilt; S.B., Sede Boqer. Blanks indicate a frequency of zero.
Examination of the position of SNP sites shows that 7 of the 16 SNPs identified are predicted to result in amino acid changes (position 64, C-T = Arg-Cys; position 151, G-T = Ala-Ser; position 194, A-T = His-Leu; position 269, T-C = Val-Ala; position 277, T-C = Ser-Pro; position 286, A-G = Ile-Val; and position 625, A-G = Ile-Val). For the 14 SNP sites on which there is complete sequence information, the first six of the seven positions (above) are predicted to result in amino acid substitutions. Random single-base mutations in codon triplets are calculated to cause amino acid changes in 76% of cases (11). For 14 SNP sites, ≈11 would be expected to result in amino acid changes under random expectations, and the observed six substitutions comprise a highly statistically significant departure from random expectations (P < 0.005, χ2 test). (For the full 16 SNP sites identified, 7 would be predicted to alter amino acid sequence, which is also highly significant with P < 0.005.) This evidence supports the hypothesis that selection has removed base substitutions (SNPs) that create detrimental amino acid changes.
Tests for a minimum number of recombination events, using the program RecMin (18), showed a lower bound of four recombination events using the algorithm of Hudson and Kaplan (19) and a lower bound of eight recombination events using the algorithm of Myers and Griffiths (18). In comparison, Bundock and Henry (11) found a single recombination event in this same region in a study of largely cultivated barley genotypes.
Correlation Between Gene Diversity and Environmental Variables.
Spearman rank correlation tests between genetic diversity (He) at the Isa locus and 16 environmental variables were carried out for the eight wild barley populations (Table 3). These tests show that gene diversity has significant (P < 0.05) positive correlation with evaporation and significant (P < 0.05) negative correlation with latitude, mean number of rainy days, mean annual rainfall, and mean annual humidity. There was highly significant (P < 0.01) negative correlation between gene diversity and mean annual rainfall and humidity. A very highly significant (P < 0.001) positive correlation was observed between genetic diversity and mean annual evaporation.
Table 3.
Correlation between genetic diversity at the Isa locus of wild barley and 16 environmental variables for six locations across Israel
| Variable | r | P | Number of sites |
|---|---|---|---|
| Long(E) | −0.0960 | 0.8200 | 8 |
| Lat(N) | −0.7950 | 0.0180 | 8 |
| Alt, m | −0.5300 | 0.1770 | 8 |
| Tm, °C | 0.6140 | 0.1060 | 8 |
| Ta, °C | 0.6750 | 0.0660 | 8 |
| Tj, °C | 0.5300 | 0.1770 | 8 |
| Td, °C | 0.1200 | 0.8220 | 6 |
| Tdd, °C | 0.8020 | 0.0550 | 6 |
| Sh, # | 0.1540 | 0.8050 | 6 |
| Trd, # | 0.7940 | 0.0590 | 6 |
| Rn, mm | -0.8920 | 0.0029 | 8 |
| Rd, # | -0.8700 | 0.0240 | 6 |
| Hu14 | -0.6750 | 0.0660 | 8 |
| Hua | -0.9280 | 0.0077 | 6 |
| Dw, # | 0.0580 | 0.9130 | 6 |
| Ev | 0.9860 | 0.0003 | 6 |
Numbers in the second column are the correlation coefficient (r) with the corresponding statistical significance (P values) in the third column. Long(E), longitude east; Lat(N), latitude north; Alt, altitude, Tm, mean annual temperature; Ta, mean temperature in August; Tj, mean temperature in January; Td, mean seasonal temperature difference; Tdd, mean diurnal temperature difference; Sh, mean number of hot and dry days; Trd, mean number of tropical days; Rn, mean annual rainfall; Rd, mean number of rainy days; Hu14, mean humidity at 1400 hours; Hua, mean annual humidity; Dw, mean number of dewy nights in summer; Ev, mean annual evaporation.
Interpretation of these results requires that the consequences of multiple testing (16 correlations) be taken into account, which can be achieved by adjusting the significance thresholds to reflect the number of tests performed. However, the number of significant correlations (Spearman rank) among the environmental variables themselves is much larger than expected by chance (data not shown), with 29 correlations significant at the 0.05 level of 120 tests. In fact, some variables were perfectly correlated, such as altitude and mean temperature in January. Thus, to correct for the multiple testing between genetic diversity and the environmental variables based on 16 tests would be overly conservative. However, even with the Bonferroni correction based on all 16 tests (experimentwise threshold = α/16), the correlation between diversity and evaporation is significant at an experimentwise 0.01 level, and that between diversity and mean annual rainfall is significant at an experimentwise 0.05 level.
