Abstract
The Sonic hedgehog (Shh) and FGF signaling pathways regulate growth and differentiation in many regions of the nervous system, but interactions between these pathways have not been studied extensively. Here, we examine the relationship between Shh and FGF signaling in granule cell precursors (GCPs), which are the most abundant neural progenitors in the cerebellum and the putative cell of origin for the childhood brain tumor medulloblastoma. In these cells, Shh induces a potent proliferative response that is abolished by coincubation with basic FGF. FGF also inhibits transcription of Shh target genes and prevents activation of a Gli-responsive promoter in fibroblasts, which suggests that it blocks Shh signaling upstream of Gli-mediated transcription. FGF-mediated inhibition of Shh responses requires activation of FGF receptors and of ERK and JNK kinases, because it can be blocked by inhibitors of these enzymes. Finally, FGF promotes differentiation of GCPs in vitro and in vivo and halts proliferation of tumor cells from patched (ptc) mutant mice, a model for medulloblastoma. These findings suggest that FGF is a potent inhibitor of Shh signaling and may be a useful therapy for tumors involving activation of the hedgehog pathway.
Keywords: brain tumor, cerebellum, FGF, medulloblastoma
Development of the nervous system is regulated by secreted signaling molecules, or morphogens, with distinct spatial and temporal expression patterns (1, 2). Elucidating the mechanisms by which these molecules act and interact is critical for understanding both normal development and the diseases that result from its dysregulation.
Among the most potent morphogens in the CNS are members of the hedgehog and FGF families. Sonic hedgehog (Shh), the most widely expressed hedgehog protein, was initially shown to regulate specification of floor plate, motor neurons, and interneurons in the developing spinal cord (3, 4). Since then, it has been implicated in a wide variety of cell fate decisions in the CNS (5), including maintenance of neural stem cells in the hippocampus and forebrain (6, 7), proliferation of neural progenitors in the cerebellum and retina (8, 9), commitment of progenitors to neuronal and oligodendrocyte lineages (10, 11), and growth and guidance of axons in the spinal cord and retina (12, 13).
Like hedgehog proteins, FGFs have diverse functions in CNS development (14, 15). During early embryogenesis, they play critical roles in neural induction (16). Once the neural tube is formed, nodes of FGF secretion serve as organizers that determine the fate of cells in adjacent regions: for example, FGF8 from the isthmus establishes the midbrain–hindbrain region that gives rise to the tectum and cerebellum (17). In addition to playing their roles in patterning, FGFs also promote proliferation of stem cells in the hippocampus and subventricular zone (18, 19) and differentiation of oligodendrocytes and neurons in the spinal cord and midbrain (20–22). Finally, a variety of studies have implicated FGF signaling in axon growth and branching in the retina, cortex, and cerebellum (23–25).
Although hedgehogs and FGFs are coexpressed in many regions of the CNS and often regulate similar biological processes, there have been few studies of the interactions between them. Where these interactions have been examined, Shh and FGF have generally been found to synergize with one another (22, 26). In contrast, we have shown that Shh and basic FGF (bFGF) exert opposing effects on granule cell precursors (GCPs) in the developing cerebellum (8): Shh acts as a mitogen, whereas bFGF potently inhibits the proliferative response to Shh. Here, we examine the mechanisms of FGF-mediated inhibition of Shh signaling and its consequences for granule cell differentiation and tumorigenesis. We show that bFGF suppresses Shh signaling in GCPs and fibroblasts. FGF-mediated inhibition requires activation of FGF receptors and the MAPKs extracellular signal regulated kinase (ERK) and c-jun N-terminal kinase (JNK). bFGF not only suppresses Shh-induced proliferation but also promotes granule cell differentiation in vitro and in vivo. Finally, by inhibiting the Shh pathway, bFGF inhibits the growth of medulloblastoma cells from patched (ptc) mutant mice. These findings may have important implications for understanding normal development and may lead to novel approaches to treatment of Shh-dependent tumors.
Results
FGF Inhibits Proliferation of GCPs.
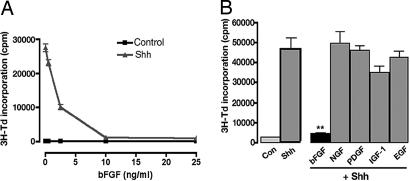
Our previous studies (8) showed that bFGF can suppress the proliferative response of GCPs to Shh. Paradoxically, some investigators have suggested that bFGF can promote proliferation of GCPs (27). Because all of these studies examined the effects of bFGF on partially purified cultures of GCPs, one explanation for these results was that the mitogenic and inhibitory effects of FGF were occurring in distinct populations of cells. To test this hypothesis we used Math1-GFP mice (28), in which GCPs express green fluorescent protein (GFP). By FACS-sorting GFP+ and GFP− cells from these animals, we demonstrated that GFP+ cells (GCPs) proliferate in response to Shh but not in response to bFGF, whereas GFP− cells proliferate in response to bFGF but not to Shh (29). Thus, the mitogenic effects of bFGF occur in non-GCPs. To determine whether the inhibitory effects of bFGF occur in GCPs, we sorted GFP+ cells, treated them with Shh in the presence or absence of bFGF, and examined their proliferation. As shown in Fig. 1A, purified GCPs proliferate robustly in response to treatment with Shh, and this proliferation is suppressed in a dose-dependent manner by bFGF. These data suggest that bFGF acts exclusively as a growth-inhibitory signal for purified GCPs.
Fig. 1.
FGF inhibits Shh-induced proliferation of purified GCPs. (A) GFP+ cells were sorted from Math1-GFP neonates and cultured for 48 h without (Control) or with recombinant Shh (3 μg/ml) and the indicated amounts of bFGF. Cells were pulsed with [3H]thymidine (3H-Td) and cultured overnight before being assayed for 3H-Td incorporation. (B) Effects of other receptor tyrosine kinase (RTK)-activating factors. GCPs were cultured with no stimuli (Con), Shh, or Shh plus bFGF (25 ng/ml), nerve growth factor (NGF) (100 ng/ml), PDGF-AA (10 ng/ml), insulin-like growth factor 1 (IGF-1) (25 ng/ml), or EGF (25 ng/ml). Con, control. Cells were assayed for 3H-Td incorporation as described above. Data represent means ± SEM of triplicate samples. ∗∗, P < 0.001 (one-way ANOVA).
