Abstract
Serine racemase (SR) generates d-serine, a coagonist with glutamate at NMDA receptors. We show that SR is physiologically S-nitrosylated leading to marked inhibition of enzyme activity. Inhibition involves interactions with the cofactor ATP reflecting juxtaposition of the ATP-binding site and cysteine-113 (C113), the site for physiological S-nitrosylation. NMDA receptor physiologically enhances SR S-nitrosylation by activating neuronal nitric-oxide synthase (nNOS). These findings support a model whereby postsynaptic stimulation of nitric-oxide (NO) formation feeds back to presynaptic cells to S-nitrosylate SR and decrease d-serine availability to postsynaptic NMDA receptors.
Keywords: neuronal nitric-oxide synthase, NMDA receptor, S-nitrosylation
Glutamate neurotransmission through NMDA receptors requires a coagonist originally thought to be glycine. Recent studies indicate that in most portions of the brain, d-serine is the physiological coagonist because selective degradation of d-serine but not glycine markedly reduces NMDA transmission (1, 2), whereas retraction of d-serine-producing glia in the hypothalamus of lactating rats also diminishes NMDA transmission (2). d-serine is formed from l-serine by serine racemase (SR), which, like d-serine, is selectively enriched in glia (2, 3), although recent studies indicate some neuronal localization (4). SR, a pyridoxal phosphate-requiring enzyme, also displays an absolute requirement for ATP, which is not hydrolyzed during SR activation (5). SR binds the glutamate receptor interacting protein, which also binds to AMPA subtypes of glutamate receptors with glutamate receptor interacting protein markedly activating SR and providing a means whereby glutamate stimulation of SR-containing cells augments d-serine formation (6).
In postsynaptic cells, NMDA signaling is mediated in part by neuronal nitric-oxide synthase (nNOS) because calcium entering through NMDA receptor channels binds to calmodulin associated with nNOS (7, 8). Extensive studies have documented a feedback from postsynaptic to presynaptic glutamatergic nerve terminals, which modulates NMDA neurotransmission, especially in long-term potentiation (9). Nitric oxide (NO) may be a retrograde messenger of long-term potentiation (10–12), although the area is controversial (12). Because SR is a component of the NMDA synaptic complex, we wondered whether it is influenced by NO. In the present study, we demonstrate that SR is physiologically S-nitrosylated leading to inhibition of enzyme activity mediated by interactions with ATP. NMDA transmission stimulates SR S-nitrosylation suggesting a feedback mechanism to diminish presynaptic formation of d-serine.
Results
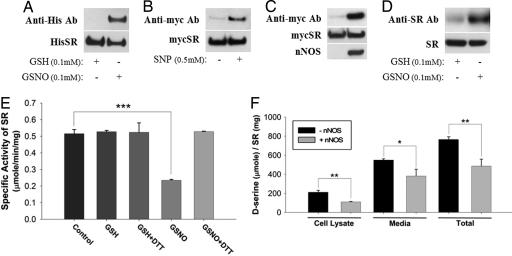
We demonstrated S-nitrosylation of SR in multiple ways. Incubation of the NO donor S-nitroso-glutathione (GSNO) with SR in vitro leads to robust nitrosylation (Fig. 1A). In HEK293 cells, treatment with the NO donor sodium nitroprusside also provides S-nitrosylation (Fig. 1B). NO produced by nNOS S-nitrosylates SR, as is evident in HEK293 cells transfected with nNOS (Fig. 1C). Mammalian brain displays SR S-nitrosylation in experiments using mixed cortical neuron-glial cultures treated with GSNO (Fig. 1D).
Fig. 1.