It should also be noted that a high proportion of the correlation tests between genetic diversity and the environmental variables produced a significant P value (5 of 16 at the 0.05 threshold). Based on the binomial distribution, the chance of five or more tests being significant at the 0.05 level from 16 tests is <8.5 × 10−4. Thus, the disproportionate number of significant correlations observed here is extremely unlikely by chance. The simplest explanation is that the significant correlations are observed, because there is an actual relationship between genetic diversity and variables influencing water availability (especially evaporation and mean annual rainfall), and that variables influencing water availability make up a large proportion of those tested.
Frequencies of individual SNP sites are shown in Table 4. Spearman rank correlation tests between the 16 environmental variables and the four widely represented SNP sites found significant correlation for all four SNPs. SNP 64: [C→T] was predicted to effect an amino acid change from arginine to cystine. A significant positive correlation (P < 0.05) was found with mean seasonal temperature difference and a highly significant (P < 0.005) negative correlation with latitude. SNP 194: [A→T] was predicted to effect an amino acid change from histidine to leucine. A significant positive correlations (P < 0.05) was found with mean diurnal temperature difference and evaporation. SNP 351: [T→ C] was not predicted to change the amino acid sequence. A significant positive correlation (P < 0.05) was found with mean seasonal temperature difference. SNP 546: [T→C] was not predicted to change the amino acid sequence. A very highly significant negative correlation (P < 0.002) was found with mean seasonal temperature difference.
Table 4.
Frequency of each SNP in the Isa gene of wild barley at each sample site
| SNP no. | EC SFS | EC NFS | Mt. Meron | Maalot | Sede Boq | Wadi Oilt | Tab TR | Tab B | Amino acid substitutions |
|---|---|---|---|---|---|---|---|---|---|
| SNP64 | 0.6296 | 0.8438 | 0.0333 | 0.0250 | 0.02666 | 0.1522 | 0.4474 | Arg–Cys | |
| SNP120 | 0.0313 | ||||||||
| SNP151 | 0.0333 | Ala–Ser | |||||||
| SNP194 | 0.0185 | 0.0313 | 0.0500 | 0.0666 | His–Leu | ||||
| SNP249 | 0.5750 | ||||||||
| SNP269 | 0.0250 | Val–Ala | |||||||
| SNP277 | 0.0185 | Ser–Pro | |||||||
| SNP286 | 0.0185 | Ile–Val | |||||||
| SNP336 | 0.0666 | ||||||||
| SNP339 | 0.9375 | 0.1750 | 0.0666 | ||||||
| SNP351 | 0.8888 | 0.1667 | 0.1429 | 0.2666 | 0.3696 | 0.7105 | |||
| SNP462 | 0.0333 | 0.1000 | |||||||
| SNP546 | 0.0313 | 0.0333 | 1.0000 | 0.6250 | 0.0666 | SNP549 |
Where relevant, predicted amino acid substitutions are shown. The four SNP sites tested for correlation with the environmental variables are indicated in bold.
Discussion
Overview.
This study found significant levels of genetic diversity in the sequence of the Isa gene. The sequences from 178 accessions of barley from eight populations in Israel comprised 16 distinct haplotypes based on the distribution of 14 single nucleotide polymorphisms in 550 bp of coding sequence. Only 6 of the 14 SNP sites are predicted to effect an amino acid change, significantly fewer than would be predicted based on random expectations. Comparative gene sequence diversity reported for adh2 and adh3 (12, 13), Dhn5, Waxy, and G3pdh (12) loci in H. spontaneum suggests that the level of sequence diversity within the Isa locus is at the higher end of the spectrum for functional genes in wild barley, although diversity levels for Isa clearly varied with population across an apparent water availability gradient.
Environmental Correlates.