To determine whether inhibition of Shh-induced proliferation is a general effect of growth factors that act through receptor tyrosine kinases (RTKs), we tested the effects of other FGFs and RTK-activating factors on Shh-induced proliferation. Among FGF family members, bFGF (FGF-2) was the most potent inhibitor of proliferation (90–95% inhibition), but substantial inhibition (50–60%) was also observed with FGF-1 and FGF-4; no significant effects were seen with FGF-5, -6, -7, -8, -9, -10, -16, or -17. Shh-induced proliferation was also not significantly affected by nerve growth factor, BDNF, neurotrophin-3, PDGF-AA, EGF, or insulin-like growth factor-1 (Fig. 1B and data not shown). Thus, inhibition of Shh-induced proliferation is specific to a subset of FGFs and is not a general property of RTK-activating growth factors.
FGF Inhibits Proliferation by Blocking Shh Signal Transduction.
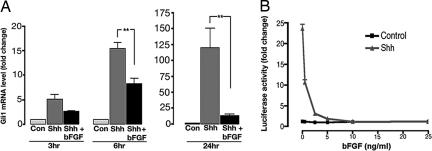
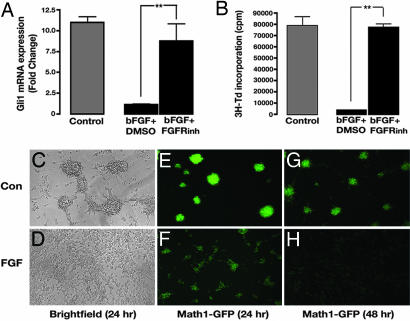
FGF could prevent Shh-induced proliferation of GCPs in at least two ways: by interfering with Shh signal transduction or by promoting cell cycle exit downstream of Shh target genes. Because gli1 is a direct target of Shh signaling in many cell types (30), we examined whether bFGF affects Shh-mediated induction of gli1 in GCPs. As shown in Fig. 2A, Shh caused a steady increase in expression of gli1 mRNA after 3, 6, and 24 h of culture. At each time point, bFGF caused marked inhibition of gli1 expression (50% inhibition at 3 and 6 h, 90% at 24 h). FGF also inhibited induction of Nmyc and cyclin D1, two other well characterized targets of the Shh pathway (data not shown). These findings suggest that bFGF interferes with Shh signaling upstream of gli-mediated transcription.
Fig. 2.
FGF prevents induction of Shh target genes. (A) Inhibition of gli1 induction in GCPs. GCPs were cultured with no stimulus (Con), Shh, or Shh plus bFGF for the indicated times. RNA was analyzed by real-time RT-PCR with primers for gli1 or actin mRNA. gli1 expression was normalized to actin mRNA and divided by levels in control GCPs to calculate fold change. (B) Inhibition of Shh responses in Shh-Light2 cells. Cells were cultured for 48 h in the absence (Control) or presence of Shh plus the indicated concentrations of bFGF and assayed for luciferase activity. Data represent means ± SEM of four samples. ∗∗, P < 0.001 (one-way ANOVA).
The observation that bFGF could inhibit Shh signaling in GCPs prompted us to ask whether it could do so in other cell types. We therefore examined the effects of bFGF in Shh-Light2 cells (NIH 3T3 fibroblasts stably expressing a Gli-responsive luciferase reporter) (31). Fig. 2B shows that Shh induces robust luciferase activity in Shh-Light2 cells. However, induction of luciferase activity is completely blocked by cotreatment with bFGF. Thus, the inhibitory action of bFGF on Shh signaling is not restricted to GCPs, but can be observed in fibroblasts as well. Together, these results suggest that bFGF can inhibit Shh signaling in a variety of cell types and imply that bFGF-mediated inhibition occurs upstream of Shh target genes.
bFGF Signals Through FGF Receptors (FGFRs) to Inhibit the Shh Pathway.
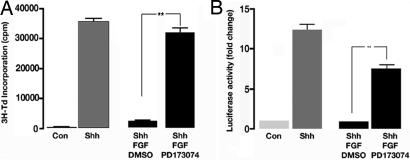
In principle, bFGF could block Shh responses by binding to FGFRs and inducing an intracellular signaling cascade or by interfering with Shh signaling in an FGFR-independent manner [e.g., by competing for extracellular heparan sulfate proteoglycans (32)]. To determine whether inhibition of Shh signaling requires activation of FGFRs, we used pharmacologic inhibitors of FGFR kinases (33). As shown in Fig. 3A, pretreatment for 1 h with the FGFR inhibitor PD173074 abolished FGF-mediated inhibition of Shh responses in GCPs. Shh-induced proliferation of GCPs was also restored by SU5402, a weaker but more commonly used FGFR inhibitor (33) [supporting information (SI) Fig. 8]. To determine whether these effects were conserved in fibroblasts, we also tested the effects of inhibiting FGFR activity in Shh-Light2 cells. As shown in Fig. 3B, PD173074 prevented bFGF from inhibiting Shh responses in Shh-Light2 cells as well. These experiments demonstrate that bFGF acts by means of FGFRs to abrogate the effects of Shh signaling.
Fig. 3.
FGF-mediated inhibition requires signaling through FGFRs. GCPs (A) or Shh-Light2 cells (B) were pretreated with vehicle (DMSO) or with the FGFR inhibitor PD173074 (0.05 μM for GCPs, 0.1 μM for Shh-Light2) for 1 h and then cultured for 48 h with no stimulus (Con) or Shh with or without bFGF. GCPs were assayed for 3H-Td incorporation, and Shh-Light2 cells were assayed for luciferase activity as described above. Data represent means ± SEM of three samples. ∗∗, P < 0.001 (one-way ANOVA).
To confirm that FGFR signaling was responsible for inhibition of Shh responses, we also tested whether the effects of bFGF could be mimicked by overexpression of an activated FGF receptor (34). Shh-Light2 cells were infected with control or constitutively active FGFR1-encoding retroviruses and then stimulated with Shh for 48 h. As shown in SI Fig. 9, control cells exhibited a robust response to Shh, whereas cells expressing activated FGFR1 showed no response. These results indicate that FGFR can mimic the effects of bFGF and lend further credence to the notion that FGF acts in a receptor-dependent fashion to inhibit the Shh pathway.
