Abstract
Chlamydia trachomatis is a bacterial pathogen that infects the eyes and urogenital tract. Ocular infection by this organism is the leading cause of preventable blindness worldwide. The infection is also a leading cause of sexually transmitted disease in the United States. As obligate intracellular pathogens, chlamydiae have evolved sophisticated, yet undefined, mechanisms to maintain a favorable habitat for intracellular growth while avoiding harm to the host. We show here that chlamydiae have the ability to interfere with the NF-κB pathway of host inflammatory response. We found that Chlamydia infection did not promote IκBα degradation, a prerequisite for NF-κB nuclear translocation/activation, nor induce p65/RelA nuclear redistribution. Instead, it caused p65 cleavage into an N terminus-derived p40 fragment and a p22 of the C terminus. The activity was specific because no protein cleavage or degradation of NF-κB pathway components was detected. Moreover, murine p65 protein was resistant to cleavage by both human and mouse biovars. The chlamydial protein that selectively cleaved p65 was identified as a tail-specific protease (CT441). Importantly, expression of either this protease or the p40 cleavage product could block NF-κB activation. A hallmark of chlamydial STD is its asymptomatic nature, although inflammatory cellular response and chronic inflammation are among the underlying mechanisms. The data presented here demonstrate that chlamydiae have the ability to convert a regulatory molecule of host inflammatory response to a dominant negative inhibitor of the same pathway potentially to minimize inflammation.
Keywords: CT441, inflammation, protease
Chlamydiae are obligate intracellular bacterial pathogens that infect a broad range of cell types, including those of the eye and genital tract epithelia. Ocular infection of Chlamydia trachomatis is the leading cause of preventable blindness worldwide, and urogenital tract infection remains the most prevalent cause of sexually transmitted diseases in the United States, resulting in pelvic inflammatory disease, ectopic pregnancy, and infertility (1–3). The production of proinflammatory chemokines, such as IL-8, has been considered as a primary factor of Chlamydia diseases (4). Tears from children with trachoma and endocervical secretions from women with Chlamydia infection have elevated IL-8 production (4). IL-8 is found in the human fallopian tube predominantly in the epithelial cells and is present in greater amounts in the distal tube, a region that coincides with severe tissue damage of Chlamydia infection. Cervical epithelial cells, a primary target of Chlamydia infection, produce IL-6 and IL-8 (5, 6).
The production of IL-6 and IL-8 is transcriptionally regulated by NF-κB activity (7). The NF-κB consists of a heterodimeric complex composed of two subunits, commonly p50/NF-κB1 and p65/RelA, which are sequestered in the cytoplasm and are rendered inactive through their association with inhibitory molecules, including IκBα. Bacterial infection or proinflammatory cytokine or LPS stimulation induces rapid degradation of IκB proteins (7, 8), resulting in the release and nuclear translocation of the NF-κB complex for gene regulation. Although chlamydial components, such as purified LPS, were reported to activate the NF-κB pathway (9, 10), no significant NF-κB activation was detected during Chlamydia infection (11). Furthermore, we found that the infection promotes IL-8 production through MAPK activation because inhibition of MAPK activity blocks IL-8 production.
Several viral and bacterial pathogens have been reported to disrupt the NF-κB signaling pathway for immune evasion (12–14). To investigate whether Chlamydia has the capacity to interfere with NF-κB signal transduction, we evaluated NF-κB activation through examination of IκBα degradation, a prerequisite of NF-κB activation and gene regulation. We found that Chlamydia infection did not induce IκBα degradation or p65 nuclear translocation; instead, it promoted p65 cleavage. In addition, the p65 cleavage activity was detected from purified Chlamydia. Here, we report the characterization of p65 cleavage by Chlamydia infection.
