Abstract
Human T cell leukemia virus type 1 (HTLV-1) has evolved a remarkable strategy to thwart the antiviral effects of the cellular cytidine deaminase APOBEC3G (hA3G). HTLV-1 infects T lymphocytes in vivo, where, like HIV-1, it is likely to encounter hA3G. HIV-1 counteracts the innate antiviral activity of hA3G by producing an accessory protein, Vif, which hastens the degradation of hA3G. In contrast, HTLV-1 does not encode a Vif homologue; instead, HTLV-1 has evolved a cis-acting mechanism to prevent hA3G restriction. We demonstrate here that a peptide motif in the C terminus of the HTLV-1 nucleocapsid (NC) domain inhibits hA3G packaging into nascent virions. Mutation of amino acids within this region resulted in increased levels of hA3G incorporation into virions and increased susceptibility to hA3G restriction. Elements within the C-terminal extension of the NC domain are highly conserved among the primate T cell leukemia viruses, but this extension is absent in all other retroviral NC proteins.
Keywords: retrovirus
Cells have evolved numerous strategies to restrict infection by pathogens such as viruses. In turn, viruses that are disseminated in a particular host have evolved mechanisms to resist cellular restriction factors. A good example of antiviral restriction emerged from studies aimed at understanding the function of HIV-1 Vif. It was known that Vif-deficient HIV-1 could replicate in some cell lines but not others, and that this phenotype depended on the virus-producer cell rather than the target cell. The cellular protein responsible for this effect was cloned and identified as CEM15, also known as APOBEC3G (1). Human APOBEC3G (hA3G) belongs to a family of cytidine deaminases and is a broadly acting antiviral restriction factor (1, 2). In addition to inhibiting the replication of Vif-deficient HIV-1, hA3G has been shown to inhibit the replication of other exogenous retroviruses, endogenous retroviruses, retrotransposons, and hepatitis B virus (3–12).
hA3G can be packaged into retrovirus particles and then inhibit virus replication in the target cell by catalyzing the conversion of cytosine to uracil in minus-strand DNA during reverse transcription (resulting in G-to-A hypermutation of the provirus) (3, 6, 13–15). Although cytosine deaminase activity appears to correlate with antiviral activity of hA3G, there is evidence that other factors may contribute to antiviral activity, and it is still unclear which stage of the virus infectious cycle is primarily affected (16–18). The exact mechanism by which hA3G is packaged into virions is also unresolved but it does appear to require both RNA and viral nucleocapsid (NC) protein (19–22). HIV-1 counteracts the effects of hA3G with the accessory protein Vif, whose primary function is to target hA3G for proteasomal degradation, thereby preventing encapsidation of hA3G into virus particles (23–28). Primate foamy viruses resist the inhibitory effects of hA3G with the accessory protein Bet, which interacts with hA3G and prevents its packaging into virus particles, but by a different mechanism than HIV-1 Vif (8). It is not clear whether or how retroviruses, which do not encode Vif homologues, resist APOBEC restriction. Human T cell leukemia virus type 1 (HTLV-1) is particularly interesting in this regard because, like HIV-1, it infects human T lymphocytes, which express hA3G.
HTLV-1 appears to resist the antiviral effects of hA3G. Mahieux et al. (10) recently reported that G-to-A mutations were not detected in the proviruses from HTLV-1-infected patients, and <0.1% of proviruses generated after in vitro infections contained G-to-A hypermutations. In addition, two other studies (11, 12) showed that HTLV-1 was relatively resistant to hA3G despite the fact that hA3G could be found in HTLV-1 particles; however, it should be noted that hA3G packaging was not quantified in those studies. We report here that HTLV-1 is less susceptible to hA3G inhibition compared with HIV-1 because HTLV-1 packages less hA3G into virus particles than HIV-1. We ruled out the possibility that HTLV-1 expresses an accessory protein that interferes with hA3G packaging or activity, and we show that the determinant for resistance to hA3G resides in the HTLV-1 structural protein Gag. Specifically, a motif in the C-terminal region of the NC domain of Gag acts in cis to exclude hA3G from HTLV-1 particles.