FGF-Mediated Inhibition of Shh Responses Is Dependent on MAPKs.
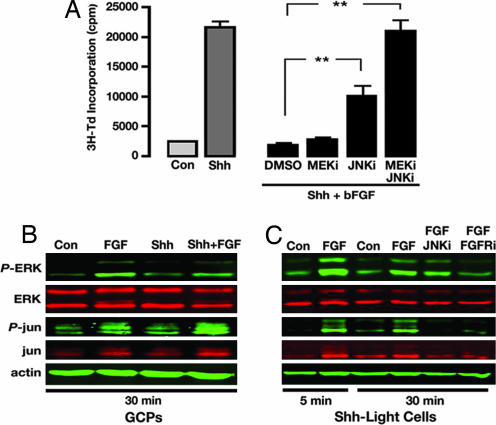
Many of the effects of FGF signaling are mediated by MAPKs (35). To determine whether MAPKs might be involved in FGF-mediated inhibition of Shh responses, we tested the effects of MAPK inhibitors on FGF-mediated inhibition in GCPs. Fig. 4A shows that U0126 (which blocks ERK activity by inhibiting the ERK-activating enzyme MEK) had little effect on FGF-mediated inhibition on its own. SP600125, an inhibitor of JNK, caused a significant restoration of Shh-induced proliferation (to 47% of the maximal Shh response). Finally, coinhibition of ERK and JNK abolished bFGF-mediated inhibition and allowed GCPs to respond to Shh to the same degree as they did in the absence of bFGF. Notably, inhibitors of Src, phospholipase Cγ, diacylglycerol kinase, protein kinase C, phosphatidylinositol 3-kinase, Akt, protein kinase A (PKA), and calcium-calmodulin kinase IV had little or no effect on bFGF-mediated inhibition (SI Table 1). These results indicate that FGF-mediated inhibition of Shh responses depends on activation of ERK and JNK kinases.
Fig. 4.
FGF-mediated inhibition depends on MAPK activity. (A) ERK and JNK inhibitors block bFGF-mediated inhibition in GCPs. Cells were pretreated with DMSO, MEK inhibitor (MEKi) (U0126, 10 μM), JNK inhibitor (JNKi) (SP600125, 10 μM), or both for 1 h before addition of Shh in the presence or absence of bFGF. After 48 h, cells were pulsed with 3H-Td, cultured for 18 h, and assayed for thymidine incorporation. Data represent means ± SEM of six samples. ∗∗, P < 0.001 (one-way ANOVA). (B) FGF activates ERK and JNK in GCPs. GCPs were cultured with no stimulus (Con) or Shh in the presence or absence of bFGF for 30 min. Cells were lysed, and expression of phospho-ERK (P-ERK), ERK, phospho-c-jun (P-jun), c-jun, and actin were examined by using Western blotting. Note the increase in phospho-ERK and phospho-jun in FGF-treated cells. (C) FGF activates ERK and JNK in Shh-Light2 cells. Cells were pretreated with DMSO, FGFR inhibitor (FGFRi) (PD173074, 0.1 μM), or JNK inhibitor (JNKi) and then treated with Shh in the presence or absence of bFGF for 5 or 30 min. Levels of phospho-ERK, ERK, phospho-c-jun, c-jun, and actin were examined by using Western blotting. Note the increase in phospho-ERK and phospho-c-jun in FGF-treated cells. ERK phosphorylation is blocked by FGFR inhibitor, whereas c-jun phosphorylation is blocked by both FGFR inhibitor and JNK inhibitor.
To verify that FGF can activate MAPKs, we examined the effects of bFGF on phosphorylation of ERK and c-jun (a substrate of JNK). Fig. 4B shows that bFGF induces phosphorylation of these proteins in GCPs within 30 min. Increased phosphorylation was also observed when cells were treated with bFGF plus Shh, but no induction was seen with Shh alone. Similar results were seen in Shh-Light2 cells (Fig. 4C). Together, these studies indicate that bFGF activates ERK and JNK and that these kinases are critical for FGF-mediated inhibition of Shh responses.
bFGF Promotes Granule Cell Cycle Exit and Differentiation.
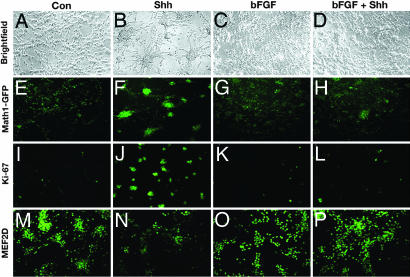
bFGF promotes differentiation of neurons and glia in many regions of the CNS (20, 22, 36). The ability of bFGF to overcome Shh-induced proliferation of GCPs suggested that it might promote differentiation of these cells as well. To investigate this possibility, we isolated GCPs from Math1-GFP mice, cultured them in Shh in the presence or absence of bFGF, and analyzed them for markers of proliferation and differentiation. As shown in Fig. 5, cells treated with Shh maintained a rounded morphology and clustered together, a behavior that is characteristic of proliferating GCPs. In contrast, bFGF-treated and bFGF plus Shh-treated cells adhered to the culture dish and extended processes, suggesting that bFGF can promote neuronal differentiation even in the presence of Shh.
Fig. 5.
bFGF promotes differentiation of GCPs in vitro. GCPs were cultured for 48–72 h with no stimulus (A, E, I, and M), Shh (B, F, J, and N), bFGF (C, G, K, and O), or bFGF plus Shh (D, H, L, and P). (A–D) Bright-field images of cells cultured for 48 h. (E–H) GFP fluorescence of Math1-GFP+ cells cultured for 48 h. (I–L) Ki-67 staining in cells cultured for 48 h. (M–P) MEF2D expression in cells cultured for 72 h. (Magnification: A–D, ×20; E–H, ×20; M–P, ×40.)
