Abstract
In the core protein-coding region of hepatitis C virus (HCV), evidence exists for both phylogenetically conserved RNA structures and a +1 alternative reading frame (ARF). To investigate its role in HCV infection, we introduced four stop codons into the ARF of a genotype 1a H77 molecular clone. The changes did not alter the core protein sequence, but were predicted to disrupt RNA secondary structures. An attenuated infection was established after inoculation of the mutant HCV RNA into an HCV naïve chimpanzee. The acute infection was atypical with low peak viremia, minimal alanine aminotransferase elevation, and early virus control by a diverse adaptive immune response. Sequencing circulating virus revealed progressive reversions at the third and then fourth stop codon. In cell culture, RNA replication of a genome with four stop codons was severely impaired. In contrast, the revertant genome exhibited only a 5-fold reduction in replication. Genomes harboring only the first two stop codons replicated to WT levels. Similarly, reversions at stop codons 3 and 4, which improved replication, were selected with recombinant, infectious HCV in cell culture. We conclude that ARF-encoded proteins initiating at the polyprotein AUG are not essential for HCV replication in cell culture or in vivo. Rather, our results provide evidence for a functionally important RNA element in the ARF region.
Keywords: alternative reading frame, RNA replication, RNA structure
Hepatitis C virus (HCV) can establish a lifelong, persistent infection that can lead to chronic liver disease. Replication in vivo is apparently noncytolytic, and liver damage is thought to result from immune-mediated inflammatory processes as the host attempts, but fails to eliminate the virus (1, 2).
The positive-sense RNA genome contains a long ORF encoding a polyprotein that is cleaved into structural proteins (C, E1, E2), p7, NS2, and proteins forming the replicase (NS3–5B) (3–5). Host cell and virus proteins cooperate with highly structured RNA elements lying outside and within the coding region to conduct virus translation and replication. Translation of the polyprotein is initiated by ribosome binding to an internal ribosome entry site (IRES), which spans most of the 5′ nontranslated region (NTR). Replication is coordinated by RNA structures in the 5′ and 3′ NTR and a cruciform located in the NS5B coding region (6, 7). The virus RNA polymerase is error-prone, yielding considerable diversity (8).
Comparison of the core gene from divergent isolates has revealed an unusually high level of nucleotide sequence conservation (9, 10). Synonymous mutations are suppressed, suggesting a functional role for the nucleic acid sequence beyond simply encoding the core protein. For all six HCV genotypes, an overlapping reading frame in the +1 frame of the core gene is present (11). In the same region, highly conserved RNA secondary structures have been predicted by using phylogenetic analyses (10, 12).
The +1 frame of the core gene has been termed the alternative reading frame (ARF) (13). Antibodies and cellular immune responses reacting to ARF-encoded peptides or recombinant protein have been detected in HCV-infected patients (11, 14–17). These observations provide evidence for expression of the ARF in vivo; however, an ARF-encoded product has yet to be detected in infected tissue. With cell-free translation and transient expression in cell culture, an ARF protein has, however, been reported for the prototype genotype 1a isolate, HCV-1. HCV-1 contains a stretch of 10 consecutive adenylate residues where a +1 frameshift event moves translation into the ARF between codons 9 and 11 of the core gene to produce a frameshift product, termed the F protein (14, 18). Another ARF protein has been reported for a genotype 1b sequence involving two frameshift events. This dual frameshift protein (DF protein) also begins translation from the AUG of the polyprotein and frameshifts further downstream after codon 42 into the ARF (19). Translation continues in the ARF until a stop codon at position 144 is reached, triggering either termination or an additional frameshift restoring translation into the reading frame of the polyprotein.
In this study, we examined the importance of the ARF for HCV replication. Because this region is dispensable for replication of subgenomic HCV replicons in cell culture, we used the chimpanzee model and a functional molecular clone of the genotype 1a H77 isolate (20). To block expression of ARF-encoded proteins, we introduced silent base substitutions in the core gene, creating four stop codons in the ARF. RNA transcripts of this mutant genome were inoculated into the liver of an HCV naïve chimpanzee. The animal became viremic, demonstrating that ARF-encoded proteins produced by the previously characterized frameshift events are not absolutely required for HCV replication. However, low levels of viremia and minimal liver damage suggested that the mutant was attenuated. Sequence analysis of circulating virus showed reversions were selected and fixed during infection. These data, together with cell culture replication studies and reversion analysis of mutant full-length genomes, indicate that the ARF harbors one or more functional RNA elements.
Results
Ablation of ARF-Encoded Gene Products by Site-Directed Mutagenesis.