S-nitrosylation of SR inhibits enzyme activity. (A) Purified His–SR is S-nitrosylated in the presence of GSNO (0.1 mM for 30 min) but not glutathione (reduced form) (0.1 mM for 30 min) as determined by biotin-switch assay. All of the S-nitrosylated proteins were precipitated with streptavidin beads, and SR was detected by Western blot analysis with anti-His antibody. SR was selectively S-nitrosylated by GSNO. (B) myc-SR expressed in transfected HEK293 cells is S-nitrosylated in the presence of sodium nitroprusside (0.5 mM for 1 h) as determined by biotin-switch assay. (C) myc-SR expressed in transfected HEK293 cells is S-nitrosylated only when coexpressed with nNOS. SR was selectively S-nitrosylated by endogenously generated NO. (D) Endogenous SR from mixed cortical neuron-glial cultures is S-nitrosylated after treatment with GSNO (0.1 mM for 30 min). SR was detected by Western blot analysis with an SR-specific antibody. (E) SR enzyme activity was measured from purified preparations of His-SR in the presence or absence of GSNO (25 μM), GSH (25 μM), and DTT (1 mM). Although GSNO robustly inhibits SR activity, GSH has no effect and DTT reverses GSNO's inhibitory effects. (F) SR enzyme activity was measured from cell lysates and media of transfected HEK293 cells expressing myc-SR alone or together with nNOS. Both cell lysates and media show reduction in d-serine production in the presence of nNOS. Bars represent the mean ± SEM of three independent experiments, each performed in triplicate (∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001).
S-nitrosylation inhibits SR. Incubation of recombinant SR with 25 μM GSNO markedly reduces enzyme activity, and activity is restored by DTT, which reverses nitrosylation (Fig. 1E). Generation of d-serine in intact cells is also inhibited by NO. In HEK293 cells, transfection of nNOS markedly reduces levels of d-serine in both cells and media. As observed earlier (6), d-serine levels are substantially higher in the media than the cells indicating that newly formed d-serine is rapidly released.
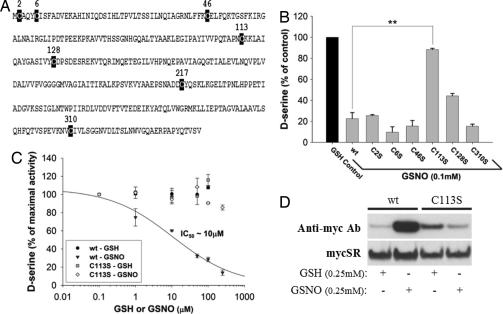
To identify the amino acid target of S-nitrosylation in SR, we selectively mutated its seven cysteines (Figs. 2A and 2B). The C113S mutation completely reverses inhibition of enzyme activity by GSNO establishing cysteine-113 (C113), a residue conserved in human, mouse, and rat SR, as the mediator for NO's regulation of enzyme activity. GSNO inhibits SR activity in a concentration-response manner with virtual abolition of enzyme activity at 0.25 mM GSNO; the C113S mutant is completely resistant to inhibitory effects of GSNO (Fig. 2C). The importance of C113 is confirmed by biotin-switch assays of SR showing a major reduction in enzyme S-nitrosylation in the C113S mutant (Fig. 2D). S-nitrosylation of C113S SR is also abolished in HEK293 cells overexpressing nNOS (data not shown).
Fig. 2.
SR is S-nitrosylated at C113. (A) Sequence of mouse SR displaying all seven cysteines as potential sites for S-nitrosylation. (B) SR enzyme activity was measured from purified preparations of His–SR, wild-type versus individual cysteine mutants, in the presence or absence of GSNO and GSH. Only C113S completely reverses the inhibitory effects of GSNO. The C217S mutant was basally inactive. (C) SR enzyme activity was measured from purified preparations of His-SR, wild-type versus C113S, in the presence or absence of increasing concentrations of GSNO and GSH. Although the wild-type protein is inhibited in a concentration-dependent manner by GSNO, C113S is completely resistant to the NO donor's effects at concentrations as high as 0.25 mM. (D) Wild-type myc-SR but not C113S mutant (expressed in transfected HEK293 cells) is S-nitrosylated in the presence of GSNO (0.25 mM for 1 h) as determined by biotin-switch assay indicating that C113 is the endogenous S-nitrosylation site on SR. Bars represent the mean ± SEM of three independent experiments, each performed in triplicate (∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001).