The morphological differentiation observed in bFGF-treated cells was also reflected in the expression of differentiation markers. In Math1-GFP-transgenic mice, GFP is expressed in proliferating GCPs in the outer external granule layer (EGL) (28, 37). When GCPs from these mice are placed in culture, they lose expression of Math1 and of the GFP transgene (Fig. 5E). Expression of Math1-GFP is maintained when cells are treated with Shh (Fig. 5F) but lost in cells treated with bFGF or bFGF plus Shh (Fig. 5 G and H). Expression of the proliferation marker Ki67 showed a similar pattern, with 40% of cells expressing Ki67 in Shh-treated cultures and only 13% expressing Ki67 in cultures treated with Shh plus bFGF (Fig. 5 I–L and SI Fig. 10). In contrast, the granule cell differentiation marker MEF2D (38) was low in Shh-treated cells (20% MEF2D+), but markedly elevated in bFGF-treated and bFGF plus Shh-treated cells (82% MEF2D+) (Fig. 5 M–P). In fact, bFGF-containing cultures showed increased expression of MEF2D compared with control cultures (82% vs. 50%). These findings suggest that bFGF can overcome Shh-induced proliferation and promote differentiation of GCPs into neurons.
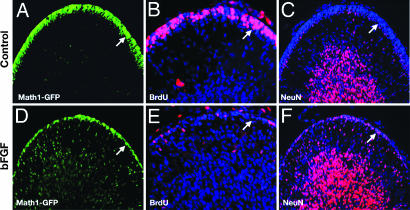
Because bFGF induced differentiation of GCPs in vitro, we wondered whether it could also do so in vivo. Delivery of FGF to the cerebellum is limited by the blood–brain barrier, so we introduced bFGF directly into the cerebrospinal fluid around the cerebellum by using intracisternal injection (39). Math1-GFP mice were injected with bFGF on postnatal day 4 (P4), P5, and P6 and analyzed on P7. Although no consistent changes were observed in the anterior lobes of the cerebellum, the posterior lobes (IX and X), which were closest to the area of injection, were decreased in size and showed a marked thinning of the EGL (SI Fig. 11). Closer examination of these lobes revealed a significant reduction in the number of immature (Math1-GFP-high) GCPs in the EGL and an increase in the number of newly differentiated (Math1-GFP-low) cells in the molecular layer and IGL (Fig. 6 A and D and SI Fig. 10). Staining with anti-BrdU antibodies showed a dramatic reduction in proliferation in the EGL of bFGF-treated mice compared with controls (Fig. 6 B and E). Finally, bFGF-treated cerebella exhibited increased expression of NeuN, a marker of postmitotic neurons, in the EGL and molecular layer (Fig. 6 C and F). Together, these data suggest that bFGF can induce cell cycle exit and differentiation of GCPs in vitro and in vivo.
Fig. 6.
bFGF accelerates differentiation of GCPs in vivo. Math1-GFP mice were given intracisternal injections of vehicle (Control, A–C) or bFGF (D–F) on P4, P5, and P6, and i.p. injections of BrdU on P6. At P7, cerebella were fixed and stained with anti-GFP (A and D), anti-BrdU (B and E), or anti-NeuN antibodies (C and F) with a DAPI counterstain (blue). Note the reduced number of immature, proliferating (GFP-high, BrdU+) cells and the abundance of differentiated (NeuN+) cells in the EGL of FGF-treated mice (see arrows in A vs. D, B vs. E, and C vs. F). Data are representative of six mice.
bFGF Halts Proliferation of Medulloblastoma Cells.
GCPs have been suggested to be the cell of origin for some types of medulloblastoma (40). Because bFGF inhibited Shh-induced proliferation of GCPs, we hypothesized that it might have similar effects on medulloblastoma cells. To test this hypothesis, we used tumor cells from an animal model of medulloblastoma, the ptc mutant mouse (41). These cells resemble GCPs and are known to depend on Shh signaling for their growth in vitro and in vivo (37, 42). To determine whether bFGF could inhibit Shh signaling in medulloblastoma cells, we cultured these cells in the presence of bFGF and measured expression of gli1. As shown in Fig. 7A, bFGF down-regulates gli1 in tumor cells, and FGFR inhibitors can reverse this effect. We also tested the effects of bFGF on proliferation of medulloblastoma cells: whereas untreated tumor cells proliferate robustly in culture, cells exposed to bFGF showed a dramatic reduction in proliferation (Fig. 6B). Consistent with this finding, bFGF treatment of tumor cells from Math1-GFP/ptc mice induces a flattened morphology (Fig. 7 C and D) and progressive loss of the GFP transgene that marks rapidly cycling tumor cells (Fig. 7 E–H). Together, these results demonstrate that bFGF suppresses growth of tumor cells from ptc mutant mice and may be a useful therapy for medulloblastomas and other tumors that depend on Shh signaling.
Fig. 7.
bFGF blocks proliferation of medulloblastoma cells from ptc mutant mice. (A and B) FGF inhibits Shh target genes and proliferation. Tumor cells from ptc+/− mice were pretreated with DMSO or FGFR inhibitor (PD173074, 100 nM), cultured in the absence (Control) or presence of bFGF for 24–48 h, and then harvested for RNA analysis (A) or pulsed and assayed for 3H-Td incorporation (B). Data represent means ± SEM of six samples. ∗, P < 0.01; ∗∗, P < 0.001 (one-way ANOVA). (C–H) FGF promotes differentiation of tumor cells. Tumor cells from Math1-GFP/ptc+/− mice were cultured with no stimulus (Con) (C, E, and G) or bFGF (D, F, and H) for 24–48 h and then photographed by using bright-field or fluorescent microscopy. (C and D) Bright-field images at 24 h. (E and F) Math1-GFP expression at 24 h. (G and H) Math1-GFP expression at 48 h. (Magnification: ×20).
Discussion
The Shh and FGF signaling pathways play critical roles in many aspects of neural development. In this study, we demonstrate that bFGF can antagonize the effects of Shh in neuronal precursors and tumor cells. We show that bFGF blocks Shh signaling in GCPs and that it exerts its effects upstream of gli-mediated transcription. We demonstrate that FGF-mediated inhibition of Shh-induced proliferation occurs in a JNK- and ERK-dependent manner. In addition, our results show that bFGF induces differentiation in vitro and in vivo. Finally, we demonstrate that bFGF markedly inhibits proliferation and promotes differentiation of medulloblastoma cells from ptc mutant mice.