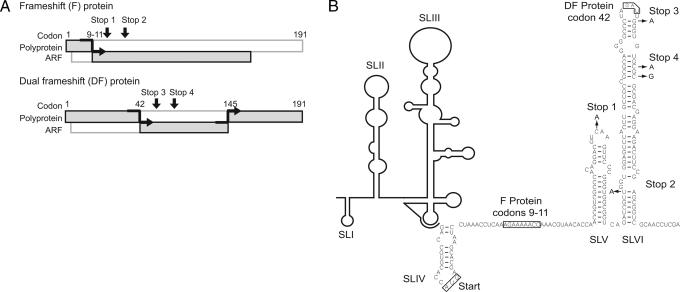
Despite its high degree of conservation, the region of the core gene encoding potential ARF proteins is dispensable for RNA replication in cell culture (21). Because ARF-encoded proteins may function in other aspects of the HCV lifecycle such as virus-immune system interactions or pathogenesis, we used the chimpanzee model to examine the behavior of a mutant HCV genome with ablated expression of both the ARF-encoded F and DF proteins (22, 23). The consensus genotype 1a H77 cDNA was chosen as parent for our studies given the wealth of information obtained from chimpanzees infected with H77 RNA transcripts or acute-phase virus (22, 23). Five nucleotide substitutions, silent for the core-coding sequence, but creating four stop codons (Stop 1,2,3,4) in the ARF were introduced (Fig. 1). To minimize leaky protein expression caused by read-through and reversion, two additional stop codons were introduced downstream from both reported frameshift sites. The frameshift sites are codons 9–11 for the F protein and codon 42 for the DF protein. Stop 1 permits expression of only the first 21 aa of the F protein. Stops 3 and 4 are predicted to block expression of the DF protein. Stops 1, 2, and 3 each required one base substitution, (C407A, U434A, and G473A, respectively), whereas two base changes were required to generate Stop 4 (C480A and C482G). Only Stop 4 created a potentially leaky opal stop codon (UGA), but rather than a C residue, which promotes translational read-through, this codon is followed by a G (24).
Fig. 1.
Strategy for blocking expression of ARF-encoded +1 frameshift proteins. (A) The F protein is produced by a +1 ribosomal frameshift near codons 9–11. Stops 1 and 2 indicate the positions of mutations creating stop codons in the +1 frame designed to block F protein expression. Stops 3 and 4 are predicted to block DF protein expression, which shifts into the +1 frame after core protein codon 42. Introduced mutations did not alter the core protein sequence. A construct that included all four stop mutations (Stop 1,2,3,4) was used for chimpanzee inoculation. (B) H77 RNA sequence and predicted structure of the core/ARF region. Downstream from the HCV IRES, the core/ARF nucleotide sequence is indicated, highlighting the two +1 frameshift regions (codons 9–11 and 42), the nucleotide substitutions used to create Stop 1,2,3,4, and the predicted RNA structures of SLV and SLVI.
An HCV Genome Without F and DF Protein Expression Is Infectious in Vivo.
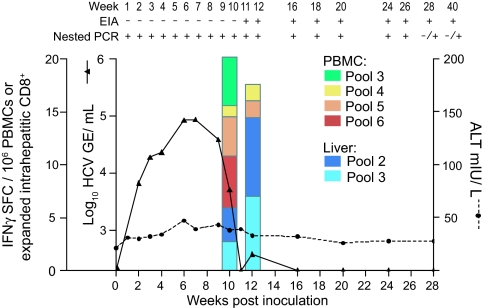
An HCV-naïve chimpanzee was inoculated with Stop 1,2,3,4 RNA by direct intrahepatic injection. The animal became infected, with circulating HCV RNA detectable by nested RT-PCR at 1 week postinoculation. By week 2, HCV RNA levels could be quantified by real-time quantitative RT-PCR. HCV RNA levels rapidly increased and peaked at weeks 6 and 7 at 8 × 104 genome equivalents (GE)/ml (Fig. 2). Circulating HCV RNA then declined rapidly and fell below the limit of detection at week 10. RNA was again measurable at week 12 (380 RNA GE/ml), but was only detectable by nested RT-PCR for the subsequent weeks analyzed (through week 40). The animal became HCV-seropositive at week 11 with no significant elevation in alanine aminotransferase (except for a slight rise of 20 milliunits/liter above baseline during the peak of viremia; Fig. 2). Compared with other monoclonal chimpanzee infections with the parental H77 sequence (20, 22, 25, 26), Stop 1,2,3,4 infection produced lower peak viremia (by 1–2 logs) and the mutant virus was controlled earlier without apparent liver damage, suggesting that the ARF mutant may be attenuated.
Fig. 2.
Infection profile of a chimpanzee inoculated with Stop 1,2,3,4. The animal was inoculated (week 0), and samples were collected weekly through week 12 and monthly thereafter. Circulating HCV RNA levels were quantified by real-time quantitative RT-PCR (sensitivity 200 GE/ml; set as 0 on the inner left y axis). Alanine aminotransferase (ALT) levels are given in milliunits per liter. Peptide pools (see Materials and Methods) spanning the H77 polyprotein and the ARF coding region were used to measure the frequency of HCV-specific lymphocytes in intrahepatic CD8+ T cells and bulk PBMCs by ELISPOT assay. Values are given as IFN-γ spot-forming cells (SFCs) per 106 PBMCs (green, yellow, orange, and red bars) or expanded CD8+ T cells (purple and blue bars) reacting to peptide pools. Positive (+) or negative (−) values indicating seroconversion [HCV-specific enzyme immunoassay (EIA)] and the presence of circulating HCV RNA (determined by nested RT-PCR) are indicated at the top.