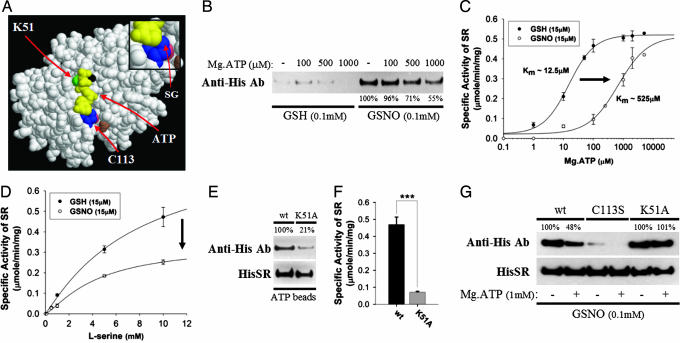
We wondered about molecular mechanisms whereby NO regulates SR. The crystal structure of SR has not been established. However, the SR sequence resembles yeast serine/alanine racemase with nearly 38% amino acid identity. Accordingly, we created a model of SR on the basis of a presumed similarity to the yeast enzyme (Fig. 3A). The putative ATP-binding site in SR lies in close proximity to the site of S-nitrosylation, C113, suggesting that S-nitrosylation influences ATP effects on SR. Moreover, the sulfur atom of C113 appears to be in close contact with the adenosine base of ATP, presumably indicating a role for C113 in coordinating the cofactor's binding to the protein. Addition of a nitroso moiety on this exposed sulfur group should interfere with ATP binding and vice versa, fitting with our finding that ATP prevents S-nitrosylation of SR (Fig. 3B). Inhibition of SR activity by S-nitrosylation is apparently competitive with ATP because the nitrosylated enzyme displays a 40-fold increase in the Km for ATP with no change in Vmax (Fig. 3C). Because S-nitrosylation is covalent, it presumably cannot directly “compete” with ATP. By contrast, S-nitrosylation does not alter SR's Km toward substrate l-serine but substantially reduces the Vmax (Fig. 3D).
Fig. 3.
S-nitrosylation inhibits SR activity by interfering with its cofactor ATP. (A) Model of mammalian SR showing ATP in yellow, magnesium ion in black, C113 in blue, lysine-51 in green, aspartic acid-318 in brown, and lysine-114 in pink. The insert shows the sulfur group (SG) of C113 in close apposition to the adenosine base of ATP. S-nitrosylation of C113 presumably interferes with ATP binding. C113 belongs to an acid–cysteine–base consensus motif for protein S-nitrosylation (13). (B) S-nitrosylation of purified His-SR is diminished by ATP in a concentration-dependent manner as determined by biotin-switch assay. (C) SR enzyme activity was measured from purified preparations of His-SR in the presence or absence of GSNO, GSH, and increasing concentration of ATP. S-nitrosylation appears to inhibit SR activity by interfering with ATP as the nitrosylated enzyme displays a 40-fold increase in the Km for ATP with no change in Vmax. (D) SR enzyme activity was measured in the presence or absence of GSNO, GSH, and increasing concentration of substrate l-serine. S-nitrosylation does not alter SR's Km but substantially reduces the Vmax and as such is noncompetitive with respect to substrate l-serine. (E) ATP–agarose beads were incubated with purified SR, wild-type versus K51A mutant, at 4°C for 30 min. The beads were washed followed by Western blot analysis with anti-His antibody. The K51A mutant shows substantially less ATP binding than the wild-type protein. (F) SR enzyme activity was measured from purified preparations of His-SR, wild-type versus K51A, showing the latter's markedly decreased basal activity. (G) Biotin-switch assay on His-SR in the presence of GSNO and ATP. Although ATP can diminish S-nitrosylation of the wild-type protein, it has no effect on the K51A mutant, which has a reduced capability to coordinate ATP. Bars represent the mean ± SEM of three independent experiments, each performed in triplicate (∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001).