Following from Turpeinen et al. (17), a central question addressed in this study is whether a relationship exists between genetic diversity at the Isa gene in H. spontaneum and ecogeographical variables. The most important outcome of the 16 correlation tests performed on haplotype diversity at the Isa locus is that five of these tests returned significant correlations with variables that relate to water availability: latitude, mean number of rainy days, mean annual rainfall, mean annual humidity, and mean annual evaporation. Similarly related variables [mean humidity at 1400 h, number of tropical (hot and humid) days, mean August temperature] exhibit correlation significance just outside the nominal (P < 0.05) value. The significance of these correlations suggests that selection may have played a role in influencing genetic diversity at the Isa locus. An alternative hypothesis is that diversity at the Isa locus across populations is related to dispersal from the center of establishment or origin of wild barley in the region. However, Turpeinen et al. (17) found that, for four populations included in their study, the average diversity (mean He) based on 18 microsatellite loci gave a different ranking of diversity (Sede Boqer > Wadi Qilt > Maalot > Mt. Meron) to that found in this study for the Isa locus (Wadi Qilt > Sede Boqer > Mt. Meron > Maalot). The ranking for the microsatellite diversity was also consistent across 12 of the 18 loci. In addition, there are local differences between Isa diversity (at Nahal Oren and Tabigha) of a scale that would be inconsistent with dispersal history as the overwhelming determinant of diversity at these sites, because the subpopulations within each site are spatially close. Thus, selection becomes a plausible explanation for the observed gradient in diversity between sites.
All four SNP sites tested for association with ecogeographical variables were found to have significant correlation with one or more variables. Three SNPs showed correlation with mean seasonal temperature difference. Of particular interest was that two SNPs predicted to effect an amino acid change showed significant correlation to environmental variables. However, synonymous SNPs with significant correlation may not necessarily be chance events but may represent positions that influence stability of the RNA message that is crucial to the expression of the Isa gene. It should be noted here that the Isa coding region is particularly GC rich (7), and it is thought that this characteristic influences stability of the RNA message.
Recombination in Isa.
The signatures of a significant number of recombination events within the coding region of the Isa gene were found in our samples. These signatures must have survived and accumulated across the evolutionary history of these wild barley populations. Using the algorithm of Myers and Griffiths (18), evidence for at least eight recombination events was found. Morrell et al. (12) found similarly high numbers of recombinations at the Dhn5 and Waxy genes of H. spontaneum. These results are surprising given that H. spontaneum has an outcrossing rate of <2% (20, 21). In a self-pollinating population, genetic diversity and heterozygosity would tend to be comparatively low. In fact, self-pollination should halve heterozygosity with every generation.
The lower gene flow and highly localized gene transfer among selfing (self-pollinating) species means that subpopulations with different allelic frequencies can exist in close proximity. Analysis of genetic composition has shown quite localized biogeographical evolution in H. spontaneum (5, 17). Accordingly, a largely self-pollinating species like H. spontaneum is severely limited in the amount of effective meiotic recombination that can occur in any generation. Of 178 accessions for which the Isa gene was sequenced, 10 were heterozygous at this locus (5.6%). Because meiotic recombination in homozygotes cannot create new haplotypes, effective recombination is limited to this small percentage of heterozygotes. One mechanism that may explain the surprising number of observed recombinant haplotypes is heterosis, which has been discussed elsewhere (11). Brown et al. (20) found that the individual population estimates varied from 0% to 9.6% outcrossing. It had also been determined that outcrossing was significantly higher in populations growing in the more mesic (average 2.1%) than the xeric regions (0.4%).
Genetic Diversity and Drought Stress.
The observed relationship between genetic diversity and water variables raises questions about what evolutionary mechanisms underlie these differences in diversity. If chance sampling is discounted, the observed differences in gene diversity among wild barley populations are due to the genetic history of the populations, in particular, the severity and timing of any population bottlenecks. Historical events may have given rise to diversity patterns that correlate coincidentally with the ecogeographical variables tested in this study. However, probability would suggest it is far more likely that the variation in genetic diversity at the Isa locus between populations is a product of selective forces. Selection pressure at this locus is likely to be caused by fungal (and/or bacterial) pathogens. If the infectivity of the pathogen(s) was directly related to water variables, as is the case with many pathogenic fungi, this could lead to more extensive selective sweeps at the sites with greater water availability and hence reduced diversity at these sites. Higher diversity in the xeric desert populations may be due to a much lower selection pressure from pathogens enabling the persistence of a wider range of Isa haplotypes through time. Alternatively, the greater diversity in xeric regions may derive from diversifying balanced selection in small and isolated populations under different pathogen stresses. Stress in the xeric south may be fragmented, whereas in the mesic north, it may be centralized. This mechanism has been proposed to explain the higher diversity of soil fungi in the desert (22). These soil fungi may influence survival of wild barley seed in the soil and the subsequent establishment of plant populations. The higher diversity of soil fungi in the dry environments may select for a higher diversity of defense proteins encoded by the Isa locus in the seed.