Given that ARF-encoded products might function in modulating host immune responses to HCV, HCV-specific T cell responses were monitored in both the periphery and the liver. At weeks 10 and 12, HCV-specific responses were detected in peripheral blood mononuclear cells (PBMCs) and from intrahepatic CD8+ T cells by ELISPOT (Fig. 2). PBMCs were stimulated from pools 3–6, representing peptides from E2 to NS4B, and HCV-specific CD8+ cell responses reacted to pools 2 and 3, which include peptides from E1, E2, p7, and NS2. Because a response could not be detected at later time points with the ELISPOT assay, we used a more sensitive approach and cloned intrahepatic CD8+ and CD4+ T cells from liver biopsies at weeks 16 and 20. Analysis of 308 independently derived CD8+ T cell clones revealed that 31% were capable of killing target cells presenting HCV proteins. We then determined the epitope of 96 CD8+ and 7 CD4+ intrahepatic T cell clones, which revealed previously unseen CD8+ epitopes spanning proteins E1 to NS5. CD4+ cells proliferated in response to epitopes in NS4B at week 16 and to epitopes in E2, NS3, NS5A by week 20 [supporting information (SI) Tables 2 and 3]. No T cell reactivity was detected against ARF-encoded epitopes. Thus, despite producing a low level of viremia, infection with Stop 1,2,3,4 stimulated a diverse intrahepatic T cell response, first detectable by week 10, and presumably controlling the virus by week 20.
The Selection of Revertants Suggests Pressure to Maintain ARF RNA Elements Rather Than F/DF Protein Expression.
The low peak viremia and early control of the mutant virus could be caused by the Stop 1,2,3,4 substitutions impeding HCV replication. Given the high mutation rate of HCV replication, reversion to the WT H77 sequence and/or accumulation of compensating, fitness-restoring mutations would provide clues to the importance of F/DF protein expression or functional RNA elements in the ARF. At weekly or monthly (after week 12) time points, the 5′-NTR-C/ARF region of circulating HCV RNA was amplified by RT-PCR. Sequences were determined for the population and multiple clones, to sample sequence heterogeneity.
At week 1, the predominant HCV sequence was identical to the inoculated Stop 1,2,3,4 RNA transcript. Because we have never detected circulating input RNA using mutant transcripts with lethal (polymerase defective, pol−) mutations (20), this finding suggests that the Stop 1,2,3,4 mutant was able to replicate in vivo, albeit inefficiently. At week 2, coincident with rise in circulating virus RNA to nearly 104 GE/ml, the parental H77 G473 was found in all HCV genomes sequenced (Fig. 3). The reversion of Stop 3 (G473) remained fixed in the population for at least 40 weeks, the last time point analyzed. This reversion removed the Stop 3 ochre codon, but the downstream Stop 4 codon was still present to block expression of DF protein.
Fig. 3.
Evolution of HCV sequences after inoculation with Stop 1,2,3,4. The sequence of Stop 1,2,3,4 in the region containing the Stop 3 and 4 codons is shown. Mutated positions (473, 480, and 482) are indicated in larger font; stop codons are underlined. The H77 parental sequence is shown at the bottom. Sequencing was performed at all time points, only the weeks where reversions were found are indicated.
At week 10, we detected a second reversion at Stop 4 restoring A480 to the WT C480 H77 sequence. It was not the dominant sequence, as virus with both reversions circulated with virus containing only the first reversion at Stop 3. The A480C reversion eliminated the Stop 4 codon, thus restoring possible expression of the DF protein. The core protein sequence was unaltered: Stop 4 preserved core Arg-47 by creating an alternative arginine codon (AGG, nucleotides 480–482); the second reversion, A480C, yielded core codon CGG. Virus with one or both reversions existed as a mixed population for the next 10 weeks. At week 20, virus with both first and second reversions was fixed in the population.
A third reversion was detected at week 40 involving the second nucleotide substitution of Stop 4, with G482 being replaced by the WT H77 nucleotide, C. This third reversion was the only sequence detected, but may not necessarily represent the virus population because we succeeded in obtaining an RT-PCR product in only one of three attempts. This third reversion did not change the core protein sequence because C482 restores the WT Arg-47 codon (CGC, nucleotides 480–482). Hence, the selective forces favoring C482 are unclear. The possibility of codon preference for Arg CGC over CGG seems unlikely because they are used at approximately equal frequency in the HCV H77 ORF (38 versus 34 times, respectively). Since week 10, the presence of the first and second reversion could restore possible DF protein expression. The third reversion would not be expected to affect DF protein production, although it would alter the amino acid at position 5 of the DF protein frame from Gly (mutant) to Ala (WT). Alternatively, the reversion to C482 may restore an RNA element in the C/ARF region important for HCV replication. The results of the sequence analyses, where Stops 1 and 2 were maintained throughout infection, clearly demonstrate that F protein expression is not required for HCV H77 replication.
Effects of ARF Mutations on Replication of HCV RNAs in Cell Culture.
