Abstract
α-Amino-3-hydroxy-5-methylisoxazole-4-propionic acid (AMPA)-type glutamate receptors undergo constitutive and ligand-induced internalization that requires dynamin and the clathrin adaptor complex AP-2. We report here that an atypical basic motif within the cytoplasmic tails of AMPA-type glutamate receptors directly associates with μ2-adaptin by a mechanism similar to the recognition of the presynaptic vesicle protein synaptotagmin 1 by AP-2. A synaptotagmin 1-derived AP-2 binding peptide competes the interaction of the AMPA receptor subunit GluR2 with AP-2μ and increases the number of surface active glutamate receptors in living neurons. Moreover, fusion of the GluR2-derived tail peptide with a synaptotagmin 1 truncation mutant restores clathrin/AP-2-dependent internalization of the chimeric reporter protein. These data suggest that common mechanisms regulate AP-2-dependent internalization of pre- and postsynaptic membrane proteins.
Keywords: endocytosis, postsynaptic, sorting signal, synaptic plasticity
Fast neurotransmission at excitatory synapses is mediated by heterotetrameric α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid (AMPA)-type glutamate receptors composed of combinations of four subunits (GluR1–4). AMPA receptors interact with different factors including the transmembrane protein stargazin (1), PDZ proteins GRIP1/ABP, SAP97, and PICK1, and NSF (2). Accumulating evidence suggests that rapid changes in functional postsynaptic AMPA receptor numbers are important means of controlling synaptic efficacy (2–5). AMPA receptors undergo constitutive and regulated clathrin- and dynamin-dependent endocytosis via distinct AMPA- or NMDA-induced signaling cascades (reviewed in refs. 2–5). How exactly AMPA receptor cargo is targeted for clathrin-mediated internalization remains an open question. One possibility is that AMPA receptors are recognized by endocytic adaptor proteins such as the clathrin adaptor complex AP-2, a major endocytic protein interaction hub (6–8). NMDA-induced AMPA receptor internalization can be blocked by overexpression of a GluR2 cytoplasmic tail (CT) peptide (pep2r) or by mutating the putative AP-2 binding motif within the GluR2 CT. Infusion of hippocampal CA1 neurons with the putative AP-2-blocking peptide prevents induction of long-term depression (LTD), suggesting that the association of GluR2 with AP-2 may be an important determinant for NMDA-induced LTD (7). Whether AP-2 directly binds to GluR2 CTs and via which of its four subunits is unknown.
Here we have identified the molecular determinants responsible for binding of AP-2 to the CTs of AMPA-type glutamate receptors. We demonstrate that the μ2 subunit of AP-2 interacts directly and with nanomolar affinity with a basic motif found in CTs of GluR1–3 and the presynaptic vesicle protein synaptotagmin 1. Our data thus suggest that common mechanisms regulate AP-2-dependent internalization of pre- and postsynaptic membrane proteins.
Results
AMPA Receptor CTs Directly Bind to AP-2μ.
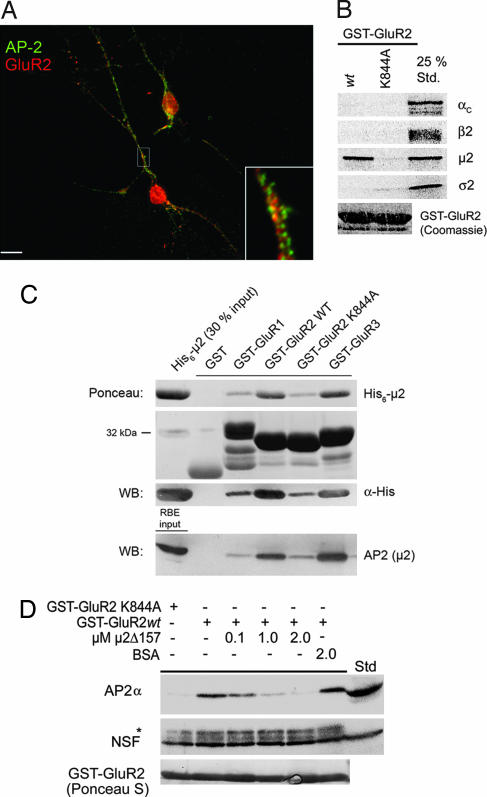
Given the importance of endocytic internalization of AMPA-type glutamate receptors for synaptic plasticity phenomena (7), we decided to dissect the putative interaction between GluR2 CT and AP-2 in molecular detail. In agreement with earlier reports (7), we found that GluR2 partially colocalizes with AP-2 within dendrites of hippocampal neurons after glutamate stimulation (Fig. 1A), and its CT specifically pulls down AP-2, but not the related endosomal adaptor complex AP-3 or other endocytic proteins including clathrin, dynamin 1, eps15, or HIP1R [supporting information (SI) Fig. 6]. A point mutant of GluR2 CT, in which a conserved lysine residue within the putative AP-2-binding sequence had been replaced by alanine (K844A), did not bind to AP-2, nor did GST.