Results
Previous studies indicated that HTLV-1 somehow resists antiviral effects of hA3G (10–12). It seemed unlikely that HTLV-1 does so by expressing a transacting accessory protein like Vif, because chronically HTLV-1-infected T cells accumulate hA3G and restrict HIVΔvif replication (29). We formally excluded a transacting mechanism by performing the following experiments. First, cotransfection of either full-length HTLV-1 or HTLV-1 Tax/Rex expression plasmids with HIVΔvif vectors and hA3G expression plasmid failed to rescue HIVΔvif replication in single-cycle infectivity assays (data not shown). Second, HTLV-1 mutants with a deletion of the pX ORFs, which encode putative accessory proteins, packaged the same amount of hA3G as wild-type virus. Also, when used as packaging plasmids for single-cycle replication assays, the pX mutants showed the same susceptibility to hA3G inhibition as wild-type vectors (data not shown). Finally, as shown below, Gag alone determines the amount of hA3G that is packaged into HTLV-1 versus HIV-1 particles.
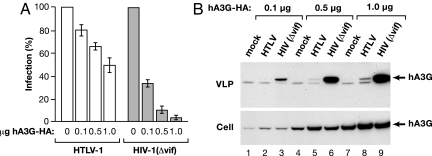
To delineate the mechanism by which HTLV-1 evades hA3G restriction, we used viral vectors in single-cycle replication assays that make it possible to quantify viral infection and replication (30). HTLV-1 or HIVΔvif virus-like particles (VLPs), pseudotyped with vesicular stomatitis virus G protein, were produced in 293T cells cotransfected with varied amounts of a hemagglutinin (HA)-tagged hA3G expression plasmid. Both HTLV-1 and HIV-1 transfer vectors encode a GFP-luciferase fusion protein so that infection of HeLa cells can be monitored by assaying for luciferase activity. As shown in Fig. 1A, and in agreement with previous studies, expression of hA3G significantly diminished infectivity of HIVΔvif, but had only a modest effect on HTLV-1 infection. When cotransfected with 1 μg of hA3G plasmid, HIVΔvif infection was reduced ≈20-fold, but HTLV-1 infection was reduced by only 2-fold. We next compared the amount of HA-tagged hA3G protein that was packaged into HTLV-1 and HIVΔvif VLPs by immunoblotting with an anti-HA antibody (Fig. 1B). Gag protein concentrations in VLP lysates were quantified by either HTLV-1 p19 (matrix, MA) or HIV-1 p24 (capsid, CA) ELISA, and equal amounts of Gag protein were loaded in each lane of SDS-protein gels. Phoshorimage analysis of immunoblots indicated that HTLV-1 particles packaged ≈20 times less hA3G than HIVΔvif VLPs. As in chronically infected cell lines, HTLV-1 expression did not diminish hA3G accumulation in transfected cells. These experiments indicated that HTLV-1 particles incorporated less hA3G compared with HIVΔvif particles and therefore HTLV-1 resists the inhibitory effects of hA3G because HTLV-1 does not efficiently package hA3G.
Fig. 1.
Differences in the inhibition of HTLV-1 versus HIV-1Δvif by hA3G correlate with the amount of hA3G incorporated into VLPs. (A) Single-cycle replication assays were performed with VLPs produced in cells cotransfected with varied amounts of hA3G-HA expression plasmid. Infection is expressed as the percent transduction of luciferase relative to no-hA3G control. Because HTLV-1 VLPs have a low specific infectivity, HIVΔvif VLPs were diluted 1:1,000 before infection to give comparable luciferase activities for the two viruses. (B) Cells were cotransfected with the indicated amount of hA3G-HA plasmid in combination with either empty vector (mock), HTLV-1 vectors, or HIVΔvif vectors. VLPs were concentrated from supernatants by centrifugation. Cell and VLP extracts were immunoblotted and probed with anti-HA antibody. Concentrations of HTLV-1 and HIV-1 Gag proteins in VLP extracts were determined by ELISA, and equal amounts of Gag were used for immunoblotting. Arrows indicate the position of the hA3G-HA, which is the upper band in VLP lysates.
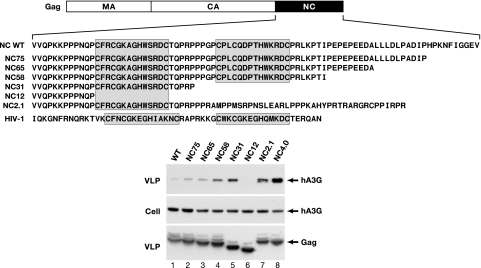
The HTLV-1 Gag protein, composed of MA, CA, and NC domains (Fig. 2), is sufficient for the assembly and release of VLPs. We compared the amount of hA3G that was packaged into VLPs produced from Gag expression plasmids that encode either wild-type, mutant, or chimeric versions of the HTLV-1 Gag protein (Fig. 2). Immunoblots of VLP lysates revealed that hA3G was not efficiently packaged into VLPs containing wild-type HTLV-1 Gag (Fig. 2, lane 1, WT). In contrast, hA3G was detected in large amounts in VLPs containing HIV-1 Gag (data not shown) and in VLPs containing a chimeric Gag composed of HTLV-1 MA/CA and HIV-1 NC domains (Fig. 2, lane 8, NC4.0). Thus, the amounts of hA3G packaged into VLPs produced from the Gag expression plasmids were similar to the amounts packaged into VLPs produced from whole-virus expression plasmids (Fig. 1B). Moreover, the difference in hA3G packaging between HIV-1 and HTLV-1 VLPs is determined by the NC domain of Gag.