Results
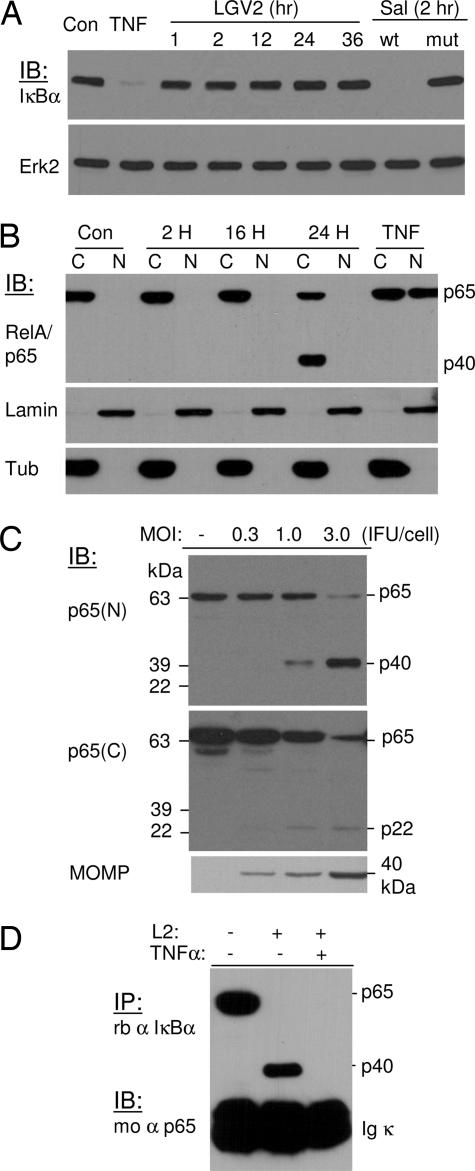
To investigate whether Chlamydia infection promoted NF-κB activation, we addressed the question of whether the infection induced IκBα degradation. To this end, monolayers of HeLa 229 cells were infected with C. trachomatis lymphogranuloma venereum 2 (LGV2) at 1 inclusion-forming unit (IFU) per cell for various time points (15). As controls, the cells were infected with invasive or noninvasive Salmonella typhimurium for 2 h or treated with TNF-α to induce IκBα degradation (16, 17). Unlike TNF-α treatment or infection with pathogenic S. typhimurium, which caused IκBα degradation, infection with C. trachomatis did not affect IκBα expression nor its degradation (Fig. 1A), suggesting that Chlamydia infection does not promote NF-κB activation.
Fig. 1.
C. trachomatis infection does not induce IκBα degradation; instead, it promotes cleavage of p65/RelA. (A) HeLa 229 cells were infected with C. trachomatis LGV2 at 1 IFU per cell for various times or with invasive (wt) or invasion-defective (mut) S. typhimurium for 2 h. TNF-α at 10 ng/ml was used as a positive control for IκBα degradation. The expression and degradation of IκBα were detected by immunoblotting analyses. Erk2 expression was used as a loading control. (B) HeLa 229 cells were infected with LGV2 at 1 IFU per cell for 2, 16, and 24 h. The cytosolic (C) and nuclear (N) fractions were separated, and p65/RelA redistribution was detected by blotting analysis. The expression of lamin A and β-tubulin were used as loading controls for nuclear and cytosolic fractions, respectively. Chlamydia infection does not promote nuclear translocation. In addition, a cleavage product of 40 kDa (p40) was detected from the cytosolic fraction of a sample of late infection. (C) Whole-cell lysates from infected and uninfected control cells were separated by SDS/PAGE, and cleavage of p65 was detected with an N or C terminus-specific antibody against p65. A 40-kDa (p40) protein band and a 22-kDa (p22) protein band were detected from samples infected for 28 h with the N and C terminus-specific antibody, respectively. The production of MOMP was monitored as an indication of infection. (D) Association of p40 with IκBα protein. HeLa 229 cells were infected with Chlamydia LGV2 at a multiplicity of infection of 3 for 28 h. The cells were then treated with 10 ng/ml TNF-α for 15 min to induce IκBα degradation, or the cells remained untreated. Lysates from infected and uninfected samples were immunoprecipitated with an anti-IκBα antibody. The association of wild-type p65 or the cleavage product with IκBα was detected by immunoblotting. Both wild-type p65 and p40 associate with IκBα. Depletion of IκBα with TNF-α treatment abolishes the association of p40 with IκBα.
To verify these findings, we examined the nuclear translocation of p65 as a measurement of NF-κB activation. The NF-κB consists of a heterodimeric complex composed of two subunits, commonly p50/NFκB1 and p65/RelA, which are sequestered in the cytoplasm and rendered inactive through their association with inhibitory molecules, including IκBα. The distribution of p65 in the cytosol and the nuclei of infected and uninfected control cells was detected after fractionation by immunoblotting analysis with an N terminus-specific anti-p65 antibody (18). Consistent with a previous study (11) and our results from the IκBα degradation studies, Chlamydia infection did not promote NF-κB activation, as determined by the absence of p65 nuclear translocation (Fig. 1B). However, a protein band of ≈40 kDa was detected from the cytosolic preparation of cells infected for 24 h, suggesting that Chlamydia infection can potentially cause p65 cleavage.