To determine the specificity of FGF-mediated inhibition, we tested the effects of various RTK-activating factors on Shh responses in GCPs. Of all of the factors tested, only bFGF, FGF-1, and FGF-4 blocked Shh-induced proliferation. The basis for this specificity is unclear, especially because we have found that FGF-mediated inhibition depends on ERK and JNK kinases, and some of the other growth factors we tested have been reported to activate these kinases. One explanation for this specificity may be differences in the kinetics of activation of MAPKs in response to distinct growth factors (43). Alternatively, FGF may activate other signals in addition to MAPKs, and these signals may be critical for inhibition of Shh signaling. Further studies of the mechanisms of FGF-mediated inhibition will be necessary to shed light on this matter.
To determine whether bFGF specifically acts to inhibit Shh signaling (as opposed to, for example, acting on the cell cycle machinery), we examined the effects of bFGF on Shh target genes. Our finding that bFGF inhibits induction of gli1 in GCPs and induction of luciferase in Shh-Light2 cells suggests that bFGF acts upstream of (or at the level of) gli transcription factors to inhibit the Shh response. It is important to note that bFGF may also inhibit proliferation of GCPs by non-Shh-dependent mechanisms. For example, FGF signaling can promote neuronal differentiation even in the absence of Shh signaling (36, 44). However, our observation of FGF-mediated inhibition of Shh responses in both fibroblasts and neurons suggests that the Shh pathway itself is an important target of FGF signaling.
To determine whether bFGF-mediated inhibition of Shh signaling is mediated by FGFRs, we used pharmacologic inhibitors of FGFR kinases. Our results demonstrate that FGF-mediated inhibition of Shh responses was completely blocked by these compounds, which suggests that inhibition of Shh responses requires activation of FGFRs. To further elucidate the mechanisms by which bFGF exerts its effects, we inhibited a number of common effectors of FGFR signaling. Because PKA can inhibit Shh responses in many tissues (45), and because pituitary adenylate cyclase activating polypeptide (PACAP) (an activator of PKA) can inhibit Shh responses in GCPs (46), it seemed possible that bFGF might exert its effects through PKA. However, whereas antagonists of PKA prevented PACAP-mediated inhibition of Shh signaling, they had little effect on bFGF-mediated inhibition, suggesting that bFGF and PACAP inhibit Shh responses through distinct mechanisms. Our observation that pharmacologic antagonists of JNK and ERK completely rescue Shh-induced proliferation in the presence of bFGF suggests that these kinases are critical mediators of bFGF-mediated inhibition.
To study the effects of FGF on granule cell differentiation we examined expression of neuronal differentiation markers in bFGF-treated cells. Our results indicate that bFGF not only promotes differentiation of GCPs on its own but also allows differentiation in the presence of Shh. Similar results were seen when bFGF was administered in vivo: despite the presence of endogenous Shh, many bFGF-treated GCPs stopped dividing and began to differentiate. The fact that GCPs are normally able to exit the cell cycle and differentiate in a Shh-rich microenvironment (the inner EGL) (47) suggests that there must be a “stop signal” that overrides the mitogenic effects of Shh and allows GCPs to differentiate. Because several FGF ligands, including the ones we have identified as inhibitors of Shh signaling, are expressed in the postnatal cerebellum (48–50), it is possible that they may function as such stop signals. However, a number of other molecules [pituitary adenylate cyclase activating polypeptide (46), bone morphogenetic proteins (51), and the extracellular matrix molecule vitronectin (47)] have also been shown to inhibit Shh-induced proliferation of GCPs in vitro and could therefore represent stop signals as well. Further experiments will be necessary to delineate the contribution of each of these signals to granule cell cycle exit and differentiation in vivo.
One of the most significant findings we have reported is the ability of bFGF to halt proliferation of medulloblastoma cells from ptc mutant mice. Our results are consistent with studies showing that bFGF can inhibit the growth of human medulloblastoma cell lines in vitro and after transplantation into the forebrain of nude mice (52, 53). In addition, gene expression analysis suggests that expression of FGF receptors is lower in medulloblastoma samples than in normal cerebellum (54), raising the possibility that FGF signaling may be dysregulated in some cases of medulloblastoma. Our study suggests that FGF inhibits growth of medulloblastoma specifically by antagonizing the Shh pathway. In addition, because Shh signaling has been shown to be important for growth of many types of tumors (55–57), our findings may have broad implications for treatment of cancer.
Materials and Methods
Animals.
C57BL/6 × CBA F1 mice were generated by crossing CBA/J males and C57BL/6J females from The Jackson Laboratory (Bar Harbor, ME). Math1-GFP mice (28) were provided by Jane Johnson (University of Texas Southwestern Medical Center, Dallas, TX). ptc+/− mice (41) were from Matthew Scott (Stanford University, Stanford, CA). Animals were bred and maintained in the animal facility at Duke Medical Center.
Growth Factors, Kinase Inhibitors, Plasmids, and Retroviruses.
Recombinant Shh-N was purchased from R & D Systems (Minneapolis, MN) or generated in Escherichia coli by using a GST-Shh-N plasmid (from Phillip Beachy, The Johns Hopkins University, Baltimore, MD). Shh supernatant was generated by transfecting 293T cells with Shh-N expression plasmid (David Robbins, Dartmouth Medical School, Hanover, NH) and harvesting supernatant for 3 days. Recombinant Shh-N was used at 3 μg/ml; supernatant was used at 20% for GCPs and 40% for Shh-Light2 cells. FGF, EGF, and insulin-like growth factor-1 (Peprotech, Rocky Hill, NJ) were all used at 25 ng/ml, except where indicated. PDGF-AA and nerve growth factor (Peprotech) were used at 10 ng/ml and 100 ng/ml, respectively.
The FGFR inhibitor PD173074 was provided by Pfizer (New York, NY); SU5402, another FGFR inhibitor, was from Yvonne Nolan (University College Cork, Cork, Ireland). Inhibitors of the following kinases were from EMD Biosciences/Calbiochem (San Diego, CA): Src-family kinases (SU6656), MEK/ERK (U0126), JNK (SP600125), phosphatidylinositol 3-kinase (wortmannin), Akt (AKT inhibitor IV), calcium/calmodulin-dependent kinase II/IV (KN-93), PKA (PKI 14–22 amide), PKC-α/β (Go6796), PKCδ (rottlerin), and PKCζ (PKCζ inhibitor). Phospholipase Cγ inhibitor (U73122) was from BIOMOL (Plymouth Meeting, PA), and diacylglycerol kinase inhibitor (R59 949) was from Axxora (San Diego, CA).