Fig. 2.
Packaging of hA3G into HTLV-1 particles is determined by the NC domain of Gag. The organization of HTLV-1 Gag is shown above the amino acid sequences of wild-type and mutated NC domains. The Zn-finger motifs in NC are shaded. The wild-type or modified HTLV-1 gag genes were subcloned into a mammalian expression plasmid. A series of C-terminal truncations of the NC domain are shown below the wild-type NC sequence. The plasmid designated as NC2.1 has a frameshift mutation between the Zn-finger motifs that replaces the C-terminal half of NC with the indicated sequence. The Gag protein designated as NC4.0 has the HTLV-1 MA and CA domains fused to the HIV-1 NC domain, whose sequence is shown. 293T cells were transfected with 1 μg of hA3G-HA expression plasmid and the indicated Gag expression plasmids. Immunoblots of cell and VLP lysates were probed with anti-hA3G antibody or anti-HTLV-1 p19(MA) antibody. Loading of equal amounts of Gag protein in VLP lysates was based on HTLV-1 p19(MA) ELISA determination.
As shown in the sequence alignments at the top of Fig. 2, HTLV-1 and HIV-1 NC proteins differ dramatically in the amino acid sequence after the two zinc finger motifs. HTLV-1 NC has a long C-terminal tail rich in proline, glutamic acid, and leucine residues. Deletions of 10–20 aa from the C terminus of the 85-residue HTLV-1 NC protein were accompanied by a slight increase in hA3G packaging into VLPs (Fig. 2, lane 2, NC75 and lane 3, NC65). The amount of hA3G incorporated into VLPs increased by ≈8-fold when 29 aa were removed from the C terminus of NC in the NC58 mutant (Fig. 2, lane 4). The amount of hA3G detected in VLPs was also high in mutant NC31, which contains only one zinc finger motif (Fig. 2, lane 5). Deletion of both zinc fingers in NC12 abolished hA3G packaging (Fig. 2, lane 6). Furthermore, hA3G was efficiently packaged by a Gag mutant (NC2.1) in which the C-terminal half of NC (including the second zinc finger) was replaced by a 38-aa-long peptide rich in proline and basic amino acid residues (Fig. 2, lane 7). These data indicate that the first zinc finger of HTLV-1 NC is sufficient for hA3G packaging and that a motif in the C terminus of NC acts to prevent hA3G packaging into the virus particle.
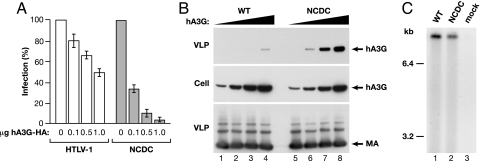
To determine how the C terminus of NC affects HTLV-1 replication and hA3G inhibition, we constructed an HTLV-1 expression plasmid with a 20-aa deletion in this region (PTIPEPEPEEDALLLDLPAD), which is designated as NCDC. In the absence of hA3G, NCDC infectivity was ≈40% of wild-type infectivity in single-cycle replication assays. VLPs expressed in cells cotransfected with increasing amounts of hA3G plasmid revealed that the NCDC mutant was ≈20 times more sensitive to hA3G than wild-type virus (Fig. 3A). Immunoblots revealed that NCDC VLPs packaged significantly higher levels of hA3G than wild-type HTLV-1 particles (Fig. 3B), consistent with the increased susceptibility of NCDC to hA3G inhibition. Because retroviral NC proteins bind to RNA and are responsible for viral RNA encapsidation, we determined the amount of viral mRNA in wild-type and NCDC VLPs by Northern blot and phosphorimage analyses (Fig. 3C). These experiments revealed that the ratio of viral mRNA-to-Gag protein [determined by HTLV-1 p19(MA) ELISA] for NCDC and WT VLPs differed by <5%, indicating that the deletion mutation did not affect viral RNA packaging. Together, these data indicate that elements in the C-terminal region of HTLV-1 NC counteract hA3G inhibition of virus replication by preventing hA3G packaging into virus particles.