Fig. 1.
AMPA receptor CTs associate directly with AP2μ. (A) Localization of AP-2α and GluR2 in K+-glutamate-stimulated hippocampal neurons (100 μM for 10 min at 37°C). GluR2 colocalizes partially with AP-2 within dendrites (yellow in the overlay). (Inset) Magnification (×5) of selected area. (Scale bar: 10 μm.) (B) GluR2 CT directly binds to AP-2μ. 35S-labeled αχ, β2, μ2, or σ2-adaptins translated in vitro were incubated with GST, GST-GluR2 CT, or an AP-2 binding-defective mutant (K844A) (100 μg). Bound material was analyzed by SDS/PAGE and autoradiography. 25% Std., 25% of the total amount of radiolabeled protein added to the assay. (C) Binding of purified His6-tagged μ2 (157–435; top blot) or AP-2 (bottom blot) to GluR1–3 CTs. Immobilized GST fusion proteins (30 μg) were incubated with recombinant His6-tagged μ2 (157–435) or Triton X-100-extracted rat brain lysates and washed extensively, and complexes were resolved by SDS/PAGE and staining with Ponceau S or analyzed by immunoblotting. RBE input was 20 μg of rat brain lysate. (D) Recombinant μ2 (157–435) competes the association of native AP-2 with GST-GluR2. Affinity purification of AP-2 using GST-GluR2 CT (20 μg) was performed in the presence of increasing concentrations of μ2 (157–435) or BSA. Samples were analyzed by immunoblotting for AP-2 or NSF. ∗, Cross-reactive band decorated with anti-NSF antibodies; Std, Triton X-100-extracted rat brain lysate (50 μg total protein).
To find out whether AP-2 was able to bind directly to GluR2 CT and, if so, via which of its four subunits, we performed pull-down experiments using 35S-labeled AP-2 subunits synthesized by coupled transcription/translation in vitro. GST-GluR2 CT wild type but not its K844A mutant specifically associated with μ2-adaptin (Fig. 1B). To confirm this result we incubated purified His6-μ2-adaptin (residues 157–435) (8) with GST-GluR1–3 CTs or GST. As seen in Fig. 1C (Ponceau; α-His) His6-μ2 (157–435) robustly bound to the CTs of GluR2 and GluR3, but not to GST. Very weak if any specific binding to GluR1 was detected perhaps because of the exchange of one lysine residue within the AP-2μ binding sequence for cysteine (C843; compare Fig. 2A). Similar results were seen for native AP-2 complexes from the brain (Fig. 1C, bottom blot). Recombinant μ2 (157–435) was also able to compete the association of native AP-2 complexes with GST-GluR2 CT in a dose-dependent manner (Fig. 1D). These data indicate that the CTs of AMPA-type glutamate receptors directly interact with AP-2μ.