FGFR1 retroviruses were generated by cloning a myristylated form of the FGFR1 kinase domain (34) (myrR1wt from Dan Donoghue, University of California at San Diego, La Jolla, CA) into Topo-pENTR (Invitrogen, Carlsbad, CA) and recombining it into Gateway-compatible MSCV-Puro-IRES-GFP (Scott Lowe, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY). Retroviral vectors were transfected into 293T cells with plasmids encoding gag-pol and vesicular stomatitis virus glycoprotein, and supernatants were concentrated by centrifugation. Retroviral stocks were tested on naive 293T cells, and equivalent titers were used for infection.
Isolation and Culture of GCPs, Tumor Cells, and Cell Lines.
GCPs and tumor cells were isolated as described in ref. 37 from 7-day-old (P7) wild-type or Math1-GFP mice and from 10- to 25-week-old ptc+/− mice displaying signs of medulloblastoma. To obtain pure GCPs, GFP+ cells were sorted from Math1-GFP pups by using a FACSVantage SE (BD Biosciences, San Jose, CA). GCPs were cultured on poly(d-lysine)-coated dishes in Neurobasal medium containing 2% B27, 1 mM sodium pyruvate, 2 mM l-glutamine, and 1% Pen/Strep (Invitrogen).
Shh-Light2 cells were cultured in DMEM with 10% calf serum, 0.4 mg/ml G418, 0.15 mg/ml Zeocin, 1% Pen/Strep, and 2 mM l-glutamine (Invitrogen).
Proliferation Assays.
GCPs were cultured in poly(d-lysine)-coated 96-well plates at 2 × 105 cells per well. Growth factors and inhibitors were added at the beginning of culture, and cells were incubated for 48 h before being pulsed with [methyl-3H]thymidine (GE Healthcare, Piscataway, NJ). After 18 h, cells were harvested by using a Mach IIIM Manual Harvester 96 (TOMTEC, Hamden, CT), and incorporated radioactivity was quantitated by using a Wallac MicroBeta microplate scintillation counter (PerkinElmer, Fremont, CA).
Luciferase Assays.
Shh-Light2 cells were plated at 4 × 102 cells per well in 24-well plates. After 24 h, cells were pretreated for 1 h with inhibitors and then stimulated with Shh in the presence or absence of bFGF for 48 h. Cells were lysed and assayed by using the Dual Luciferase Assay system (Promega, Madison, WI). Firefly luciferase activity was normalized to Renilla luciferase activity and then divided by values for control cultures to calculate fold change in luciferase activity.
RNA Isolation and RT-PCR.
RNA was isolated by using the RNAqueous-Micro kit (Ambion, Austin, TX). Lysates were treated with DNA-free DNase treatment and removal reagents (Ambion), and RNA was quantitated on a TD-700 fluorometer (Turner BioSystems, Sunnyvale, CA) by using RiboGreen (Invitrogen). Real-time PCR for gli1 was performed as described in ref. 37.
Western Blotting.
Cells were lysed in buffer containing 50 mM Hepes, 1 mM EDTA, 1% IGEPAL CA-630, 1 mM DTT, 1 mM sodium orthovanadate (all from Sigma, St. Louis, MO), and protease inhibitor mixture (Pierce, Rockford, IL). Clarified lysates were quantitated by using a protein assay kit from Bio-Rad (Hercules, CA). Proteins were resolved by SDS/PAGE and transferred to PVDF membranes (Millipore, Bedford, MA). Membranes were probed with antibodies against actin (Sigma), ERK, phospho-ERK, c-jun, or phospho-jun (all from Santa Cruz Biotechnology, Santa Cruz, CA), followed by goat anti-mouse or anti-rabbit antibodies conjugated to Alexa Fluor 680 (Invitrogen, Eugene, OR) or IRdye 800 (Rockland, Gilbertsville, PA). Proteins were detected by using the Odyssey imaging system (LI-COR, Lincoln, NE).
Immunofluorescence Staining and Analysis.
GCPs were plated on poly(d-lysine)-coated glass coverslips in 24-well plates at 5 × 105 cells per well and cultured for 24–72 h before fixation in 2% paraformaldehyde and permeabilization with 0.2% Triton X-100. Coverslips were stained overnight with antibodies specific for Ki-67 or MEF2D (BD-PharMingen, San Diego, CA) and for 1 h with Alexa Fluor 488-anti-mouse IgG (Invitrogen) and then mounted by using Vectashield and DAPI (Vector Laboratories, Burlingame, CA).
For cerebellar sections, permeablization was carried out as described above and sections were stained overnight with antibodies specific for BrdU (Abcam, Cambridge, MA) or NeuN (Millipore) and for 1 h with Alexa Fluor 568-anti-rat IgG or Alexa Fluor-594-streptavidin (Invitrogen). Images were acquired by using a Nikon TE200 inverted fluorescent microscope and Openlab software (Improvision, Lexington, MA).
Intracisternal Injections.
Mice were given daily intracisternal injections of bFGF (200 ng/μl; 4 μl) or vehicle [5 mM Tris (pH 7.5); 4 μl] from P4 to P6, followed by an i.p. injection of BrdU (10 mg/ml; 70 μl) on P6, to detect proliferating cells. At P7 pups were perfused with 4% paraformaldehyde, their cerebella were dissected, cryoprotected in 30% sucrose, embedded in OCT compound, and cryosectioned at a thickness of 12 μm. Immunofluorescence staining was carried out as described in Immunofluorescence Staining and Analysis.
Supplementary Material
Acknowledgments
We thank Dan Donoghue for FGFR1 cDNA, Alison Meloni for sharing unpublished research, Rasi Wickramasinghe for contributions to early studies, Mike Cook for help with FACS, Judy Johnston and Jack Dutton for help with intracisternal injections, Marc Caron and Ann Marie Pendergast for critical review of the manuscript, and Pfizer for PD173074. This work was supported by the Pediatric Brain Tumor Foundation and by National Institute of Mental Health Grant MH067916-03 (to R.J.W.-R). R.J.W.-R. is a Kimmel Foundation Scholar.