The cleavage of p65 was confirmed by the detection of a 22-kDa fragment with a C terminus-specific antibody against p65 (Fig. 1C). Enhanced detection of p22 was observed in cells treated with MG-132 (data not shown), suggesting that this cleavage product was subjected to proteasomal degradation. The 40-kDa fragment was stable and was subjected to further characterization after purification. To this end, 293T cells transfected for GST-p65 protein expression were infected with LGV2 for 28 h. After purification, the cleavage product was subjected to trypsin digestion followed by MALDI-TOF identification. Seven proteolytic fragments were detected, which covered the N-terminal GST tag and the RHD region of p65 (data not shown). These findings are consistent with results from immunoprecipitation studies. Cell lysates from Chlamydia-infected or uninfected control cells were immunoprecipitated with an anti-IκBα antibody. The presence of p65 or p40 in the immunocomplexes was detected by immunoblotting. As shown in Fig. 1D, both p65 and p40 could associate with IκBα. The association of p40 with IκBα was specific because depletion of IκBα in the infected cells by TNF-α treatment resulted in the disappearance of p40 in the anti-IκBα immunocomplexes. Together, these results demonstrated that Chlamydia infection causes p65 cleavage into a p22 fragment of the C terminus and a p40 fragment for IκBα binding.
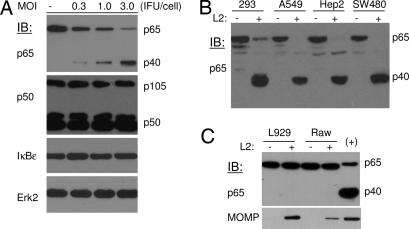
The cleavage of p65 was selective because expression of the p50/105, IκBα, and IκBε proteins of the NF-κB signaling pathway was not affected by Chlamydia infection (Fig. 2A). In addition, p65 cleavage was not restricted to HeLa 229 cells. We observed that Chlamydia infection caused p65 cleavage in human cell lines of different origins (Fig. 2B). However, no p65 cleavage by C. trachomatis infection was detected in murine cells (Fig. 2C), suggesting the cleavage is species-specific.
Fig. 2.
Specificity and selectivity of p65 cleavage by Chlamydia. (A) HeLa 229 cells were infected with LGV2 at various multiplicities of infection for 28 h. The expression of p50/105, IκBε, and IκBα, molecules of the NF-κB signaling pathway, in infected and uninfected control cells was determined by blotting analysis. Erk2 expression was used as a loading control. (B and C) The cleavage of p65 protein is species-specific. Monolayers of human and mouse cells were infected with LGV2 for 28 h. The expression and cleavage of p65 was detected by immunoblot analysis. No p65 cleavage was detected from Chlamydia-infected murine cells. MOMP content in murine cells was determined as an indication of productive infection. +, A sample of Chlamydia-infected 293 cell lysate as a positive control.
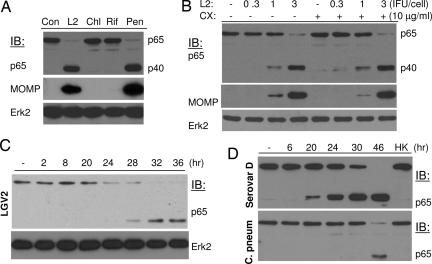
To preliminarily investigate whether a host or bacterial factor contributed to p65 cleavage, the infected cells were treated with antibiotics to inhibit bacterial growth or with cycloheximide to block host protein synthesis. Treatment with bacteria-static chloramphenicol or rifampin blocked p65 cleavage (Fig. 3A). In contrast, treatment with penicillin G, an antibiotic that inhibits Chlamydia maturation but not its growth (19) or with cycloheximide (Fig. 3B) did not prevent p65 cleavage, suggesting p65 cleavage depends on bacterial protein synthesis.
Fig. 3.
Chlamydia cleavage of p65 depends on bacterial growth. Inhibition of Chlamydia growth with antibiotics, but not host protein synthesis with cycloheximide, blocks p65 cleavage. (A) HeLa 229 cells were infected with Chlamydia LGV2 at a multiplicity of infection of 1. The infected cells remained untreated or were treated with 0.1 μg/ml rifampin (Rif) or 60 μg/ml chloramphenicol (Chl), reagents that inhibit Chlamydia growth by blocking transcription or translation, respectively. Penicillin G (Pen; 100 μg/ml), which does not block Chlamydia replication but its maturation, was included as a control. Inhibition of Chlamydia growth, as determined by MOMP production, blocks p65 cleavage activity. (B) In a similar experiment, the cells were infected with LGV2 at various multiplicities of infection for 18 h before treatment with 10 μg/ml cycloheximide (CX). The cells were harvested 28 h after infection, and p65 cleavage as well as MOMP production in cycloheximide-treated and untreated samples were detected by immunoblotting. Erk2 and MOMP was detected as loading controls for cellular and bacterial growth. (C and D) Time course of p65 cleavage during Chlamydia infection. HeLa 229 cells were infected with C. trachomatis LGV2 (C), serovar D, or C. pneumoniae (D) at 1 IFU per cell for various times. Protein expression and cleavage of p65 were detected by blotting analyses. The p65 cleavage activity is associated with the late stages of Chlamydia growth. HK, heat-inactivated Chlamydia EB.