Although the ARF region is dispensable for replication of subgenomic replicons, mutations in this region have not been examined in the context of full-length replication-competent genome RNAs. Because our in vivo results with Stop 1,2,3,4 suggested a possible deleterious effect on HCV replication, we made a series of constructs using a cell culture adapted version of the H77 consensus cDNA (27). The parental clone, H/L+I, harbors two cell adaptive mutations, one in NS3 (P1496L) and another in NS5A (S2204I). Replication was assayed by electroporating highly permissive, hepatoma Huh-7.5 cells (28) with in vitro-transcribed RNA and measuring HCV RNA levels at successive time points. RNA levels for the mutant constructs were compared with the WT (H/L+I) parent or a replication defective (pol−) control. For the H/L+I parent, RNA levels declined slightly as input RNA was degraded and plateaued by day 4 as new RNA was generated (Fig. 4A). By day 3, pol− RNA remained detectable, but at levels >1,000-fold lower than the H/L+I parent. The Stop 1,2,3,4 mutant was highly impaired with transient, low-level replication observed at day 3, which then decreased to pol− levels by day 5. To determine whether the reversions observed in vivo restored replication fitness in cell culture, genomes harboring the revertant mutations were compared (Fig. 4B). Removal of Stop 3 in the Stop 1,2,4 mutant restored replication to nearly 18% of the H/L+I WT parent. Eliminating both Stops 3 and 4 (Stops 1 and 2; the equivalent to the triple revertant found in vivo at week 40) restored replication to WT levels. These results suggest that the reversions observed in vivo were selected because they improved the replication ability of the virus. Furthermore, mutations in Stops 3 and 4 seemed to be largely responsible for the replication defect. Consistent with this, Stop 3,4 genomes exhibited a 44-fold reduction in HCV replication.
Fig. 4.
Replication phenotypes of ARF mutants in cell culture. (A) RNA levels in Huh-7.5 cells at 3, 4, or 5 days posttransfection with WT H/L+I parental RNA, a pol− control, or the Stop 1,2,3,4 mutant. (B) RNA levels of genomes in A, along with additional mutants Stop 1,2,4, Stop 1,2, and Stop 3,4 measured at 5 days postelectroporation. Values (mean and SD) are given as the log10 HCV RNA molecules per nanogram of cellular GAPDH RNA. The data shown represent four independent transfections using two different preparations of transcript RNAs. (C) As described for B, replication of Stop 1,2,3,4 was analyzed on the genotype 2a J6/JFH genetic background. RNA levels were compared at 3 days posttransfection to the parental construct J6/JFH and a pol− negative control.
Phenotype and Reversion of ARF Mutations in the HCV Cell Culture (HCVcc) System.
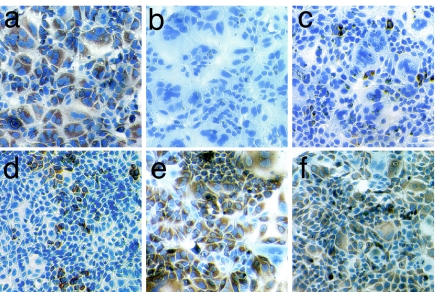
For genotype 1a (H77), the Stop 1,2,3,4 mutations impair HCV replication in cell culture and are subject to selective pressure in vivo. To examine the phenotype of this mutant in the HCVcc system, Stop 1,2,3,4 substitutions were engineered into an H/JFH chimeric genome consisting of the H77 5′NTR-NS2 region with the remaining sequences (NS3–3′NTR) derived from JFH. Two adaptive mutations, I348V in E1 and S1103T in NS3, allow this recombinant to produce infectious virus (M.J.E. and C.M.R., unpublished work). Initially, H/JFH Stop 1,2,3,4 was impaired for replication with few cells expressing NS5A (Fig. 5). In the first week, low levels of replication persisted and no sequence change was detected (Table 1). As cells were passaged, a mixture of reversions at Stop 3 and at 480 of Stop 4 was detected in the population with a concomitant increase in the number of NS5A-positive cells. Sequencing individual clones revealed a mixed population of the original input genome, and genomes with a reversion either at Stop 3 (G473A) or at A480C within Stop 4. By the third week, the original input sequence was no longer detected, and all clones contained either of the two reversions with the A480C reversion being more prevalent (Table 1).
Fig. 5.
Replication analysis of H/JFH in Huh-7.5 cells. (a–c) Immunohistochemistry staining of NS5A showing replication of H/JFH (a), a pol− control (b), and H/JFH Stop 1,2,3,4 (c) in Huh-7.5 cells at day 2 postelectroporation. (d–f) Cells harboring H/JFH Stop 1,2,3,4 were passaged and stained for NS5A (brown) and nuclei (blue) at days 7 (d), 21 (e), and 41 (f). (Magnification: × 20.)
Table 1.
Reversion analysis of H/JFH in Huh-7.5 cells
| Day postelectroporation | Reversion |
|||
|---|---|---|---|---|
| A473G | A480C | A473G + A480C | None | |
| 1–7 | 27/27 | |||
| 10 | 1/6 | 1/6 | 4/6 | |
| 11 | 1/8 | 1/8 | 6/8 | |
| 15 | 1/10 | 3/10 | 6/10 | |
| 21 | 3/13 | 9/13 | 1/13 | |
| 23 | 9/29 | 19/29 | 1/29 | |
| 38 | 3/9 | 6/9 | ||
| 41 | 1/10 | 9/10 | ||
| 50 | 1/10 | 9/10 | ||
Individual clones were sequenced from HCV RNA isolated from passaged cells, and the number of clones possessing the indicated reversions are listed.