Fig. 3.
A peptide region near the C terminus of HTLV-1 NC acts to exclude hA3G from VLPs. (A) A 20-aa region (PTIPEPEPEEDALLLDLPAD) near the C terminus of NC was deleted in pCMVHT-ΔEnv to yield the NCDC mutant. Single-cycle replication assays were performed with VLPs produced from cells transfected with HTLV-1 or NCDC vectors and the indicated amounts of hA3G-HA plasmid. Infection is expressed as percent transduction relative to no-hA3G control. (B) Immunoblots of cell and VLP lysates from 293T cells transfected with HTLV-1 or NCDC vectors in combination with 0 μg (lanes 1 and 5), 0.1 μg (lanes 2 and 6), 0.5 μg (lanes 3 and 7), or 1.0 μg (lanes 4 and 8) of hA3G expression plasmid. Immunoblots were probed with anti-hA3G antibody or anti-HTLV-1 p19(MA) monoclonal antibody. (C) Viral poly(A)+ mRNA was prepared from virions produced from cells transfected with wild-type or NCDC versions of the infectious clone of HTLV-1. Northern blots were probed with a 32P-labeled gag gene fragment. The positions of RNA size markers, which were run on the same gel, are indicated.
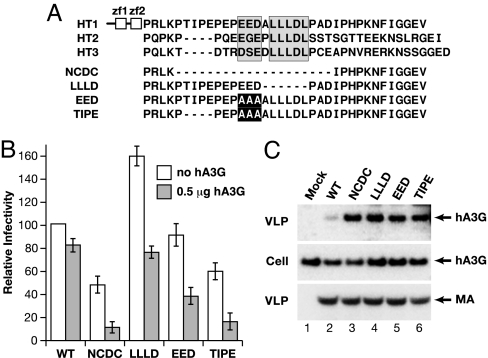
To define amino acids in the C terminus of NC that mediate susceptibility to hA3G, we constructed site-directed mutants. An alignment of HTLV-1, HTLV-2, and HTLV-3 (31) NC proteins revealed several highly conserved motifs within otherwise divergent C termini (Fig. 4A). These consisted of a cluster of acidic amino acids and an adjacent LLLDL sequence, which were contained within the region deleted in the NCDC mutant. The mutated viral vectors were transfected without or with 0.5 μg of hA3G-HA plasmid, and VLPs were tested in single-cycle replication assays (Fig. 4B). In the absence of hA3G, specific infectivity ranged between 60% above (LLLD), about equal to (EED), or 40% below wild type. In these experiments, cotransfection with 0.5 μg of hA3G-HA plasmid inhibited wild-type HTLV-1 infectivity by ≈20% and inhibited NCDC by ≈80%. All of the site-directed mutants were more susceptible to hA3G compared with wild type. Cotransfection with 0.5 μg of hA3G-HA plasmid inhibited replication of the LLLD mutant by 50%; the EED mutant was inhibited by 54%; and the TIPE mutant was inhibited by 66% (Fig. 4B). The relative amounts of hA3G that were incorporated into VLPs produced by the mutated viral vectors were higher than wild-type VLPs (Fig. 4C), and the amounts of VLP-associated hA3G correlated with the antiviral effect. Furthermore, wild-type and mutated viruses packaged equivalent amounts of viral RNA (data not shown). In summary, site-directed mutagenesis indicates that the cluster of acidic amino acids and the LLLDL motif contribute to the ability of the C-terminal region of HTLV-1 NC to resist hA3G.
Fig. 4.
Conserved motifs in the C-terminal region of the NC domain mediate HTLV-1 resistance to hA3G. (A) Amino acid sequence alignment of the C-terminal portions of HTLV-1, HTLV-2, and HTLV-3 (31) NC domains is shown; the highly conserved N terminus, containing two zinc finger motifs (zf1 and zf2) is depicted for HTLV-1 at the top. A cluster of acidic amino acids and a conserved LLLDL motif are shaded. Shown below the sequence alignment are the mutated HTLV-1 NC domains that were cloned into the pCMVHT-ΔEnv packaging vector. Deleted amino acids are indicated by dashed lines, and alanine substitutions are highlighted. (B) Wild-type and mutated packaging vectors were tested in single-cycle replication assays. VLPs were produced in the absence or presence of 0.5 μg of hA3G-HA plasmid. Infectivity, normalized to p19(MA) levels, is plotted relative to the value determined for wild-type vector in the absence of hA3G set at 100. Error bars indicate standard deviations of duplicate samples from at least two experiments. (C) 293T cells were cotransfected with the indicated wild-type or mutant HTLV-1 packaging vectors or without HTLV-1 expression plasmid (lane 1, mock). All samples were cotransfected with 1 μg of hA3G expression plasmid. Cell and VLP extracts were immunoblotted as in Fig. 1.