C. trachomatis has a unique biphasic developmental cycle (20, 21). After internalization, the infectious elementary body (EB) differentiates into a metabolically active reticulate body for replication within 8–12 h after infection. The active reticulate body begins to redifferentiate into EB at ≈20–24 h after infection. No p65 cleavage was detected during the Chlamydia replication phase (Fig. 3C). The cleavage was detected at ≈24 h after infection and persisted thereafter. In fact, serovar D exhibited a similar pattern of p65 cleavage, whereas cleavage by Chlamydia pneumoniae was delayed (Fig. 3D) but was consistent with the slower growth rate of this organism, indicating the cleavage activity was associated with EB.
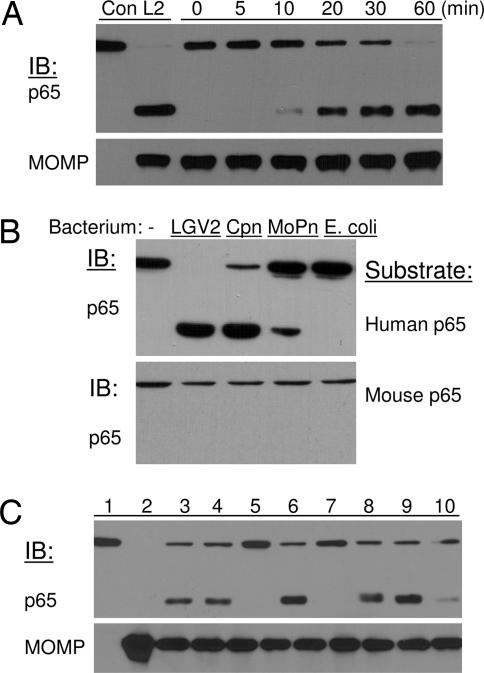
This conclusion was substantiated by results from in vitro protease activity assays. Gradient-purified LGV2 EB was solubilized with a buffer containing 1% Nonidet P-40. The fraction containing soluble bacterial proteins was found to have p65 cleavage activity (Fig. 4A). In parallel experiments, we also tested p65 cleavage activity of C. pneumoniae and C. trachomatis of mouse pneumonitis (MoPn). Similar to the LGV2 serovar, C. pneumoniae and MoPn also displayed p65 cleavage activity. No p65 cleavage was detected from a lysate of purified Escherichia coli (Fig. 4B). Consistent with the results from in vivo studies, these chlamydial lysates showed no activity against murine p65 protein (Fig. 4B Lower). In addition, we found that the activity was independent of multivalent cations because inclusion of EDTA (Fig. 4C, lane 6) had no effect on p65 cleavage. Instead, lactacystin inhibited the p65 cleavage activity of Chlamydia (Fig. 4C, lane 7), suggesting potential involvement of serine proteases (22).
Fig. 4.
Purified Chlamydia elementary bodies contain p65 cleavage activity. (A) Gradient-purified EBs were solubilized with a buffer containing 1% Nonidet P-40. One microgram of soluble bacterial proteins was incubated with p65 from 293T cells at 37°C for various times as indicated. (Upper) The two lanes on the far left, labeled con and L2, represent samples from uninfected and LGV2 infected cells, respectively. (Lower) The content of MOMP was used as a measure of EB input. (B) Soluble proteins from different Chlamydia species contain p65 cleavage activity against human, but not mouse, p65 proteins. (C) Treatment with serine protease inhibitors blocks p65 cleavage activity. In parallel experiments, protease inhibitors were included in the in vitro p65 cleavage assay. Lanes: 1, 293T lysate; 2, solubilized EB; 3, p65 and EB proteins incubated at 37°C for 60 min; 4, Roche protease inhibitor mixture at 10×; 5, heat-inactivated EB; 6, EDTA at 5 mM; 7, lactacystin at 10 μM; 8, MG132 at 20 μM; 9, tosylphenylalanylchloromethane at 20 μM; 10, tosyl-lysylchloromethane at 20 μM. MOMP was used as a loading control.