Abbreviations
- bFGF
basic fibroblast growth factor
- EGL
external granule layer
- ERK
extracellular signal regulated kinase
- FGFR
fibroblast growth factor receptor
- GCP
granule cell precursor
- 3H-Td
[3H]thymidine
- JNK
c-jun N-terminal kinase
- Pn
postnatal day n
- PKA
protein kinase A
- RTK
receptor tyrosine kinase
- Shh
Sonic hedgehog.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0605770104/DC1.
References
- 1.Cayuso J, Marti E. J Neurobiol. 2005;64:376–387. doi: 10.1002/neu.20169. [DOI] [PubMed] [Google Scholar]
- 2.Fogarty MP, Kessler JD, Wechsler-Reya RJ. J Neurobiol. 2005;64:458–475. doi: 10.1002/neu.20166. [DOI] [PubMed] [Google Scholar]
- 3.Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, McMahon AP. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- 4.Roelink H, Augsburger A, Heemskerk J, Korzh V, Norlin S, Ruiz i Altaba A, Tanabe Y, Placzek M, Edlund T, Jessell TM, et al. Cell. 1994;76:761–775. doi: 10.1016/0092-8674(94)90514-2. [DOI] [PubMed] [Google Scholar]
- 5.Marti E, Bovolenta P. Trends Neurosci. 2002;25:89–96. doi: 10.1016/s0166-2236(02)02062-3. [DOI] [PubMed] [Google Scholar]
- 6.Machold R, Hayashi S, Rutlin M, Muzumdar MD, Nery S, Corbin JG, Gritli-Linde A, Dellovade T, Porter JA, Rubin LL, et al. Neuron. 2003;39:937–950. doi: 10.1016/s0896-6273(03)00561-0. [DOI] [PubMed] [Google Scholar]
- 7.Ahn S, Joyner AL. Nature. 2005;437:894–897. doi: 10.1038/nature03994. [DOI] [PubMed] [Google Scholar]
- 8.Wechsler-Reya RJ, Scott MP. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 9.Jensen AM, Wallace VA. Development (Cambridge, UK) 1997;124:363–371. doi: 10.1242/dev.124.2.363. [DOI] [PubMed] [Google Scholar]
- 10.Hynes M, Porter JA, Chiang C, Chang D, Tessier-Lavigne M, Beachy PA, Rosenthal A. Neuron. 1995;15:35–44. doi: 10.1016/0896-6273(95)90062-4. [DOI] [PubMed] [Google Scholar]
- 11.Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I, Chan J, McMahon AP, Stiles CD, Rowitch DH. Neuron. 2000;25:317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- 12.Charron F, Stein E, Jeong J, McMahon AP, Tessier-Lavigne M. Cell. 2003;113:11–23. doi: 10.1016/s0092-8674(03)00199-5. [DOI] [PubMed] [Google Scholar]
- 13.Trousse F, Marti E, Gruss P, Torres M, Bovolenta P. Development (Cambridge, UK) 2001;128:3927–3936. doi: 10.1242/dev.128.20.3927. [DOI] [PubMed] [Google Scholar]
- 14.Reuss B, von Bohlen und Halbach O. Cell Tissue Res. 2003;313:139–157. doi: 10.1007/s00441-003-0756-7. [DOI] [PubMed] [Google Scholar]
- 15.Dono R. Am J Physiol. 2003;284:R867–R881. doi: 10.1152/ajpregu.00533.2002. [DOI] [PubMed] [Google Scholar]
- 16.Streit A, Berliner AJ, Papanayotou C, Sirulnik A, Stern CD. Nature. 2000;406:74–78. doi: 10.1038/35017617. [DOI] [PubMed] [Google Scholar]
- 17.Martinez S, Crossley PH, Cobos I, Rubenstein JL, Martin GR. Development (Cambridge, UK) 1999;126:1189–1200. doi: 10.1242/dev.126.6.1189. [DOI] [PubMed] [Google Scholar]
- 18.Gage FH, Coates PW, Palmer TD, Kuhn HG, Fisher LJ, Suhonen JO, Peterson DA, Suhr ST, Ray J. Proc Natl Acad Sci USA. 1995;92:11879–11883. doi: 10.1073/pnas.92.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gritti A, Parati EA, Cova L, Frolichsthal P, Galli R, Wanke E, Faravelli L, Morassutti DJ, Roisen F, Nickel DD, et al. J Neurosci. 1996;16:1091–1100. doi: 10.1523/JNEUROSCI.16-03-01091.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandran S, Kato H, Gerreli D, Compston A, Svendsen CN, Allen ND. Development (Cambridge, UK) 2003;130:6599–6609. doi: 10.1242/dev.00871. [DOI] [PubMed] [Google Scholar]
- 21.Dasen JS, Liu JP, Jessell TM. Nature. 2003;425:926–933. doi: 10.1038/nature02051. [DOI] [PubMed] [Google Scholar]
- 22.Ye W, Shimamura K, Rubenstein JL, Hynes MA, Rosenthal A. Cell. 1998;93:755–766. doi: 10.1016/s0092-8674(00)81437-3. [DOI] [PubMed] [Google Scholar]
- 23.Webber CA, Hyakutake MT, McFarlane S. Dev Biol. 2003;263:24–34. doi: 10.1016/s0012-1606(03)00435-4. [DOI] [PubMed] [Google Scholar]
- 24.Szebenyi G, Dent EW, Callaway JL, Seys C, Lueth H, Kalil K. J Neurosci. 2001;21:3932–3941. doi: 10.1523/JNEUROSCI.21-11-03932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams EJ, Furness J, Walsh FS, Doherty P. Development (Cambridge, UK) 1994;120:1685–1693. doi: 10.1242/dev.120.6.1685. [DOI] [PubMed] [Google Scholar]
- 26.Kessaris N, Jamen F, Rubin LL, Richardson WD. Development (Cambridge, UK) 2004;131:1289–1298. doi: 10.1242/dev.01027. [DOI] [PubMed] [Google Scholar]