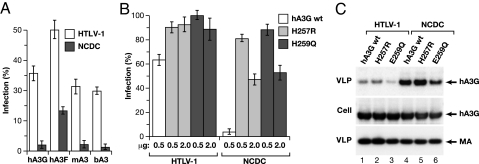
To determine whether HTLV-1 resistance to hA3G mediated by the C terminus of NC extends to other APOBEC proteins, we tested human A3F (hA3F), mouse A3 (mA3), and bovine A3 (bA3) proteins against wild-type or NCDC HTLV-1 vectors. In all cases, NCDC was much more sensitive to APOBEC 3 compared with wild-type HTLV-1 vector. In general, hA3F was less inhibitory than hA3G, whereas mA3 and bA3 were slightly more active than hA3G (Fig. 5A). These results indicate that the C terminus of HTLV-1 NC acts to resist the APOBEC 3 proteins in a general manner and also suggest that these APOBEC proteins are packaged into HTLV-1 VLPs by a common mechanism.
Fig. 5.
Single-cycle replication of HTLV-1 vectors is inhibited by various APOBEC proteins and inhibition requires cytidine deaminase activity. (A) HTLV-1 or NCDC vectors were produced in the absence or presence of 1 μg of the indicated APOBEC expression plasmids; hA3G, hA3F, mouse A3 (mA3), or bovine A3 (bA3). The data are plotted as the percent activity remaining in the presence of APOBEC relative to no-APOBEC controls. (B) HTLV-1 and NCDC vectors were produced in the presence of the indicated amounts of wild-type hA3G, H257R, or E259Q. The data are plotted as the percent activity remaining in single-cycle replication assays compared with no-hA3G controls. (C) Packaging of wild-type or mutated forms of hA3G into HTLV-1 or NCDC VLPs. 293T cells were cotransfected with HTLV-1 or NCDC expression vectors and 2 μg of the indicated hA3G expression plasmids. Cell and VLP extracts were immunoblotted as in Fig. 1.
To determine whether cytidine deaminase activity of hA3G is required for the inhibition of HTLV-1 infection, we mutated conserved amino acids in the second zinc coordination motif of hA3G, which is essential for enzymatic activity. The H257R and E259Q mutations were previously shown to diminish anti-HIV-1 activity of hA3G (11, 16, 18). hA3G, H257R, and E259Q were tested as inhibitors in single-cycle replication assays with either HTLV-1 or NCDC vectors (Fig. 5B). With HTLV-1 vectors, 0.5 μg of hA3G plasmid inhibited replication by ≈40%, whereas cotransfection with 0.5 μg of either H257R or E259Q plasmids had no significant inhibitory effect. Increasing the amounts of H257R or E259Q plasmids to 2 μg did not result in significant inhibition. With NCDC vectors, cotransfection with 0.5 μg of hA3G inhibited replication by >95% and 0.5 μg of either H257R or E259Q resulted in 10% to 20% inhibition (Fig. 5B). Increasing the amounts of H257R and E259Q plasmids to 2 μg resulted in ≈50% inhibition of NCDC replication. Both H257R and E259Q were incorporated into HTLV-1 and NCDC VLPs, although E259Q was packaged less efficiently than wild-type hA3G or H257R (Fig. 5C). As before, NCDC VLPs contained more of the wild-type or mutated hA3G protein than HTLV-1 VLPs. The second Zn-coordination motif, and hence the cytidine deaminase activity of hA3G, is required to achieve complete inhibition of HTLV-1 infectivity.
Discussion
Although HTLV-1 and HIV-1 both infect human T lymphocytes, the viruses differ in many ways and have evolved different strategies to circumvent cellular restriction mechanisms. HTLV-1 provides the first clear example of a virus that evades hA3G by a cis-acting exclusion mechanism. In contrast, other retroviruses evade hA3G by expressing transacting factors, such as HIV-1 Vif (23–28) or primate foamy virus Bet proteins (8). A previous study indicated that HTLV-1 resists G-to-A hypermutation elicited by hA3G (10). Two other groups reported that HTLV-1 was weakly susceptible to hA3G inhibition (11, 12), but they did not address the basis for this apparent resistance. The resistance of HTLV-1 to hA3G restriction and the ability of the virus to prevent hA3G packaging are not absolute. As shown here and before (11, 12), hA3G can be detected in HTLV-1 particles when it is expressed at high levels. Nonetheless, by quantifying the relative levels of hA3G and Gag, we showed that hA3G is packaged much less efficiently into HTLV-1 compared with HIVΔvif VLPs. We also found that elements in the C terminus of HTLV-1 NC inhibit hA3G packaging into HTLV-1 particles. When these elements were deleted or mutated, the virus was more susceptible to hA3G inhibition and incorporated more hA3G into viral particles than wild-type virus.