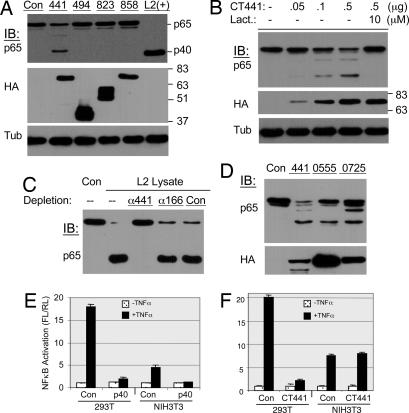
The chlamydial genome encodes 16 proteases (23), including four putative proteases that contain a leader sequence for periplasmic secretion and potential translocation by the chlamydial type III secretory system. The mature peptides of those putative proteases were therefore screened for their ability to cleave p65 after expression in 293T cells. As shown in Fig. 5A, expression of CT441 caused p65 cleavage. No activity was detected from overexpressed CT494, CT823, and CT858 proteins. The cleavage by CT441 was dose-dependent and relied on protease activity because lactacystin treatment prevented p65 cleavage (Fig. 5B). CT858 also encodes CPAF, a tail-specific protease (Tsp) of Chlamydia sp., which has previously been reported to promote protein degradation of RFX5 transcriptional factor (24) and Bim protein of apoptosis (25). In addition to lack of p65 cleavage activity, it is worth noting that CPAF also differs from CT441 in that CPAF activity was not detected from purified Chlamydia (26), whereas p65 cleavage activity was found from purified bacteria as well as during an infection.
Fig. 5.
Identification and characterization of a chlamydial protease that cleaves p65 protein. (A) Mature peptides of putative chlamydial proteases were expressed as N-terminal, HA-tagged proteins in 293T cells and screened for p65 cleavage ability. Expression of CT441, a tail-specific chlamydial protease, resulted in selective cleavage of p65. No activity was detected from CT494, CT823, or CT858. (B) Increased expression of CT441 promoted more profound cleavage of p65. (C) Immunodepletion assay. Cellular preparations containing p65 cleavage activity were depleted by incubation with a polyclonal antibody that was immobolized on protein G Sepharose beads or a serum from a preimmunized animal. The supernatants were assayed for p65 cleavage activity. Depletion with anti-CT441 (α441) but not a control serum from the same animal (con) or anti-CT166 (α166) ablated p65 cleavage activity. (D) Homologous proteins from C. pneumoniae (Cpn0555) and MoPn (TC0725) also exhibited p65 cleavage activity when expressed in 293T cells. (E and F) Expression of CT441 or p40, the cleavage product derived from the N terminus of p65, inhibited NF-κB activity. Monolayers of 293T or NIH 3T3 cells were cotransfected with pIL8-Luc (27) and a plasmid for the expression of chlamydial protease CT441 or the p40 cleavage product of p65. pSV40-RL was included as an internal control for transfection reactions. The cells were treated with TNF-α (10 ng/ml) 18 h after transfection to induce luciferase expression. Luciferase activities were determined with a Dual-Glo reagent kit (Promega) 18 h after TNF-α treatment. The ratio of firefly and Renilla luciferase activities (FL/RL) was plotted. All transfections were performed in duplicate, and the data are presented as means ± error.
The essential role of CT441 in p65 cleavage was further demonstrated by immunodepletion (Fig. 5C). Fractions of soluble proteins that were used for in vitro p65 cleavage assay were incubated with a polyclonal antibody immobolized on protein G Sepharose beads. Depletion with anti-CT441 anti-serum, but not with a serum from the same animal of preimmunization or from a CT166-immunized animal, ablated p65 cleavage activity in the supernatant.
CT441 is a Tsp. A database search indicates that this protease shares high homology among Chlamydia species, although not with other organisms. Indeed, the protein from serovar A of ocular infection is identical to CT441 of serovar D. The corresponding Tsp (Cpn0555) of C. pneumoniae and TC0725 of murine C. trachomatis (MoPn) also displayed p65 cleavage activity (Fig. 5D). These Tsp, however, exhibited no cleavage activity when expressed in murine cells, further demonstrating the species specificity of p65 cleavage by Chlamydia infection.
Chlamydia infection induces proinflammatory cytokine production. Unlike most invasive microorganisms that promote rapid but transient inflammatory response, cytokine production by Chlamydia infection is significantly delayed and is dependent on bacterial growth. Furthermore, IL-8 production by Chlamydia infection is regulated by MAPK activity. We hypothesized that Chlamydia utilizes Tsp to suppress host inflammatory response. The lack of genetic mutants, or tools to generate such mutants, prevented a direct demonstration of CT441 as a virulence factor in an infection setting. We therefore investigated whether expression of CT441 interfered with NF-κB signal transduction. To this end, 293T or NIH 3T3 cells were transfected with pIL8-luciferase (27) and a plasmid for p40 or CT441 expression. The cells were then treated with TNF-α to induce luciferase expression. Expression of p40 (amino acid residues 1–351 of p65) inhibited luciferase expression in both 293T and NIH 3T3 cells (Fig. 5E). However, such inhibitory effect was detected only in 293T but not in murine NIH 3T3 cells that were transfected for CT441 expression (Fig. 5F). These data directly link p40 production by CT441 or Chlamydia infection to inhibition of NF-κB activity.