We also examined the phenotype of the Stop 3 and 4 mutations in a genotype 2a J6/JFH chimera capable of replication and virus production without cell adaptive mutations (29–31). Genotype 2a differs from genotype 1a by ≈30% at the nucleotide level. As observed in the genotype 1a background, the J6/JFH Stop 3,4 mutant exhibited a dramatic defect in RNA replication, >5-fold decrease relative to the parent (Fig. 4C). Thus, the ability of C/ARF nucleotide sequence to modulate HCV replication is conserved across genotypes. Furthermore, the reversion analyses suggest that common selective pressures operate during animal infection and virus replication in cell culture to restore ARF function and replicative fitness.
Discussion
We have shown that an HCV genome containing four stop codons in the +1 frame overlapping the core gene is capable of establishing an infection in a chimpanzee. The infection was, however, atypical with low peak viremia, short duration of the acute phase, and no appreciable liver pathology. A multispecific adaptive immune response was elicited in both the liver and the periphery that coincided with a decline in circulating HCV RNA. During infection, a series of reversions emerged, indicating a selective pressure for RNA sequence and predicted RNA structure rather than a frameshift protein. Replication in cell culture recapitulated the in vivo results. Base substitutions creating the two downstream stop codons, the same bases to revert during the infection, were detrimental to HCV replication in Huh-7.5 cells. These substitutions disrupt base-pairing of a predicted RNA secondary structure in the core gene, implicating this RNA element in the modulation of HCV replication.
Features of this infection differed from previous young naïve chimpanzee infections launched with this consensus H77 genome. The titer reached only 8 × 104 GE/ml, 1–2 logs lower than previous studies (25, 32). Also, circulating HCV RNA peaked at week 6 and was detectable only by nested RT-PCR after week 12, several weeks earlier than a typical acute infection. The low level of circulating HCV RNA during acute infection may indicate that few hepatocytes became infected, low replication fitness of the mutant virus on a per-cell basis, or both. Virus levels declined without elevated liver transaminases in the serum, signifying little liver damage. Again, this result is atypical where an increase in serum alanine aminotransferase (usually to 100–200 milliunits/liter) coincides with immune-mediated hepatocellular injury and a rapid decrease in circulating HCV RNA. We did, however, find a diverse adaptive immune response in both the liver and periphery at the time of virus decline that persisted as virus disappeared from circulation. HCV-specific CD8+ and CD4+ T cells detectable at low frequencies were capable of recognizing HCV peptides in 5 of 10 pools representing the entire H77 polyprotein. No T cells targeting epitopes from the ARF were found. Thus, HCV lacking expression of an ARF protein established an infection, albeit with low levels of circulating virus and short viremia. Infection with the mutant virus elicited a diverse adaptive immune response, although virus-specific CD4+ and CD8+ T cells were detected at a lower frequency.
During infection, we detected a series of sequence reversions in the virus population. The predominant sequence in the first week of infection was the input mutant Stop 1,2,3,4 sequence. Because we have not detected carry over RNA from inoculation in the circulation, this finding suggests that the input mutant was able to replicate (20). The dominant virus during the acute phase harbored a reversion that eliminated the Stop 3 codon. Stops 1, 2, and 4 remained intact, however, blocking the expression of both the F and DF proteins. A second reversion restored one of two nucleotide substitutions in Stop 4 and was selected at a time of immune stimulation. Finally, a reversion of the second substitution creating Stop 4 was selected as the dominant virus. Because the DF protein expression was possible after the second reversion, and at no time was there a change in the core amino acid sequence, these results are consistent with selection to maintain the core gene nucleotide sequence rather than DF protein expression. Taken together, these data indicate that neither the F or DF proteins are required for HCV H77 replication in vivo.
Given that ARF sequences are dispensable for replication of subgenomic HCV replicons in cell culture (21, 27), the ARF stop mutant phenotypes in cell culture were surprising. We used a parental H77 genome, identical to the one in our in vivo study with the exception of two cell culture adaptive mutations. The Stop 1,2,3,4 mutant was severely impaired for replication in Huh-7.5 cells. In contrast, genomes with Stop 1,2,4 could replicate, but only with 18% efficiency compared with WT. Replication of the Stop 1,2 mutant was unimpaired, whereas replication of Stop 3,4 was reduced to 2% of the parental level. We also tested the effect of selected ARF mutations in the HCVcc sytem (29–31). In the context of the genotype 2a HCVcc J6/JFH, mutations for Stops 3 and 4 were deleterious, reducing replication to 18% of the parent. In contrast, an HCVcc J6/JFH mutant harboring Stop 1 was unimpaired for RNA replication and infectious virus production (data not shown), demonstrating that F protein is dispensable for complete replication in cell culture and reinforcing the in vivo results. A chimeric H77/JFH construct was used to mimic our chimpanzee experiment in cell culture. Replication of HCVcc H77/JFH Stop 1,2,3,4 was severely impaired. Upon passaging, identical reversions arose to those seen in vivo. Replication increased when either the reversion of Stop 3 or the reversion of 480 in Stop 4 occurred. These results strongly suggest that the reversions first identified in vivo act by restoring replication fitness, not by allowing translation of the F or DF proteins, but rather by restoring a functional RNA element conserved across divergent HCV genotypes.