This study identifies a pattern of distribution of diversity at the DNA sequence level in wild barley that confirms earlier results at the protein and DNA marker levels in other organisms. Polymorphism in allozymes increased southward in Israel toward the desert in 21 species of plants and animals involving 142 populations and 5,474 individuals (23). All these species largely share a geographically short (260-km) and ecologically stressful gradient of increasing aridity in Israel both eastward and (mainly) southward. Observed heterozygosity, H, and gene diversity, He, were positively and overall significantly correlated with rainfall variation. The trends of increasing diversity toward the desert are true at both the protein and DNA levels, suggesting that natural selection appears to be an important evolutionary force for both the coding and noncoding genomic regions. These regional trends follow global (24, 25) trends. Thus, molecular diversity at global, regional, and local scales is nonrandom and structured and is positively correlated with, and partly predictable by, abiotic and biotic environmental heterogeneity and stress.
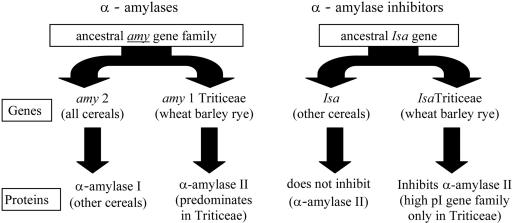
The evolution of resistance of starchy seeds to pest α-amylases has involved the expression of α-amylase inhibitors in the seed. These proteins inhibit the pest's α-amylases. The production of gibberellic acid (GA) by some pathogens [e.g., fungi (26)] may induce expression of the plant's α-amylases as a mechanism to overcome the specific inhibition of pathogen α-amylase. In turn, specific inhibition of plant α-amylase by BASI in the peripheral tissues of the seed may be a response to this evolutionary strategy (11). The evolution of α-amylases and α-amylase inhibitors in cereals is depicted in Fig. 1. The divergence of both enzyme and inhibitor genes in the Triticeae is an example of coevolution of two interacting genes. The high pI α-amylases found specifically in the Triticeae have evolved apparently by a duplication event from the more widespread low pI family of α-amylases that are found in other grasses. The α-amylase inhibitor activity of the protein encoded by the Isa gene in the Triticeae specifically inhibits only the high pI α-amylases found in this tribe (22). The function of this activity has been unknown. However, Furtado et al. (10) discovered that the promoter of this gene directed expression to the pericarp in barley. Specific expression in the peripheral tissues of the seed suggested a primary defense role for the protein (11). The gene may be involved in blocking pathogen access to plant starch. BASI may act by preventing the pathogen from using GA to stimulate plant α-amylase expression to achieve digestion of starch in the seed. As such, this may represent an unprecedented level of evolution of host pathogen interaction.
Fig. 1.
Evolution of α-amylases and α-amylase inhibitors in cereals.
Materials and Methods
Plant Materials.
Seeds from plants of wild barley [H. spontaneum (Earl K. Koch, Cleveland, OH)] were collected from eight populations in Israel with 5–10 seeds collected from each plant and 18–30 accessions collected from each population. The eight populations include two from opposing aspects in “Evolution Canyon,” Nahal Oren (28). Of these, one faces directly north and one south, providing starkly contrasting yet proximal microclimates. Two sites are included from Tabigha (29) representing two contrasting soil types, terra rosa and basalt. The Meron and Maalot sites are classified as mesic (moderately moist) climate, and Sede Boqer and Wadi Qilt are classified as xeric (dry) climate (30).
DNA Extraction.
Leaf tissue was collected from individual seedlings at 10 days from germination, and extraction was performed using the MWG Theonyx Liquid Performer robot (MWG Biotech, Ebersberg, Germany). The extraction protocol used was MagAttract 96 DNA Plant Protocol (Qiagen, Frankfurt, Germany), which was modified to include two RPW washes and three ethanol washes at steps 6 and 8, respectively.
PCR and Sequencing.