Discussion
Chlamydia infection is the most common cause of the notifiable diseases in the United States (1). Most patients are not aware of an infection because the disease generally remains asymptomatic. Chlamydiae are obligate intracellular bacterial pathogens that have evolved sophisticated mechanisms to secure a favorable habitat for progeny production while avoiding harm to their host. We found that chlamydiae contain a Tsp that selectively cleaves the p65/RelA subunit of NF-κB to potentially interfere with host inflammatory response. Indeed, the p40 cleavage product that contains an intact RHD of p65 for IκBα interaction and DNA binding functions as a dominant negative regulator of NF-κB activation. This postulation was substantiated with the use of cloned CT441 in cleavage susceptible as well as resistant cells. We found that expression of CT441 inhibited TNF-α-induced NF-κB activation in human cells but not in mouse fibroblast cells because murine p65 protein is resistant to Chlamydia and CT441 cleavage activity.
Several limited examples demonstrate that viral and bacterial pathogens can exploit the NF-κB pathway to regulate cellular response (12–14, 28, 29). Nonpathogenic Salmonella sp. was found to inhibit NF-κB translocation by blocking ubiquitination of IκBα (17) and a commensal anaerobic bacterium to attenuate inflammation by causing efflux of nuclear p65 (30). In addition to promoting p65 cleavage, Chlamydia sp. may apply multiple mechanisms to interfere with host inflammatory response. Chlamydiae are the only bacterial pathogens with sequenced genomes that encode a protease that is homologous to A20 of protein ubiquitination/deubiquitination activity (31–33). Chlamydial components, such as LPS, were reported to activate the NF-κB pathway. However, Chlamydia infection does not induce NF-κB activation. Instead, it protects IκBα from TNF-α induced degradation. It remains uninvestigated whether chlamydial deubiquitin-like proteases play a role in protecting IκBα from degradation. Together, these studies define molecular mechanisms that are used by microorganisms to evade the immune system.
The cleavage of p65 protein by Chlamydia sp. is species-specific. We found that different serovars and biovars of chlamydiae cleave human p65 protein, but not murine p65 protein, effectively. Murine models of genital tract infection are widely used for the studies of host response to Chlamydia infection because these model systems in many aspects closely mimic acute genital tract infection in women (34). However, one of the major differences between the model system and human infection is the duration of an infection (2, 34). Chlamydia infection in mice generally resolves in a few weeks without antimicrobial treatment, whereas the infection can last several months in humans before spontaneous clearance. Moreover, repeat and persistent Chlamydia infection occurs in humans, whereas mice tend to develop a long-lasting immunity that protects against reinfection (34). A key event in triggering immune response for innate and adaptive immunity involves NF-κB pathway activation (35–37). Disruption of NF-κB signal transduction with the encoded Tsp may allow for persistent Chlamydia infection in humans and prevent the establishment of a long-lasting immunity against the infection.
A safe and effective vaccine has been identified as essential in controlling infection with C. trachomatis, but progress toward this goal has proven to be very challenging with disappointedly modest results (2, 34). The development of Chlamydia vaccines have historically been focused on the induction of humoral immunity (2). Recent studies have confirmed the importance of both B and T cells in resolving chlamydial genital infection (38). Chlamydiae are obligate intracellular bacteria that normally infect the single-cell columnar layer of the genital tract epithelia, which express MHC class I molecules. In addition to mucosal transmission for infection, adding difficulties for vaccine development, these organisms have also evolved mechanisms for immune evasion. The identification of a Tsp that selectively targets the NF-κB pathway for immune evasion may be selected as a candidate for vaccine development. Cell-mediated immunity targeting genes critical for Chlamydia intracellular survival and immune evasion may be advantageous in eliminating Chlamydia-harboring cells before the production of progeny.
Methods
Bacterial strains.
C. trachomatis serovars C (TW-3), D (UW-3/Cx), and F (IC-Cal-3) and C. pneumoniae CWL-029 were obtained from the American Type Culture Collection (Manassas, VA). C. trachomatis lymphogranuloma venereum (LGV2) was obtained from G. Zhong (University of Texas Health Science Center, San Antonio, TX) and C. trachomatis of MoPn Nigg was obtained from L. M. de la Maza (University of California, Irvine, CA). The titers of these bacteria were determined in HeLa 229 cells and are expressed as IFU per milliliter.
Infection Assays.
Monolayers of HeLa 229 cells, or cells as specified, were infected with LGV2 at 1 IFU per cell or as otherwise stated. To improve infection efficiency by serovars or biovars other than LGV2, the cells were treated with 30 μg/ml DEAE-dextran before inoculation. No cycloheximide was used during infection assays.
Antibodies and Western Blots.