Two RNA helix-forming stem loop (SL) structures, SLV and SLVI, have been proposed in the region containing the ARF stop codons and confirmed by enzymatic structure probing (33) (Fig. 1A). The base change conferring Stop 1 is not predicted to alter a base-pair interaction; however, substitutions for Stops 2–4 could disrupt the upper stem in a large RNA helix. A phylogenetic study of covariant base pairing shows high sequence conservation for the structure SLVI for all six genotypes (SI Fig. 6). The WT nucleotide of Stop 3, G473, is conserved in 1,272 of 1,274 sequences, and the Stop 4 mutations, 480 and 482, are conserved in 1,256 of 1,274 sequences. Covariant base pairs are detected in 18 and 14 sequences for Stop 4 mutant positions 480 and 482, respectively. The first reversion at Stop 3 restores a base-pair interaction at the base of the loop of SLVI, and the second and third reversions would repair two base-pair disruptions in the stem of SLVI.
RNA elements in both the noncoding and coding regions regulate translation and replication, but the functions of SLV and SLVI are unknown. The downstream boundary of the IRES does extend into the core-coding region, and IRES function can be influenced by both the core protein and RNA structures immediately downstream of the AUG start codon (34, 35). The 5′ base of the SLVI stem in the core gene has also been shown to base-pair with a complementary region in the 5′ NTR between SLI and the IRES, down-modulating IRES function (36). The base substitutions we used to create stop codons in the ARF do not affect the sequence proposed to base-pair with the 5′ NTR, but would disrupt the stem of SLVI itself. Also, a liver specific microRNA, miR122, can base-pair with the same region in the 5′ NTR and enhance HCV replication (37). This interplay between cellular miR122, the 5′ NTR, and SLVI may indicate that HCV translation and replication are regulated by competing higher-order RNA structures.
Our results clearly show that the F and DF proteins are dispensable for HCV replication in vivo and in vitro. We cannot exclude that functionally important ARF products are expressed by internal initiation from regions downstream of the Stop 4 codon (ARF codon 46). In this regard, internal initiation has been proposed to occur between ARF codons 80 and 86 (38), and the importance of such ARF-encoded products remains to be examined.
In summary, a study designed to test the importance of the ARF-encoded F and DF proteins, instead revealed a functionally important RNA element in the HCV protein-coding region. This SL structure (SLVI) resides in the ARF and core-protein coding region and may be part of an assembly of higher-order RNA structures that regulate HCV translation and replication.
Materials and Methods
Plasmid Constructions.
The H77 full-length HCV consensus sequence (GenBank accession no. AF009606; ref. 20), the cell culture-adapted H77 full-length genome, pH/FL (L+I) [(P1496L) and (S2204I)] (27), and genomes containing the JFH replicase, J6/JFH and H77/JFH (I348V/S1103T) (M.J.E. and C.M.R., unpublished work), which permit infectious virus (HCVcc) production in cell culture were used in all experiments (29, 31, 39). Base substitutions for stop codons in the ARF were created by site-directed mutagenesis. Mutations generating the ARF stop codons are: Stop 1, C407A; Stop 2, T434A; Stop 3, G473A; and Stop 4, C480A and C482G. Polymerase-defective genomes (20, 27), pol−, were used as a replication-defective controls.
In Vitro RNA Transcription.
For the chimpanzee study, pH/FL Stop 1,2,3,4 was linearized by digestion with BsmI, and 3′ overhangs were converted to blunt ends with T4 DNA polymerase (40). Parental and mutant derivates of pH/L+I were linearlized with HpaI and pJ6/JFH and pH/JFH (I348V/S1103T) linearized with XbaI. RNA was transcribed and purified as described (6). The yield of RNA was determined by A at 260 nm, and integrity was verified by agarose gel electrophoresis.
Chimpanzee Inoculation and Sample Collection.
Chimpanzee 1602 was inoculated with 0.6 mg of transcript RNA by direct intrahepatic injection (20). Peripheral blood was collected in acid citrate dextrose tubes for isolation of PBMCs, expansion of CD8+ T cells, and plasma. Liver tissue obtained by hepatic needle biopsy was placed in RPMI medium 1640 for T cell studies or flash-frozen for later RNA analysis. Peripheral blood and liver needle biopsies were collected weekly for the first 12 weeks and monthly thereafter. Animal housing, maintenance, and care met the National Institutes of Health's requirements for the humane use of animals in scientific research.
Immune Response Analyses.
HCV-specific antibodies were determined by using the HCV EIA-2 assay (Abbott, Abbott Park, IL). PBMC and intrahepatic lymphocytes were isolated and expanded as described (41, 42). Peripheral and intrahepatic HCV-specific T cell responses were assessed by proliferation (CD4+), ELISPOT, and cytotoxicity (CD8+) assays (41, 42). HCV recombinant antigens C22–3 (core; amino acids 2–120), C33c (NS3; amino acids 1192–1457), C100 (NS4; amino acids 1569–1931), C200 (NS3-NS4; amino acids 1192–1931), and NS5A (amino acids 2054–2995), expressed and purified as fusion proteins with human superoxide dismutase, were a gift from M. Houghton (Chiron, Emeryville, CA). Peptide stimulation was conducted by using 10 pools of 40–50 peptides (Genemed Synthesis, San Francisco, CA) prepared from 426 overlapping peptides (15- to 20-mers overlapping by 10 residues) encompassing the entire H77 polyprotein (including the ARF coding region).
Detection and Sequencing of HCV RNA.