HTLV NC proteins are different from those expressed by all other retroviruses in that they contain a 35-aa extension on the C-terminal side of the zinc finger domains. By comparison, NC proteins from HIV-1 and the closely related deltaretrovirus, bovine leukemia virus, have only 6-aa extensions. Site-directed mutations in this region of HTLV-1 NC indicated that two highly conserved motifs were responsible for hA3G resistance, which consisted of a small cluster of acidic amino acids and an adjacent leucine-rich motif (LLLDL). Mutations in either of these elements resulted in increased hA3G incorporation into virions and increased susceptibility to hA3G inhibition. The resistance conferred by the C-terminal region of HTLV-1 NC was observed only in the context of HTLV-1 Gag. When expressed in trans, a peptide containing the 35 aa from the C terminus of HTLV-1 NC (with a myc-epitope tag) did not prevent hA3G packaging into NCDC or HIV-1Δvif VLPs nor did it interfere with hA3G restriction (data not shown). Furthermore, the same 35-aa peptide did not diminish hA3G packaging into VLPs when appended to the C terminus of the chimeric Gag protein containing HTLV-1 MA-CA and HIV-1 NC (NC4.0 in Fig. 2). Unfortunately, in-frame insertion of this 35-aa peptide into the NC domain of HIV-1 expression vectors abolished VLP assembly and release, making it impossible to assess effects on replication (data not shown).
The inhibition of HTLV-1 replication by hA3G required cytidine deaminase activity. Mutations in the second zinc coordination motif that were previously reported to abolish catalytic activity (11, 16, 18) failed to inhibit HTLV-1 replication. The difference between wild-type and mutated forms of hA3G as inhibitors of HTLV-1 were small because HTLV-1 resists hA3G inhibition, which probably explains why a previous report failed to detect differences between wild-type and mutated hA3G with HTLV-1 (12). The effects of hA3G were more dramatic on the NCDC mutant, and differences between wild-type (95% inhibition) and mutated hA3G (10–20% inhibition) were significant. These experiments showed that the cytidine deaminase activity of hA3G significantly contributes to antiviral restriction; but when expressed at high levels, the H257R and E259Q mutations retained some antiviral activity. HTLV-1 also was inhibited weakly by human A3F, bovine A3, and mouse A3 proteins. Deletion of the C-terminal extension of NC rendered the NCDC virus highly susceptible to restriction by all APOBECs tested. The resistance conferred by the C terminus of NC to the various APOBEC proteins suggests that these APOBECs are packaged into virions by a common mechanism.
It is likely that the mechanism of cis-acting exclusion of hA3G by HTLV-1 NC is closely related to the ways that NC, RNA, and hA3G interact. It is relevant that the C terminus of HTLV-1 NC contributes to the exclusion of hA3G from VLPs, because it is NC that brings viral and cellular RNAs into the virion. It is currently believed that both NC and RNA are necessary for incorporation of hA3G into virions (19, 20, 22, 32–34), but it is unclear whether hA3G is simply tethered to RNA (21, 22, 34) or interacts with a NC–RNA complex (11). Deleting 20 aa near the C terminus of HTLV-1 NC (NCDC mutant) did not alter the amount of viral RNA in virions but did increase the amount of hA3G that was packaged. The C terminus of HTLV-1 NC could contribute to this phenomenon in several different ways by altering the structure or nucleic acid binding properties of NC. For example, if hA3G is bound to cellular RNAs that are nonspecifically incorporated into virions, a low affinity of HTLV-1 NC for these RNAs would result in diminished hA3G packaging. This outcome would be consistent with previous physical studies showing that HTLV-1 NC binds nucleic acids with lower affinity compared with other NC proteins (35). It is also possible that HTLV-1 NC binds to RNA in such a way that hA3G is physically prevented from binding (i.e., hA3G is occluded). Another possibility is that the C terminus of NC prevents the interaction between hA3G and an RNA–NC complex, which is suggested by the deduced interaction between the C terminus and the zinc fingers of HTLV-1 NC (35). Future in vitro analyses of the RNA binding and chaperone activity of HTLV-1 NC and characterization of the molecular interactions among hA3G, RNA, and NC will resolve this issue and shed light on the general mechanism by which hA3G is packaged into virus particles.