Except as noted, extracts were prepared by lysis of cells with a buffer containing 143 mM NaCl, 50 mM Tris·HCl (pH 7.5), 1 mM DTT, 1% Nonidet P-40, 0.1% SDS, and protease inhibitors (Roche Diagnostics, Indianapolis, IN). Soluble proteins were separated by SDS/PAGE or Tris-glycine gels (Invitrogen, Carlsbad, CA). Blots were probed with specific antibodies. Rabbit anti-CT441 (amino acids 353–492) was generated by following protocol approved by an Institutional Animal Care and Use Committee. This antibody recognizes a single protein band of 70 kDa from purified EB or total lysate of Chlamydia-infected cells. Antibody to chlamydial major outer membrane protein (MOMP) was from L. M. de la Maza. Antibodies to p65 (N), IκBα, and Erk2 were from Santa Cruz Biotechnology (Santa Cruz, CA), p50/105 and p65 (C) were from eBiosciences (San Diego, CA), IκBε was from Active Motif (Carlsbad, CA), and β-tubulin and the HA tag were from Sigma (St. Louis, MO).
In Vitro p65 Cleavage Assays.
Gradient-purified chlamydial EB were solubilized with Tris-buffered saline supplemented with 1% Nonidet P-40. The soluble proteins were collected and used for p65 cleavage assays. To assay for p65 cleavage ability, 1 μg of bacterial proteins was resuspended in 30 μl of reaction buffer containing 20 mM Tris·HCl and 143 mM NaCl, with or without p65 protein expressed in 293T cells or the endogenous p65 from 293T cell lysate. For inhibition assays, an inhibitor was added 30 min before the addition of p65 protein. All assays were performed at 37°C for 60 min or as otherwise indicated.
Constructs for Expression of Bacterial Proteases.
cDNAs of the putative proteases (23) were PCR-amplified by using purified bacteria as templates and were inserted into pRK5/HA for N-terminal HA-tagged protein expression. The proteins were expressed in 293T cells by transient transfection with FuGene-6 (Roche) and assayed for their ability to cleave the endogenous p65 protein with Western blot analyses.
Reporter Gene Assay.
Monolayers of 293T or NIH 3T3 cells in 24-well plates were transfected in duplicates with a mixture of DNA preparations (0.06 μg of pIL8-GL, 0.01 μg of pSV40-RL, and 0.13 μg of pRK5/Myc or a plasmid DNA for protein expression). To construct a plasmid for p40 expression, we first mapped the cleavage site of p65 to amino acid residue 351 with standard molecular biology protocols (unpublished data). The DNA corresponding to amino acids 1–351 was inserted into pRK5/Myc for Myc-p40 protein expression [p22 was not included due to its instability; see supporting information (SI) Fig. 6]. The transfected cells were treated with human 10 ng/ml TNF-α 18 h after transfection to induce firefly luciferase expression, or the cells remained untreated. The cells were harvested 18 h later, and luciferase activities were assayed with a Dual-Glo reagent kit (Promega, Madison, WI). The results were presented as the ratio of firefly luciferase activity against Renilla luciferase activity (mean ± error of duplicate samples), with 1 as a ratio for the corresponding controls.
Supplementary Material
Acknowledgments
We thank E. Fukuda for technical assistance; C. Wrank for proteomic studies; P. Rutledge for administrative assistance; and J. Mathison, D. Stupack, and J. D. Lee for scientific discussions. This work was supported by a grant from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health.
Abbreviations
- EB
chlamydial elementary body
- LGV2
lymphogranuloma venereum 2
- IFU
inclusion-forming unit
- MoPn
Chlamydia trachomatis of mouse pneumonitis
- MOMP
chlamydial major outer membrane protein
- Tsp
tail-specific protease.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0608393104/DC1.