Evidence that S-nitrosylation inhibits SR primarily by displacing ATP supports the model with C113 adjacent to the ATP-binding site. The phosphate groups of ATP typically interact with critical lysines in proteins, and K51 appears close to the putative ATP-binding site. Mutation of K51 to alanine reduces by 80% the binding of ATP to recombinant SR (Fig. 3E). Because SR catalytic activity is absolutely dependent on ATP, we explored the influence of K51A mutation on SR activity and observed a 90% reduction (Fig. 3F). Assuming that S-nitrosylation of SR interferes with ATP binding, one might expect the inhibition of S-nitrosylation elicited by ATP to be lost after disruption of the ATP-binding site. As predicted, although ATP inhibits S-nitrosylation of wild-type SR ≈50%, it fails to influence S-nitrosylation of the K51A mutant (Fig. 3G). These findings substantiate a model of SR in which physiological S-nitrosylation and ATP binding occur in close proximity and thus mutually interact in regulating the enzyme.
C113 is surrounded by an acid, D318, and a base, K114, brought into close apposition to the cysteine residue by tertiary structure (Fig. 3A). This observation is consistent with the acid-cysteine-base consensus motif proposed for protein S-nitrosylation by Stamler and colleagues (13). However, this consensus sequence is not definitive because some nitrosylated proteins such as endothelial NOS (14), fail to display the sequence.
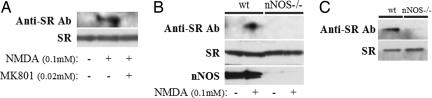
NMDA receptor activation is well known to stimulate nNOS catalytic activity. We wondered whether such stimulation elicits a feedback with the generated NO S-nitrosylating SR in adjacent glia or neurons. We explored this question using cerebellar slices treated with 0.1 mM NMDA for 30 min, conditions typically used to elicit physiological NMDA neurotransmission. In these experiments, NMDA strikingly augments SR S-nitrosylation in a receptor-specific fashion being prevented by the NMDA antagonist MK801 (Fig. 4A). The NMDA activation reflects an influence on endogenous neuronally derived NO because it is not detected in brain preparations of nNOS-deleted mice (Fig. 4B). In these experiments, S-nitrosylation of SR is not evident in untreated brain slices. However, with longer exposure, we observe basal S-nitrosylation of SR, which is abolished in nNOS-deleted mice establishing that S-nitrosylation of SR is a physiological process elicited by neuronally derived NO (Fig. 4C).
Fig. 4.
NMDA receptor stimulation in vivo promotes the S-nitrosylation of SR through activation of nNOS. (A) Cerebellar slices prepared from 3-week-old mice were equilibrated in 95% oxygen/5% CO2 at 37°C for 30 min in artificial cerebrospinal fluid buffer. The slices were then treated with NMDA (0.1 mM) with or without MK801 (0.02 mM) for another 30 min at 37°C after which biotin-switch assays were performed. Marked S-nitrosylation of SR occurs after NMDA stimulation, an effect abolished by the NMDA receptor antagonist, MK801. (B) Cerebellar slices prepared from 3-week-old wild-type versus nNOS knockout mice were subjected to NMDA (0.1 mM) treatment for 30 min at 37°C followed by the biotin-switch assay. The S-nitrosylation of SR observed in the wild-type samples after NMDA stimulation was absent in tissues obtained from nNOS knockout animals. (C) Whole-brain slices prepared from 3-week-old wild-type versus nNOS knockout mice were subjected to the biotin-switch assay. Basal S-nitrosylation of SR is observed in the wild-type but not in the nNOS knockout brain.
Discussion
The main finding of our study is that NO physiologically S-nitrosylates SR decreasing catalytic activity. In a glioblastoma cell line, Shoji et al. (15, 16) observed inhibition of SR activity by an NO donor.
S-nitrosylation appears to be a physiological process reflecting the actions of neuronally derived NO because it occurs under basal conditions and is lost in nNOS knockout mice. NMDA neurotransmission also stimulates SR S-nitrosylation, a process mediated by nNOS. Because basal NO formation depends on NMDA receptor activation (17), we assume that under basal conditions, NMDA neurotransmission regulates SR.