Quantitative real-time RT-PCR (detection limit 200 RNA GE/ml) was performed with total RNA extracted from 100 μl of serum (43). For nested RT-PCR, total RNA was isolated with a QIAamp Virus RNA mini kit (Qiagen, Valencia, CA). RT-PCR was performed by using SuperScript III and Platinum High Fidelity One Step system (Invitrogen, Carlsbad, CA). RNA was denatured at 60°C for 5 min followed by reverse transcription at 55°C for 40 min. PCR cycling conditions were 94°C for 30 s, 55°C for 30 s, and 68°C for 1 min for 25 cycles. For samples requiring nested RT-PCR, 2 μl of the first reaction was used for seminested PCR with Pfx polymerase (Invitrogen). The cycling conditions were 94°C for 15 s, 55°C for 30 s, and 68°C for 1 min for 20 cycles. Reverse primer 5′-CCGCCTCGTACACAATACTCG (nucleotides 970–990) and forward primer 5′-GTGCCCCCGCAAGACTGC (nucleotides 233–250) were used. Seminested PCR combined the forward primer above and an internal primer, 5′-GGTGACATGGTAAAGCCCCG (nucleotides 934–953). PCR products were either sequenced directly or cloned into pCR2.1 (Invitrogen), and individual clones were sequenced.
Cell Culture Replication.
Huh-7.5 cell culture and electroporation of transcribed HCV RNA have been described (6, 28). HCV-positive Huh-7.5 cells were visualized by immunohistochemical staining for NS5A. All washes and diluents consisted of PBS with 0.1% Tween20. Endogenous peroxide was quenched with 3% H2O2. Cells were incubated for 1 h at 37°C with mAb 9E10 diluted 1:200. After washing, bound antibody was detected with ImmPRESS peroxidase-conjugated anti-mouse (Vector Laboratories, Burlingame, CA) diluted 1:4 and incubated for 30 min at 37°C. Peroxidase was detected with diaminobenzidine substrate (Vector Laboratories), and nuclei were counterstained with hematoxylin 2.
Supplementary Material
Acknowledgments
We thank Merna Torres and Maryline Panis for technical support, Pat Holst for laboratory management, and Shihyun You and Mike Flint for helpful discussions and critical reading of the manuscript. This work was funded by National Institutes of Health Grant CA57973, Public Health Service Grant CA85883, Public Health Service Grant AI40034, National Institute on Drug Abuse Grant 016156 (to A.D.B.), Public Health Service Grant DDK-066936 (to A.D.B.), the Ellison Medical Foundation (C.M.R.), the Starr Foundation, and the Greenberg Medical Research Institute (C.M.R.). C.M.R. is an Ellison Medical Foundation Senior Scholar in Global Infectious Diseases.
Abbreviations
- HCV
hepatitis C virus
- HCVcc
HCV cell culture
- IRES
internal ribosome entry site
- ARF
alternative reading frame
- NTR
nontranslated region
- pol−
polymerase defective
- F protein
frameshift protein
- DF protein
dual frameshift protein
- GE
genome equivalents
- SL
stem loop
- PBMC
peripheral blood mononuclear cell.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611267104/DC1.
References
- 1.Bowen DG, Walker CM. Nature. 2005;436:946–952. doi: 10.1038/nature04079. [DOI] [PubMed] [Google Scholar]
- 2.Thimme R, Bukh J, Spangenberg HC, Wieland S, Pemberton J, Steiger C, Govindarajan S, Purcell RH, Chisari FV. Proc Natl Acad Sci USA. 2002;99:15661–15668. doi: 10.1073/pnas.202608299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartenschlager R, Frese M, Pietschmann T. Adv Virus Res. 2004;63:71–180. doi: 10.1016/S0065-3527(04)63002-8. [DOI] [PubMed] [Google Scholar]
- 4.Appel N, Schaller T, Penin F, Bartenschlager R. J Biol Chem. 2006;281:9833–9836. doi: 10.1074/jbc.R500026200. [DOI] [PubMed] [Google Scholar]
- 5.Lindenbach BD, Rice CM. In: Fields Virology. Knipe DM, Howley PM, editors. Vol. 1. Philadelphia: Lippincott–Raven; 2001. pp. 991–1041. [Google Scholar]
- 6.You S, Stump DD, Branch AD, Rice CM. J Virol. 2004;78:1352–1366. doi: 10.1128/JVI.78.3.1352-1366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friebe P, Boudet J, Simorre JP, Bartenschlager R. J Virol. 2005;79:380–392. doi: 10.1128/JVI.79.1.380-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simmonds P. J Gen Virol. 2004;85:3173–3188. doi: 10.1099/vir.0.80401-0. [DOI] [PubMed] [Google Scholar]
- 9.Ina Y, Mizokami M, Ohba K, Gojobori T. J Mol Evol. 1994;38:50–56. doi: 10.1007/BF00175495. [DOI] [PubMed] [Google Scholar]
- 10.Smith DB, Simmonds P. J Mol Evol. 1997;45:238–246. doi: 10.1007/pl00006226. [DOI] [PubMed] [Google Scholar]