References
- 1.Centers for Disease Control and Prevention. MMWR Morb Mortal Wkly Rep. 2003;52:16–18. [PubMed] [Google Scholar]
- 2.Brunham RC, Rey-Ladino J. Nat Rev Immunol. 2005;5:149–161. doi: 10.1038/nri1551. [DOI] [PubMed] [Google Scholar]
- 3.Belland R, Ojcius DM, Byrne GI. Nat Rev Microbiol. 2004;2:530–531. doi: 10.1038/nrmicro931. [DOI] [PubMed] [Google Scholar]
- 4.Stephens RS. Trends Microbiol. 2003;11:44–51. doi: 10.1016/s0966-842x(02)00011-2. [DOI] [PubMed] [Google Scholar]
- 5.Rasmussen SJ, Eckmann L, Quayle AJ, Shen L, Zhang YX, Anderson DJ, Fierer J, Stephens RS, Kagnoff MF. J Clin Invest. 1997;99:77–87. doi: 10.1172/JCI119136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukuda EY, Lad SP, Mikolon DP, Iacobelli-Martinez M, Li E. Infect Immun. 2005;73:4017–4024. doi: 10.1128/IAI.73.7.4017-4024.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richmond A. Nat Rev Immunol. 2002;2:664–674. doi: 10.1038/nri887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karin M, Ben-Neriah Y. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 9.Ingalls RR, Rice PA, Qureshi N, Takayama K, Lin JS, Golenbock DT. Infect Immun. 1995;63:3125–3130. doi: 10.1128/iai.63.8.3125-3130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heine H, Muller-Loennies S, Brade L, Lindner B, Brade H. Eur J Biochem. 2003;270:440–450. doi: 10.1046/j.1432-1033.2003.03392.x. [DOI] [PubMed] [Google Scholar]
- 11.Molestina RE, Miller RD, Lentsch AB, Ramirez JA, Summersgill JT. Infect Immun. 2000;68:4282–4288. doi: 10.1128/iai.68.7.4282-4288.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyer L, Lemichez E. Nat Rev Micro. 2004;2:779–788. doi: 10.1038/nrmicro1005. [DOI] [PubMed] [Google Scholar]
- 13.Akira S, Uematsu S, Takeuchi O. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Santoro MG, Rossi A, Amici C. EMBO J. 2003;22:2552–2560. doi: 10.1093/emboj/cdg267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lad SP, Fukuda EY, Li J, de la Maza LM, Li E. J Immunol. 2005;174:7186–7193. doi: 10.4049/jimmunol.174.11.7186. [DOI] [PubMed] [Google Scholar]
- 16.Jones BD, Ghori N, Falkow S. J Exp Med. 1994;180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neish AS, Gewirtz AT, Zeng H, Young AN, Hobert ME, Karmali V, Rao AS, Madara JL. Science. 2000;289:1560–1563. doi: 10.1126/science.289.5484.1560. [DOI] [PubMed] [Google Scholar]
- 18.Joseph TW, Moll UM. Methods Mol Biol. 2003;234:211–217. doi: 10.1385/1-59259-408-5:211. [DOI] [PubMed] [Google Scholar]
- 19.Moulder JW. Infect Agents Dis. 1993;2:87–99. [PubMed] [Google Scholar]
- 20.Moulder JW. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stephens RS. Infect Agents Dis. 1992;1:279–293. [PubMed] [Google Scholar]
- 22.Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber SL. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 23.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, et al. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 24.Zhong G, Liu L, Fan T, Fan P, Ji H. J Exp Med. 2000;191:1525–1534. doi: 10.1084/jem.191.9.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pirbhai M, Dong F, Zhong Y, Pan KZ, Zhong G. J Biol Chem. 2006;281:31495–31501. doi: 10.1074/jbc.M602796200. [DOI] [PubMed] [Google Scholar]
- 26.Zhong G, Fan P, Ji H, Dong F, Huang Y. J Exp Med. 2001;193:935–942. doi: 10.1084/jem.193.8.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim SW, Hayashi M, Lo JF, Fearns C, Xiang R, Lazennec G, Yang Y, Lee JD. Cancer Res. 2005;65:8784–8791. doi: 10.1158/0008-5472.CAN-04-4422. [DOI] [PubMed] [Google Scholar]
- 28.Tato CM, Hunter CA. Infect Immun. 2002;70:3311–3317. doi: 10.1128/IAI.70.7.3311-3317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neznanov N, Chumakov KM, Neznanova L, Almasan A, Banerjee AK, Gudkov AV. J Biol Chem. 2005;280:24153–24158. doi: 10.1074/jbc.M502303200. [DOI] [PubMed] [Google Scholar]
- 30.Kelly D, Campbell JI, King TP, Grant G, Jansson EA, Coutts AG, Pettersson S, Conway S. Nat Immunol. 2004;5:104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 31.Makarova KS, Aravind L, Koonin EV. Trends Biochem Sci. 2000;25:50–52. doi: 10.1016/s0968-0004(99)01530-3. [DOI] [PubMed] [Google Scholar]
- 32.Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, et al. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 33.Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, et al. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 34.Morrison RP, Caldwell HD. Infect Immun. 2002;70:2741–2751. doi: 10.1128/IAI.70.6.2741-2751.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janeway CA., Jr Proc Natl Acad Sci USA. 2001;98:7461–7468. doi: 10.1073/pnas.131202998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pulendran B, Ahmed R. Cell. 2006;124:849–863. doi: 10.1016/j.cell.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 37.Holmgren J, Czerkinsky C. Nat Med. 2005;11:S45–S53. doi: 10.1038/nm1213. [DOI] [PubMed] [Google Scholar]
- 38.Loomis WP, Starnbach MN. Curr Opin Microbiol. 2002;5:87–91. doi: 10.1016/s1369-5274(02)00291-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.