Materials and Methods
Plasmids.
HTLV-1 and HIV-1 vectors for single-cycle replication assays have been described (30). Briefly, the systems consist of packaging plasmids for HTLV-1 (pCMVHTΔEnv) and HIV-1 (pCMV-Δ8.2R), transfer vectors that encode a GFP-luciferase fusion protein (pHTC-GFPLuc and pUCHR-GFPLuc for HTLV-1 and HIV-1, respectively), and VSV-G expression plasmid, pCMV-VSVG for pseudotyping. The HTLV-1 mutant, pCMVHTΔX-NCDC, was derived from pCMVHTΔEnv and encodes a NC protein lacking amino acids (PTIPEPEPEEDALLLDLPAD) near the C terminus of the protein. Other pCMVHTΔEnv vectors with mutations in the NC domain of gag were constructed by site-directed mutagenesis. HTLV-1 Gag expression plasmids were constructed by cloning portions of the gag gene into an expression plasmid, pCMVREM, containing the HIV-1 Rev responsive element. pCMVREM2.0 contains the WT HTLV-1 gag gene. pCMVREM2-NC58 and other Gag expression plasmids with deletions at the 3′ end of NC were made by PCR amplification; the number at the end designates the number of amino acids remaining after deletions from the 85-aa NC domain. pCMVREM4.0 contains HTLV-1 MA and CA domains fused to HIV-1 NC. pRS-HRev encodes HIV-1 Rev under the control of a Rous sarcoma virus promoter and has been described (36). HA-tagged human A3G and mouse A3 expression plasmids were provided by Nathaniel Landau (The Salk Institute, San Diego, CA). Human A3F and bovine A3 expression plasmids were a gift from Reuben Harris (University of Minnesota, Minneapolis, MN). Mutations in the catalytic domains of hA3G to generate hA3G-H257R and hA3G-E259Q were made by site-directed mutagenesis, cloned into mammalian expression plasmids, and confirmed by nucleotide sequence analysis.
Transfections and Infections.
Human 293T and HeLa cells were maintained in DMEM supplemented with 10% FBS. Single-cycle replication assays were performed as described (30). Briefly, 6-cm plates of 293T cells were transfected with 1 μg of packaging plasmid, 3 μg of transfer vector, 0.25 μg of pCMV-VSVG plasmid, and varying amounts of hA3G-HA expression plasmid or empty vector. Supernatants were collected 48 h after transfection, filtered, and applied to HeLa cell targets for infection. Luciferase activity was measured in HeLa cell extracts 72 h after infection. Aliquots of the filtered supernatants were saved for Gag ELISA and immunoblotting. Both HIV-1 (p24-CA) and HTLV-1 (p19-MA) ELISA kits from Zeptometrix (Buffalo, NY) include a purified protein antigen as a reference standard. Infection experiments were performed in duplicate and repeated at least two times.
Immunoblotting.
VLPs from transfected 293T cells were concentrated by ultracentrifugation through 20% glycerol cushions as described (30). Cell and VLP extracts were fractionated by SDS/PAGE and transferred to Immobilon membranes. Blots were probed with anti-HA monoclonal antibody (Covance, Richmond, CA), anti-hA3G rabbit polyclonal antisera (generously provided by Klaus Strebel, National Institutes of Health, Bethesda, MD), and anti-HTLV-1 p19 (MA) monoclonal antibody (Zeptometrix).
RNA and Northern Blot Analysis.
Viral mRNA was prepared from virions using RNA-STAT 60 reagent (Tel-Test, Friendswood, TX) and fractionated on agarose-formaldehyde gels. After transfer to nylon membrane, blots were probed with a 32P-labeled DNA fragment from the HTLV-1 gag gene (30). Hybridization was quantified on a Storm phosphorimager (Applied Biosystems, Foster City, CA).
Acknowledgments
We thank Richard Frederickson for graphics. This work was supported by the Intramural Research Program of the National Institutes of Health and the National Cancer Institute, Center for Cancer Research.