Abundant research supports a model of synaptic plasticity whereby postsynaptic neurons activated by NMDA neurotransmission send a molecular messenger in a retrograde fashion to diminish glutamatergic activity (9). NO has been proposed as a retrograde messenger for such synaptic modulation, especially for long-term potentiation (10–12), although there exists conflicting evidence (12). Because d-serine generated by glia or neurons is an endogenous coactivator of NMDA receptors, SR inhibition by retrogradely transmitted NO would constitute a feedback signal impacting synaptic plasticity. According to this model, glutamate activates receptors on closely adjacent astrocytes or neurons (Fig. 5). Glutamate receptor interacting protein, bound to glutamate receptors under basal conditions, dissociates and binds to SR activating the enzyme. d-serine is then released to participate with Glutamate in activation of postsynaptic neurons. Within these neurons, calcium entering the NMDA receptor channel binds to calmodulin associated with nNOS to stimulate NO generation. This NO then diffuses into adjacent astrocytes or neurons to S-nitrosylate and inactivate SR. Because NO appears to mediate both postsynaptic NMDA actions and a feedback inhibition of d-serine generation, it is difficult to predict how nNOS deletion should impact overall NMDA transmission. This fits with conflicting, generally minimal effects reported of nNOS deletion on NMDA transmission (18).
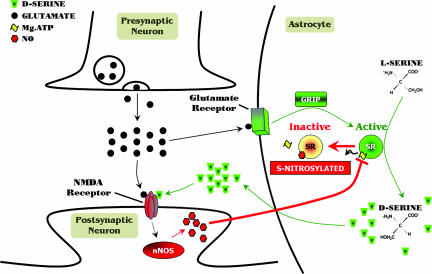
Fig. 5.
Model for d-serine signaling in the brain. d-serine is synthesized from l-serine by SR and stored primarily within astrocytes ensheathing neuronal synapses containing NMDA receptors. SR and d-serine may also occur in neurons (4). When the presynaptic neuron releases glutamate, it acts not only on the postsynaptic neuron, but also on the surrounding astrocyte resulting in the activation of glutamate receptors and the subsequent synthesis and release of d-serine presumably through SR's association with glutamate receptor interacting protein. In the synaptic cleft, d-serine binds to the glycine/d-serine-binding site on the NMDA receptor and, in conjunction with l-glutamate, results in the opening of the receptor channel. Calcium entering the postsynaptic neuron, together with calmodulin (not shown), activates nNOS leading to the production of NO, which can then diffuse into adjacent astrocytes or neurons to S-nitrosylate SR. S-nitrosylation of SR at C113 prevents ATP from binding to SR, thus inhibiting enzyme activity. This model affords a potential negative feedback loop to regulate NMDA neurotransmission.
SR is uniquely stimulated by ATP, which appears to be absolutely required for enzyme activity and to display high affinity for SR with a Km of ≈12 μM, far lower than endogenous ATP levels in most cells (19). This activation of SR is independent of ATP hydrolysis being elicited by nonmetabolized ATP analogs (5). Our structural model places the ATP-binding site in close proximity to the cysteine, which is S-nitrosylated to inhibit SR by interfering with ATP binding. Evidently, nitrosylation displaces ATP from its binding site. Thus, ATP and NO reciprocally activate and inhibit the enzyme by acting at the same locus on the protein. The rationale for regulation of SR by ATP is unclear. Conceivably, it reflects a means whereby the energy state of the cell signals to SR, although this would seem inconsistent with the complete occupation of the ATP site at concentrations substantially lower than endogenous cellular ATP levels. Of course, the microenvironment of SR may contain low ATP levels and be susceptible to modulation by micromolar concentrations. The 10 μM IC50 of GSNO toward SR underestimates NO's inhibitory capacity because GSNO does not instantaneously decompose into NO but is in the range of GSNO levels reported in brain tissue (20). As such, the effective NO levels are lower and the actual IC50 is also less than what we observed. Thus, NO might be more potent than ATP in modifying SR. Additionally, NO levels may also become locally elevated near the synapse after NMDA receptor activation leading to robust S-nitrosylation and inhibition of SR. The exact species of NO that modifies SR might be NO or GSNO. GSNO could S-nitrosylate through release of NO or by transnitrosylation.
Materials and Methods
Animals.
Biochemical experiments involving animals were performed on brains removed from 3-week-old wild-type and nNOS knockout animals. Animals were maintained on a 12-h light/dark cycle at a room temperature of 23°C with free access to food and water. All animal use procedures were in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Cells.