Primers for both PCR amplification and terminator sequencing reactions were designed to the Isa coding region based on the available sequence from both cultivated and wild barley using the program MacVector (MacVector v6.5; Oxford Molecular, Campbell, CA). PCR amplification of the fragments used the following reaction components per sample: Primer IsacodF1 (gCCTCCTCCTCCTCTCCCTTAT) 0.2 μM, primer IsacodR1 (ACgCCCTTSACggATggA) 0.2 μM, dNTPs (each) 200 μM, 0.05 units/μl Taq polymerase (Roche, Indianapolis, IN), 10% vol/vol glycerol, and 30 ng of template DNA. PCR amplification was performed by using a Corbett Research thermal palm cycler, model 9600–000 (Corbett Research, Sydney, Australia) using an initial denaturing step of 95°C per 5 min followed by 37 cycles of 95°C for 30 s, 58°C for 35 s, and 72°C for 45 s.
Samples of the resulting fragments underwent gel electrophoresis to check for purity and quantity of PCR product. After cycling, generated PCR products were purified by using QIAquick PCR purification columns (Qiagen). Sequencing reactions were performed by using ABI BigDye Terminator version 3.1 (Applied Biosystems, Foster City, CA) in forward and reverse directions using 2 μl of cleaned PCR product at ≈10 ng/μl of sample per 12-μl reaction. Nested sequencing primers were TCCTCTCCCTTATTCTggC (forward) and CCTTCTTgAACACgACgA (reverse). Sequencing reactions used an initial denaturing step of 96°C per 1 min followed by 30 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. Purification of sequencing products was performed by using a standard sodium acetate/ethanol precipitation protocol. Samples were sequenced by using an Applied Biosystems 3730 genetic analyzer.
Identification of SNPs, Haplotypes, and Recombination.
Sequences were aligned by using Sequencher software (Gene Codes, Ann Arbor, MI), and SNP sites were identified in Sequencher from the sequence chromatograms, using both forward and reverse sequences. Heterozygous individuals were identified from either mismatches to the consensus or ambiguous base calls and were confirmed by viewing chromatogram peaks. The patterns of SNP sites were used to identify discrete haplotypes within the populations, which were then used in subsequent analyses. Constituent haplotypes of heterozygotes were identified through analysis of additional seeds from the same accession. A test for a minimum number of recombination events in the gene was performed by using the RecMin program (18). This program also returns the more conservative lower bound of Hudson and Kaplan (19).
Population Genetics Analysis.
Each sample site was compared with all other sites to determine whether there was a significant difference in haplotype representation between sites. Fisher's exact test, as implemented by the software package SPSS (31), was used to compare counts for each haplotype recorded for each site. The null hypothesis being tested was that the representation of each haplotype is equivalent between each pair of sites. A χ2 test was performed to determine whether the observed ratio of synonymous sites to nonsynonymous substitutions was significantly different from random expectations. A random base substitution has a probability of 76%, resulting in an amino acid change for that codon (11), so an amino acid substitution rate significantly different from that figure would suggest nonneutral selection events to be in effect. Haplotype frequencies and Nei's genetic diversity index (32) were calculated for each population. This index is defined as He =
where x is allele frequency and i is number of alleles. This information was used to perform a Spearman rank correlation test of haplotype diversity between populations and 16 environmental variables by using SPSS (31). The presence of association among four widely represented SNP sites (SNP 64, SNP 194, SNP 351, and SNP 546) and the environmental variables (SI Table 13) was also tested by using Spearman rank correlations. Allele frequencies from all sites were included for these correlations, including those from sites that were fixed for the SNP allele.
Supplementary Material
Abbreviation
- BASI
α-amylase/subtilisin inhibitor.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611226104/DC1.