- 11.Walewski JL, Keller TR, Stump DD, Branch AD. RNA. 2001;7:710–721. doi: 10.1017/s1355838201010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tuplin A, Wood J, Evans DJ, Patel AH, Simmonds P. RNA. 2002;8:824–841. doi: 10.1017/s1355838202554066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Branch AD, Stump DD, Gutierrez JA, Eng F, Walewski JL. Semin Liver Dis. 2005;25:105–117. doi: 10.1055/s-2005-864786. [DOI] [PubMed] [Google Scholar]
- 14.Xu Z, Choi J, Yen TS, Lu W, Strohecker A, Govindarajan S, Chien D, Selby MJ, Ou J. EMBO J. 2001;20:3840–3848. doi: 10.1093/emboj/20.14.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komurian-Pradel F, Rajoharison A, Berland JL, Khouri V, Perret M, Van Roosmalen M, Pol S, Negro F, Paranhos-Baccala G. Hepatology. 2004;40:900–909. doi: 10.1002/hep.20406. [DOI] [PubMed] [Google Scholar]
- 16.Varaklioti A, Vassilaki N, Georgopoulou U, Mavromara P. J Biol Chem. 2002;277:17713–17721. doi: 10.1074/jbc.M201722200. [DOI] [PubMed] [Google Scholar]
- 17.Bain C, Parroche P, Lavergne JP, Duverger B, Vieux C, Dubois V, Komurian-Pradel F, Trepo C, Gebuhrer L, Paranhos-Baccala G, et al. J Virol. 2004;78:10460–10469. doi: 10.1128/JVI.78.19.10460-10469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi J, Xu Z, Ou JH. Mol Cell Biol. 2003;23:1489–1497. doi: 10.1128/MCB.23.5.1489-1497.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boulant S, Becchi M, Penin F, Lavergne JP. J Biol Chem. 2003;278:45785–45792. doi: 10.1074/jbc.M307174200. [DOI] [PubMed] [Google Scholar]
- 20.Kolykhalov AA, Agapov EV, Blight KJ, Mihalik K, Feinstone SM, Rice CM. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 21.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 22.Lanford RE, Lee H, Chavez D, Guerra B, Brasky KM. J Gen Virol. 2001;82:1291–1297. doi: 10.1099/0022-1317-82-6-1291. [DOI] [PubMed] [Google Scholar]
- 23.Bukh J. Hepatology. 2004;39:1469–1475. doi: 10.1002/hep.20268. [DOI] [PubMed] [Google Scholar]
- 24.Li G, Rice CM. J Virol. 1993;67:5062–5067. doi: 10.1128/jvi.67.8.5062-5067.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Major ME, Mihalik K, Puig M, Rehermann B, Nascimbeni M, Rice CM, Feinstone SM. J Virol. 2002;76:6586–6595. doi: 10.1128/JVI.76.13.6586-6595.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Major ME, Mihalik K, Fernandez J, Seidman J, Kleiner D, Kolykhalov AA, Rice CM, Feinstone SM. J Virol. 1999;73:3317–3325. doi: 10.1128/jvi.73.4.3317-3325.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blight KJ, McKeating JA, Marcotrigiano J, Rice CM. J Virol. 2003;77:3181–3190. doi: 10.1128/JVI.77.5.3181-3190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blight KJ, McKeating JA, Rice CM. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, et al. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindenbach BD, Meuleman P, Ploss A, Vanwolleghem T, Syder AJ, McKeating JA, Lanford RE, Feinstone SM, Major ME, Leroux-Roels G, Rice CM. Proc Natl Acad Sci USA. 2006;103:3805–3809. doi: 10.1073/pnas.0511218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Major ME, Dahari H, Mihalik K, Puig M, Rice CM, Neumann AU, Feinstone SM. Hepatology. 2004;39:1709–1720. doi: 10.1002/hep.20239. [DOI] [PubMed] [Google Scholar]
- 33.Tuplin A, Evans DJ, Simmonds P. J Gen Virol. 2004;85:3037–3047. doi: 10.1099/vir.0.80141-0. [DOI] [PubMed] [Google Scholar]
- 34.Wang TH, Rijnbrand RC, Lemon SM. J Virol. 2000;74:11347–11358. doi: 10.1128/jvi.74.23.11347-11358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boni S, Lavergne JP, Boulant S, Cahour A. J Biol Chem. 2005;280:17737–17748. doi: 10.1074/jbc.M501826200. [DOI] [PubMed] [Google Scholar]
- 36.Kim YK, Lee SH, Kim CS, Seol SK, Jang SK. RNA. 2003;9:599–606. doi: 10.1261/rna.2185603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 38.Vassilaki N, Mavromara P. J Biol Chem. 2003;278:40503–40513. doi: 10.1074/jbc.M305504200. [DOI] [PubMed] [Google Scholar]
- 39.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 40.Han JQ, Barton DJ. RNA. 2002;8:512–525. doi: 10.1017/s1355838202020617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shoukry NH, Grakoui A, Houghton M, Chien DY, Ghrayeb J, Reimann KA, Walker CM. J Exp Med. 2003;197:1645–1655. doi: 10.1084/jem.20030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grakoui A, Shoukry NH, Woollard DJ, Han JH, Hanson HL, Ghrayeb J, Murthy KK, Rice CM, Walker CM. Science. 2003;302:659–662. doi: 10.1126/science.1088774. [DOI] [PubMed] [Google Scholar]
- 43.Puig M, Mihalik K, Yu MY, Feinstone SM, Major ME. J Virol Methods. 2002;105:253–263. doi: 10.1016/s0166-0934(02)00119-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.