HEK293 cells were obtained from the American Type Culture Collection (Manassas, VA). They were grown in a humid atmosphere of 95% air and 5% CO2 at 37°C in DMEM supplemented with 10% FBS, l-glutamine (300 μg/ml), penicillin (100 units/ml), and streptomycin (100 μg/ml).
Reagents.
l-serine, d-serine, NMDA, and MK801 were purchased from Sigma (St. Louis, MO). All of the antibodies were from BD Biosciences (San Jose, CA).
Plasmid Constructions.
Full-length mouse SR-encoding gene (GeneBank sequence NM_013761) was amplified by PCR by using primers harboring SalI/NotI restriction sites and cloned into the pCMV Myc (Clontech, Palo Alto, CA) and pET-28c+ His vectors (Novagen, San Diego, CA). Forward primer sequence is 5′- ACGCGTCGACATGTGTGCTCAGTACTGCATCTCCTTTGC, and the backward primer is 5′- ATAAGAATGCGGCCGCTTAAACAGAAACCGTCTGGTAAGG. The murine neuronal nitric-oxide synthase (GeneBank sequence NM_008712) was cloned into the pcDNA3.1+ vector (Invitrogen, Carlsbad, CA).
Site-Directed Mutagenesis.
The QuikChange site-directed mutagenesis system (Stratagene, La Jolla, CA) was used per the manufacturer's instructions to alter the seven cysteine residues in SR to serine as well as the lysine-51 residue to alanine. Each mutant was verified through automated sequencing by the Johns Hopkins Core Facility.
Protein Purification.
Purification of SR was carried out as described in ref. 21. In brief, mouse full-length SR DNA was subcloned into pET-28c+, which encodes a 6x-histidine tag and introduced into BL21 codon plus bacteria (Stratagene). Expression of SR was induced by 0.5 mM isopropyl 1-thio-β-d-galactopyranoside (Sigma) for 12 h at 30°C. Cells were collected by centrifugation and disrupted by sonication in medium containing 20 mM Tris·HCl (pH 7.4)/15 μM PLP/10 mM imidazole/400 mM NaCl. After the addition of 1% Triton X-100, the suspension was cleared by centrifugation (40,000 × g for 15 min), and SR was purified from the supernatant by binding to Talon resin (Clontech) according to the manufacturer's instructions.
Serine Racemase Activity Assay.
To determine SR activity, reactions with purified SR, pretreated with GSNO or GSH for 30 min or cells expressing SR with or without nNOS were started with the addition of 10 mM l-serine unless indicated otherwise and incubated at 37°C for 1 h or 5 h, respectively. To stop the reaction, the samples were boiled for 5 min. Half of the samples were incubated for 30 min at 37°C with Escherichia coli d-serine deaminase, an enzyme known to specifically convert d-serine to pyruvate, and the other half incubated without the enzyme. Pyruvate levels were then measured by spectrophotometrically monitoring the rate of NADH to NAD+ conversion by lactate dehydrogenase at 340 nm. Total pyruvate levels in the samples with d-serine deaminase were then subtracted from basal pyruvate in the absence of the enzyme to determine d-serine levels. HPLC analysis for d-serine, as described in ref. 21, was also performed in addition to the spectrophotometric assays.
S-Nitrosylation (Biotin-Switch) Assay.
Cells were homogenized by 26-gauge needle in HEN (250 mM Hepes·NaOH, pH 7.7/1 mM EDTA/0.1 mM neocuproine) buffer and then centrifuged at 1,000 × g for 10 min at 4°C. Cell lysates (240 μg) or pure His–SR protein (0.3 μg) was added to 4 vol of blocking buffer (9 vol of HEN buffer plus 1 vol 25% SDS, adjusted to 20 mM MMTS with a 2 M stock prepared in dimethylformamide) at 50°C for 20 min with frequent vortexing. The MMTS was then removed by adding 4 vol acetone and the proteins precipitated at −20°C for 20 min. After removal of the acetone, the proteins were resuspended in HENS buffer. To the suspension was added biotin–HPDP prepared fresh as a 4 mM stock in DMSO from a 50 mM stock suspension in DMF. Sodium ascorbate was added to a final concentration of 1 mM. After incubation for 1 h at 25°C, biotinylated proteins were precipitated by streptavidin–agarose beads. The streptavidin–agarose was then pelleted and washed five times with HENS buffer. The biotinylated proteins were eluted by SDS/PAGE sample buffer and subjected to Western blot analysis.