Abbreviations
- HTLV-1
human T cell leukemia virus type 1
- hA3G
human APOBEC3G
- VLP
virus-like particle
- MA
matrix
- CA
capsid
- NC
nucleocapsid.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 2.Harris RS, Liddament MT. Nat Rev Immunol. 2004;4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 3.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 4.Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, Bollman B, Munk C, Nymark-McMahon H, Landau NR. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi M, Takaori-Kondo A, Shindo K, Abudu A, Fukunaga K, Uchiyama T. J Virol. 2004;78:8238–8244. doi: 10.1128/JVI.78.15.8238-8244.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 7.Doehle BP, Schafer A, Wiegand HL, Bogerd HP, Cullen BR. J Virol. 2005;79:8201–8207. doi: 10.1128/JVI.79.13.8201-8207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell RA, Wiegand HL, Moore MD, Schafer A, McClure MO, Cullen BR. J Virol. 2005;79:8724–8731. doi: 10.1128/JVI.79.14.8724-8731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turelli P, Mangeat B, Jost S, Vianin S, Trono D. Science. 2004;303:1829. doi: 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- 10.Mahieux R, Suspene R, Delebecque F, Henry M, Schwartz O, Wain-Hobson S, Vartanian JP. J Gen Virol. 2005;86:2489–2494. doi: 10.1099/vir.0.80973-0. [DOI] [PubMed] [Google Scholar]
- 11.Navarro F, Bollman B, Chen H, Konig R, Yu Q, Chiles K, Landau NR. Virology. 2005;333:374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Sasada A, Takaori-Kondo A, Shirakawa K, Kobayashi M, Abudu A, Hishizawa M, Imada K, Tanaka Y, Uchiyama T. Retrovirology. 2005;2:32. doi: 10.1186/1742-4690-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lecossier D, Bouchonnet F, Clavel F, Hance AJ. Science. 2003;300:1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Nat Struct Mol Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 16.Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. Curr Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 17.Chiu YL, Soros VB, Kreisberg JF, Stopak K, Yonemoto W, Greene WC. Nature. 2005;435:108–114. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- 18.Shindo K, Takaori-Kondo A, Kobayashi M, Abudu A, Fukunaga K, Uchiyama T. J Biol Chem. 2003;278:44412–44416. doi: 10.1074/jbc.C300376200. [DOI] [PubMed] [Google Scholar]
- 19.Alce TM, Popik W. J Biol Chem. 2004;279:34083–34086. doi: 10.1074/jbc.C400235200. [DOI] [PubMed] [Google Scholar]
- 20.Cen S, Guo F, Niu M, Saadatmand J, Deflassieux J, Kleiman L. J Biol Chem. 2004;279:33177–33184. doi: 10.1074/jbc.M402062200. [DOI] [PubMed] [Google Scholar]
- 21.Svarovskaia ES, Xu H, Mbisa JL, Barr R, Gorelick RJ, Ono A, Freed EO, Hu WS, Pathak VK. J Biol Chem. 2004;279:35822–35828. doi: 10.1074/jbc.M405761200. [DOI] [PubMed] [Google Scholar]
- 22.Zennou V, Perez-Caballero D, Gottlinger H, Bieniasz PD. J Virol. 2004;78:12058–12061. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conticello SG, Harris RS, Neuberger MS. Curr Biol. 2003;13:2009–2013. doi: 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 24.Sheehy AM, Gaddis NC, Malim MH. Nat Med. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 25.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 26.Marin M, Rose KM, Kozak SL, Kabat D. Nat Med. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 27.Stopak K, de Noronha C, Yonemoto W, Greene WC. Mol Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- 28.Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. J Biol Chem. 2004;279:7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- 29.Sova P, Volsky DJ. J Virol. 1993;67:6322–6326. doi: 10.1128/jvi.67.10.6322-6326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Derse D, Hill SA, Lloyd PA, Chung HK, Morse BA. J Virol. 2001;75:8461–8468. doi: 10.1128/JVI.75.18.8461-8468.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Switzer WM, Qari SH, Wolfe ND, Burke DS, Folks TM, Heneine W. J Virol. 2006;80:7427–7438. doi: 10.1128/JVI.00690-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Douaisi M, Dussart S, Courcoul M, Bessou G, Vigne R, Decroly E. Biochem Biophys Res Commun. 2004;321:566–573. doi: 10.1016/j.bbrc.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 33.Luo K, Liu B, Xiao Z, Yu Y, Yu X, Gorelick R, Yu XF. J Virol. 2004;78:11841–11852. doi: 10.1128/JVI.78.21.11841-11852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schafer A, Bogerd HP, Cullen BR. Virology. 2004;328:163–168. doi: 10.1016/j.virol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Morcock DR, Kane BP, Casas-Finet JR. Biochim Biophys Acta. 2000;1481:381–394. doi: 10.1016/s0167-4838(00)00181-3. [DOI] [PubMed] [Google Scholar]
- 36.Martarano L, Stephens R, Rice N, Derse D. J Virol. 1994;68:3102–3111. doi: 10.1128/jvi.68.5.3102-3111.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]