References
- 1.Zohary D, Hopf M. Domestication of Plants in the Old World. New York: Oxford Univ Press; 2000. [Google Scholar]
- 2.Harlan JR, Zohary D. Science. 1966;153:1074–1080. doi: 10.1126/science.153.3740.1074. [DOI] [PubMed] [Google Scholar]
- 3.Nevo E, Baum B, Beiles A, Johnson DA. Genet Res Crop Evol. 1998;45:151–159. [Google Scholar]
- 4.Hawkes JG. Isr J Bot. 1991;40:529–536. [Google Scholar]
- 5.Nevo E. Plant Genet Res. 2006;4:36–46. [Google Scholar]
- 6.Hejgaard J, Bjorn S, Nielsen G. Theor Appl Genet. 1984;68:127–130. doi: 10.1007/BF00252327. [DOI] [PubMed] [Google Scholar]
- 7.Leah R, Mundy J. Plant Mol Biol. 1989;12:673–682. doi: 10.1007/BF00044158. [DOI] [PubMed] [Google Scholar]
- 8.Mundy J, Svendsen I, Hejgaard J. Carlsberg Res Commun. 1983;48:81–90. [Google Scholar]
- 9.Sancho A, Faulds C, Svensson B, Bartolme B, Williamson G, Juge N. Biochim Biophys Acta. 2003;1650:136–144. doi: 10.1016/s1570-9639(03)00209-7. [DOI] [PubMed] [Google Scholar]
- 10.Furtado A, Henry R, Scott K, Meech S. Plant Mol Biol. 2003;52:787–799. doi: 10.1023/a:1025097218768. [DOI] [PubMed] [Google Scholar]
- 11.Bundock PC, Henry RJ. Theor Appl Genet. 2004;109:543–551. doi: 10.1007/s00122-004-1675-z. [DOI] [PubMed] [Google Scholar]
- 12.Morrell PL, Lundy KE, Clegg MT. Proc Natl Acad Sci USA. 2003;100:10812–10817. doi: 10.1073/pnas.1633708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin JZ, Brown AHD, Clegg MT. Proc Natl Acad Sci USA. 2001;98:531–536. doi: 10.1073/pnas.011537898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turpeinen T, Vanhala T, Nevo E, Nissila E. Theor Appl Genet. 2003;106:1333–1339. doi: 10.1007/s00122-002-1151-6. [DOI] [PubMed] [Google Scholar]
- 15.Vanhala TK, van Rijn CPE, Buntjer J, Stam P, Nevo E, Poorter H, van Eeuwijk FA. Euphytica. 2004;137:297–309. [Google Scholar]
- 16.Baek HJ, Beharav A, Nevo E. Theor Appl Genet. 2003;106:397–410. doi: 10.1007/s00122-002-1029-7. [DOI] [PubMed] [Google Scholar]
- 17.Turpeinen T, Tenhola T, Manninen O, Nevo E, Nissila E. Mol Ecol. 2001;10:1577–1591. doi: 10.1046/j.1365-294x.2001.01281.x. [DOI] [PubMed] [Google Scholar]
- 18.Myers SR, Griffiths RC. Genetics. 2003;163:375–394. doi: 10.1093/genetics/163.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hudson R, Kaplan N. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown A, Zohary D, Nevo E. Heredity. 1978;41:49–62. [Google Scholar]
- 21.Abdel-Ghani AH, Parzies HK, Omary A, Geiger HH. Theor Appl Genet. 2004;109:588–595. doi: 10.1007/s00122-004-1657-1. [DOI] [PubMed] [Google Scholar]
- 22.Grishkan I, Beharav A, Nevo E. Biol J Linn Soc. 2007 in press. [Google Scholar]
- 23.Nevo E, Beiles A. Biol J Linn Soc. 1998;35:229–245. [Google Scholar]
- 24.Nevo E. Proc Natl Acad Sci USA. 2001;98:6233–6240. doi: 10.1073/pnas.101109298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nevo E, Bieles A, Ben-Shlomo R. In: Lecture Notes in Biomathematics. Levin S, editor. Vol 53. Berlin: Springer; 1984. pp. 13–213. [Google Scholar]
- 26.Bruckner B, Blechschmidt D. Crit Rev Biotechnol. 1991;11:163–192. [Google Scholar]
- 27.Henry RJ, Battershell VG, Brennan PS, Oono K. J Sci Food Agric. 1992;58:281–284. [Google Scholar]
- 28.Nevo E. Theor Popul Biol. 1977;52:231–243. doi: 10.1006/tpbi.1997.1330. [DOI] [PubMed] [Google Scholar]
- 29.Nevo E, Brown AHD, Zohary D, Storch N, Beiles A. Syst Evol. 1981;138:287–292. [Google Scholar]
- 30.Nevo E, Zohary D, Brown AHD, Haber M. Evolution (Lawrence, Kans) 1979;33:815–833. doi: 10.1111/j.1558-5646.1979.tb04737.x. [DOI] [PubMed] [Google Scholar]
- 31.SPSS for Windows. Chicago: SPSS; 2002. Anonymous. Release 11.5.0. [Google Scholar]
- 32.Nei M. Proc Natl Acad Sci USA. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.