Modeling of Mammalian Serine Racemase.
A working model of the mouse SR (target) was built by using the crystallographic structure of the Schizosaccharomyces pombe enzyme with bound β,γ-methyleneadenosine 5′-triphosphate as a template (Protein Data Bank accession code 1WTC). Alignment of the sequences of the target and the template with the program BLAST showed that the two molecules had 121 of 313 identical residues (38%); 188 of 313 (60%) when including conservative mutations with only eight gaps. An initial model was built with the program Modeller. This model was modified by manual inspection and optimized by conjugate gradients energy minimization by using the software package QUANTA (Accelrys, San Diego, CA). Because the model of the mouse enzyme was slightly rotated and translated with respect to the template as a result of the minimization, it was realigned with the template with the alignment tools of the program O. The coordinates of the PLP and of the β,γ-methyleneadenosine 5′-triphosphate of the template were transferred to the realigned model and the conformation of the side chain of lysine-56 was manually adjusted to make the covalent bond to PLP. The modeling image was generated by using RasMol.
ATP-Binding Assays.
ATP-agarose beads (Sigma) were reconstituted as per the manufacturer's protocol and incubated with purified SR at 4°C for 30 min. The beads were washed three times with wash buffer containing 20 mM Tris (pH 7.7), 0.05% Triton X-100, and 250 mM NaCl followed by Western blot analysis.
Biotin-Switch Assay From Brain Slices.
Three-week-old wild-type and nNOS knockout animals were killed by cervical dislocation. The brain was immediately removed and sliced into 400-μm stripes with a Mcllwain Tissue Chopper (Brinkmann Instruments, Westbury, NY). The tissue was then equilibrated with 95% oxygen/5% CO2 at 37°C for 30 min in preoxygenated artificial cerebrospinal fluid buffer containing 125 mM NaCl, 3 mM KCl, 1.6 mM CaCl2, 0.2 mM arginine, 25 mM Hepes, pH 7.4, 10 mM d-glucose, and 1.25 mM Na2HPO4 with or without 1.8 mM MgCl2. After equilibration, the tissue was split into equal portions by weight and incubated with NMDA (0.1 mM) and MK801 (0.02 mM) in artificial cerebrospinal fluid buffer at 37°C for 30 min. Biotin-switch assay was then carried out as described earlier.
Statistical Analysis.
Add data are expressed as mean ± SEM of three independent experiments, each performed in triplicate. Data were analyzed by unpaired Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Acknowledgments
We thank Dr. Jonathan Stamler for his valuable suggestions as well as Drs. Rod Welch (University of Wisconsin, Madison, WI) and Herman Wolosker (Technion University, Haifa, Israel) for providing the d-serine deaminase construct. This work was supported by National Institutes of Health Grant MH18501 and Research Scientist Award DA00074 (to S.H.S.).
Abbreviations
- SR
serine racemase
- nNOS
neuronal nitric-oxide synthase
- GSNO
S-nitroso-glutathione
- C113
cysteine-113.
Footnotes
Conflict of interest statement: Patents related to this work have been licensed to Guilford Pharmaceuticals, now MGI Pharma. S.H.S. and The Johns Hopkins University are entitled to receive royalties from product sales. This matter is being handled by The Johns Hopkins Committee on Conflict of Interest in accordance with its policies.