- 27.Cheng Y, Tao Y, Black IB, DiCicco-Bloom E. J Neurobiol. 2001;46:220–229. doi: 10.1002/1097-4695(20010215)46:3<220::aid-neu1004>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 28.Lumpkin EA, Collisson T, Parab P, Omer-Abdalla A, Haeberle H, Chen P, Doetzlhofer A, White P, Groves A, Segil N, et al. Gene Expr Patterns. 2003;3:389–395. doi: 10.1016/s1567-133x(03)00089-9. [DOI] [PubMed] [Google Scholar]
- 29.Lee A, Kessler JD, Read TA, Kaiser C, Corbeil D, Huttner WB, Johnson JE, Wechsler-Reya RJ. Nat Neurosci. 2005;8:723–729. doi: 10.1038/nn1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai CB, Stephen D, Joyner AL. Dev Cell. 2004;6:103–115. doi: 10.1016/s1534-5807(03)00394-0. [DOI] [PubMed] [Google Scholar]
- 31.Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 32.Rubin JB, Choi Y, Segal RA. Development (Cambridge, UK) 2002;129:2223–2232. doi: 10.1242/dev.129.9.2223. [DOI] [PubMed] [Google Scholar]
- 33.Skaper SD, Kee WJ, Facci L, Macdonald G, Doherty P, Walsh FS. J Neurochem. 2000;75:1520–1527. doi: 10.1046/j.1471-4159.2000.0751520.x. [DOI] [PubMed] [Google Scholar]
- 34.Hart KC, Robertson SC, Kanemitsu MY, Meyer AN, Tynan JA, Donoghue DJ. Oncogene. 2000;19:3309–3320. doi: 10.1038/sj.onc.1203650. [DOI] [PubMed] [Google Scholar]
- 35.Tsang M, Dawid IB. Sci STKE. 2004;2004:pe17. doi: 10.1126/stke.2282004pe17. [DOI] [PubMed] [Google Scholar]
- 36.Vicario-Abejon C, Johe KK, Hazel TG, Collazo D, McKay RD. Neuron. 1995;15:105–114. doi: 10.1016/0896-6273(95)90068-3. [DOI] [PubMed] [Google Scholar]
- 37.Oliver TG, Read TA, Kessler JD, Mehmeti A, Wells JF, Huynh TT, Lin SM, Wechsler-Reya RJ. Development (Cambridge, UK) 2005;132:2425–2439. doi: 10.1242/dev.01793. [DOI] [PubMed] [Google Scholar]
- 38.Lin X, Shah S, Bulleit RF. Brain Res Mol Brain Res. 1996;42:307–316. doi: 10.1016/s0169-328x(96)00135-0. [DOI] [PubMed] [Google Scholar]
- 39.Fukui Y, Hoshino K, Kameyama Y. Exp Neurol. 1987;98:54–66. doi: 10.1016/0014-4886(87)90071-9. [DOI] [PubMed] [Google Scholar]
- 40.Read TA, Hegedus B, Wechsler-Reya R, Gutmann DH. Ann Neurol. 2006;60:3–11. doi: 10.1002/ana.20912. [DOI] [PubMed] [Google Scholar]
- 41.Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 42.Romer JT, Kimura H, Magdaleno S, Sasai K, Fuller C, Baines H, Connelly M, Stewart CF, Gould S, Rubin LL, et al. Cancer Cell. 2004;6:229–240. doi: 10.1016/j.ccr.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 43.Marshall CJ. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 44.Hatten ME, Lynch M, Rydel RE, Sanchez J, Joseph-Silverstein J, Moscatelli D, Rifkin DB. Dev Biol. 1988;125:280–289. doi: 10.1016/0012-1606(88)90211-4. [DOI] [PubMed] [Google Scholar]
- 45.Hammerschmidt M, Bitgood MJ, McMahon AP. Genes Dev. 1996;10:647–658. doi: 10.1101/gad.10.6.647. [DOI] [PubMed] [Google Scholar]
- 46.Nicot A, Lelievre V, Tam J, Waschek JA, DiCicco-Bloom E. J Neurosci. 2002;22:9244–9254. doi: 10.1523/JNEUROSCI.22-21-09244.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pons S, Trejo JL, Martinez-Morales JR, Marti E. Development (Cambridge, UK) 2001;128:1481–1492. doi: 10.1242/dev.128.9.1481. [DOI] [PubMed] [Google Scholar]
- 48.Matsuda S, Ii Y, Desaki J, Yoshimura H, Okumura N, Sakanaka M. Neuroscience. 1994;59:651–662. doi: 10.1016/0306-4522(94)90184-8. [DOI] [PubMed] [Google Scholar]
- 49.Wilcox BJ, Unnerstall JR. Neuron. 1991;6:397–409. doi: 10.1016/0896-6273(91)90248-x. [DOI] [PubMed] [Google Scholar]
- 50.Ozawa K, Uruno T, Miyakawa K, Seo M, Imamura T. Brain Res Mol Brain Res. 1996;41:279–288. doi: 10.1016/0169-328x(96)00108-8. [DOI] [PubMed] [Google Scholar]
- 51.Rios I, Alvarez-Rodriguez R, Marti E, Pons S. Development (Cambridge, UK) 2004;131:3159–3168. doi: 10.1242/dev.01188. [DOI] [PubMed] [Google Scholar]
- 52.Duplan SM, Theoret Y, Kenigsberg RL. Clin Cancer Res. 2002;8:246–257. [PubMed] [Google Scholar]
- 53.Vachon P, Girard C, Theoret Y. J Neurooncol. 2004;67:139–146. doi: 10.1023/b:neon.0000021824.41701.e5. [DOI] [PubMed] [Google Scholar]
- 54.Park PC, Taylor MD, Mainprize TG, Becker LE, Ho M, Dura WT, Squire J, Rutka JT. J Neurosurg. 2003;99:534–541. doi: 10.3171/jns.2003.99.3.0534. [DOI] [PubMed] [Google Scholar]
- 55.Watkins DN, Berman DM, Burkholder SG, Wang B, Beachy PA, Baylin SB. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 56.Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY, Qi YP, Gysin S, Fernandez-del Castillo C, Yajnik V, et al. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karhadkar SS, Bova GS, Abdallah N, Dhara S, Gardner D, Maitra A, Isaacs JT, Berman DM, Beachy PA. Nature. 2004;431:707–712. doi: 10.1038/nature02962. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.