References
- 1.Mothet JP, Parent AT, Wolosker H, Brady RO, Jr, Linden DJ, Ferris CD, Rogawski MA, Snyder SH. Proc Natl Acad Sci USA. 2000;97:4926–4931. doi: 10.1073/pnas.97.9.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Panatier A, Theodosis DT, Mothet JP, Touquet B, Pollegioni L, Poulain DA, Oliet SH. Cell. 2006;125:775–784. doi: 10.1016/j.cell.2006.02.051. [DOI] [PubMed] [Google Scholar]
- 3.Wolosker H, Blackshaw S, Snyder SH. Proc Natl Acad Sci USA. 1999;96:13409–13414. doi: 10.1073/pnas.96.23.13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kartvelishvily E, Shleper M, Balan L, Dumin E, Wolosker H. J Biol Chem. 2006;281:14151–14162. doi: 10.1074/jbc.M512927200. [DOI] [PubMed] [Google Scholar]
- 5.De Miranda J, Panizzutti R, Foltyn VN, Wolosker H. Proc Natl Acad Sci USA. 2002;99:14542–14547. doi: 10.1073/pnas.222421299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim PM, Aizawa H, Kim PS, Huang AS, Wickramasinghe SR, Kashani AH, Barrow RK, Huganir RL, Ghosh A, Snyder SH. Proc Natl Acad Sci USA. 2005;102:2105–2110. doi: 10.1073/pnas.0409723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bredt DS, Snyder SH. Proc Natl Acad Sci USA. 1990;87:682–685. doi: 10.1073/pnas.87.2.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bredt DS, Hwang PM, Glatt CE, Lowenstein C, Reed RR, Snyder SH. Nature. 1991;351:714–718. doi: 10.1038/351714a0. [DOI] [PubMed] [Google Scholar]
- 9.Bliss TV, Collingridge GL. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 10.Boehning D, Snyder SH. Annu Rev Neurosci. 2003;26:105–131. doi: 10.1146/annurev.neuro.26.041002.131047. [DOI] [PubMed] [Google Scholar]
- 11.Arancio O, Kiebler M, Lee CJ, Lev-Ram V, Tsien RY, Kandel ER, Hawkins RD. Cell. 1996;87:1025–1035. doi: 10.1016/s0092-8674(00)81797-3. [DOI] [PubMed] [Google Scholar]
- 12.Medina JH, Izquierdo I. Brain Res Brain Res Rev. 1995;21:185–194. doi: 10.1016/0165-0173(95)00013-5. [DOI] [PubMed] [Google Scholar]
- 13.Stamler JS, Toone EJ, Lipton SA, Sucher NJ. Neuron. 1997;18:691–696. doi: 10.1016/s0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 14.Erwin PA, Lin AJ, Golan DE, Michel T. J Biol Chem. 2005;280:19888–19894. doi: 10.1074/jbc.M413058200. [DOI] [PubMed] [Google Scholar]
- 15.Shoji K, Mariotto S, Ciampa AR, Suzuki H. Neurosci Lett. 2006;394:163–167. doi: 10.1016/j.neulet.2005.10.064. [DOI] [PubMed] [Google Scholar]
- 16.Shoji K, Mariotto S, Ciampa AR, Suzuki H. Neurosci Lett. 2006;392:75–78. doi: 10.1016/j.neulet.2005.08.063. [DOI] [PubMed] [Google Scholar]
- 17.Bredt DS, Snyder SH. Proc Natl Acad Sci USA. 1989;86:9030–9033. doi: 10.1073/pnas.86.22.9030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mashimo H, Goyal RK. Am J Physiol. 1999;277:G745–G750. doi: 10.1152/ajpgi.1999.277.4.G745. [DOI] [PubMed] [Google Scholar]
- 19.Silver IA, Deas J, Erecinska M. Neuroscience. 1997;78:589–601. doi: 10.1016/s0306-4522(96)00600-8. [DOI] [PubMed] [Google Scholar]
- 20.Kluge I, Gutteck-Amsler U, Zollinger M, Do KQ. J Neurochem. 1997;69:2599–2607. doi: 10.1046/j.1471-4159.1997.69062599.x. [DOI] [PubMed] [Google Scholar]
- 21.Foltyn VN, Bendikov I, De Miranda J, Panizzutti R, Dumin E, Shleper M, Li P, Toney MD, Kartvelishvily E, Wolosker H. J Biol Chem. 2005;280:1754–1763. doi: 10.1074/jbc.M405726200. [DOI] [PubMed] [Google Scholar]