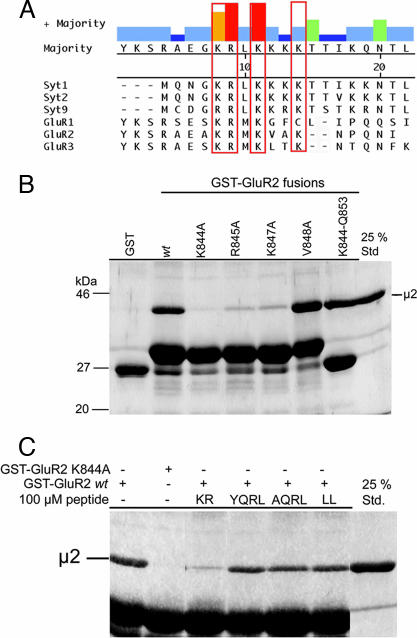
Fig. 2.
Binding of μ2-adaptin to the GluR2 CT involves basic residues and can be competed by a synaptotagmin 1-derived AP-2 binding peptide. (A) Multiple sequence alignment of the AP-2μ binding motifs from synaptotagmin 1 (residues 317–335), synaptotagmin 2 (318–336), synaptotagmin 9 (397–415) (C2B domains from rat), GluR1 (residues 830–850), GluR2 (residues 837–856), and GluR3 (residues 842–861) CTs (rat) using the Clustal W algorithm. Conserved basic residues are boxed (red). (B) GluR2 CT or point mutants thereof were used for pull-downs with His6-tagged μ2 (157–435) (see legend to Fig. 1C) and analyzed by Coomassie blue staining. K844-Q853 is a fusion protein between GST and amino acids 844–853 of GluR2. (C) Direct binding of His6-tagged μ2 (157–435) to GST-GluR2 can be competed by a synaptotagmin 1 C2B domain-derived AP-2 binding peptide, but not by tyrosine (YQRL) or dileucine (LL) sorting motif peptides (100 μM each). A nonfunctional tyrosine motif peptide (AQRL) was used as a control. Samples were analyzed as described (see legend to Fig. 1C).
AP-2μ Binds to GluR2 via an Atypical Basic Motif also Found in the Presynaptic Vesicle Protein Synaptotagmin.
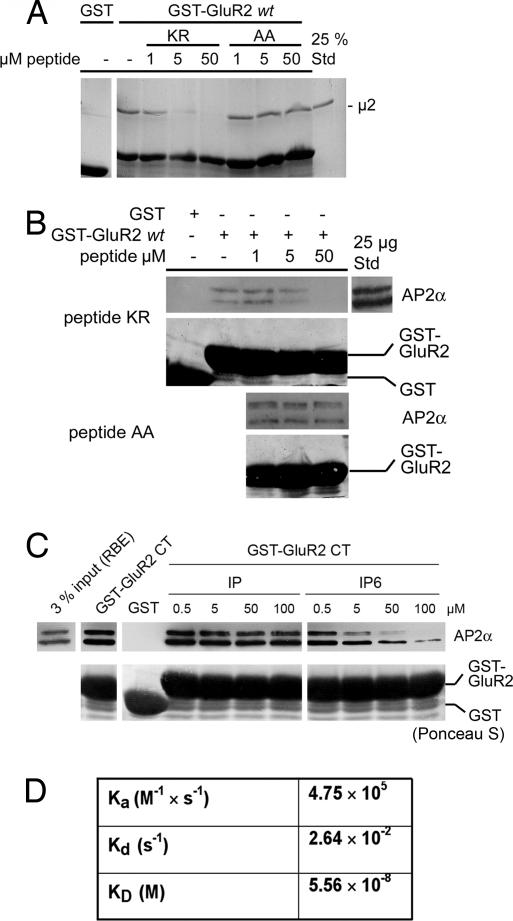
Because the association of GluR2 CT with AP-2 appears to require the presence of several basic lysine and arginine residues within a conserved sequence of its CT (7), we wanted to find out whether similar characteristics hold true for the direct binding of GluR2 CT to purified μ2-adaptin. Mutation of one of several basic residues (i.e., K844, R845, and K847) contained within the putative AP-2 binding motif of GluR2 CT to alanine significantly diminished binding to recombinant μ2 (157–435) (Fig. 2B). Because the atypical AP-2μ interacting motif is not only conserved between different AMPA receptor subunits but also bears similarity to the AP-2 binding site of synaptotagmins (Fig. 2A) (9), we hypothesized that both protein families may use a common recognition mechanism for their clathrin/AP-2-dependent internalization. Hence, one would expect that synaptotagmin and GluR2 bind to AP-2μ via the same site. To test this we performed affinity chromatography experiments using GST-GluR2 CT and μ2 in the presence of different endocytic internalization motifs. Strikingly, binding of μ2 to GST-GluR2 CT was inhibited by an AP-2 binding peptide derived from the C2B domain of synaptotagmin 1 (KR) but not by classical tyrosine-(YQRL) or dileucine-based (LL) endocytic sorting motif peptides (Fig. 2C). Similar results were seen for the association of native AP-2 with GluR2 CT (SI Fig. 7). Small amounts of AP-2 coimmunoprecipitated with GluR2 from synaptosomal rat brain lysates, and this interaction was inhibited by the synaptotagmin 1-derived basic AP-2 binding peptide (SI Fig. 7B). A mutant version of the peptide in which two of the conserved lysine residues had been replaced by alanines (AA) was unable to interfere with binding of μ2-adaptin (157–435) (Fig. 3A) or native AP-2 (Fig. 3B) to GST-GluR2 CT. As seen for the interaction between AP-2 and synaptotagmin 1 (10), the binding of GST-GluR2 CT could be competed by increasing concentrations of inositol(1,2,3,4,5,6)-hexakisphosphate, but not by inositol(1)-monophosphate (Fig. 3C), further strengthening the hypothesis that synaptotagmin 1 and GluR2 bind to AP-2 via similar mechanisms. Quantitative analysis of the association of μ2 (157–435) with the GluR2 CT-derived basic motif peptide by surface plasmon resonance revealed a high-affinity interaction with a KD of ≈56 nM (Fig. 3D and SI Fig. 8). Much weaker binding was seen for a mutated peptide in which two of the basic residues corresponding to K844 and R845 (see Fig. 2A) were replaced by alanines (data not shown).
Fig. 3.
Direct high-affinity binding of μ2-adaptin to the GluR2 CT. (A and B) The inhibitory effect of the synaptotagmin 1-derived peptide on the association of μ2 (157–435; A) or AP-2 (B) with GST-GluR2 requires basic residues. Affinity purification was performed in the presence of increasing concentrations of wild-type (KR) or mutant (AA) synaptotagmin 1 C2B domain peptides (see legend to Fig. 2). Coomassie blue (A) and Ponceau S (B) staining were used to verify equal loading of GST or GST-GluR2 fusion proteins. (C) Affinity purification from rat brain extracts was done in the presence of increasing concentrations of inositol(1)-monophosphate (IP) or inositol(1,2,3,4,5,6)-hexakisphosphate. Immunoblotted material was stained with Ponceau S or antibodies against AP-2αA/C. (D) Surface plasmon resonance analysis of the binding of purified μ2 (157–435) to immobilized GluR2-derived AP-2 binding peptide (pep2R). Rate constants and KD were derived from sensograms shown in SI Fig. 8.
The GluR2-Derived AP-2 Binding Peptide Is Able to Target a Chimeric Synaptotagmin 1 ΔC2B Reporter Membrane Protein for AP-2μ-Dependent Internalization.
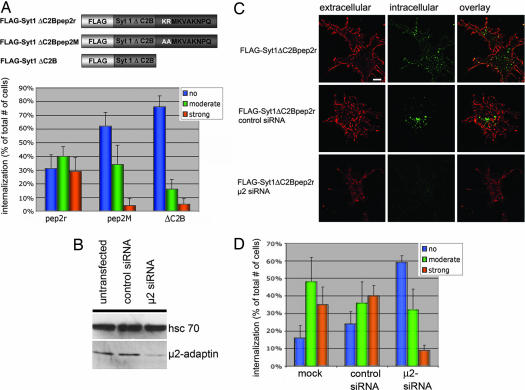
To obtain insights into the physiological relevance of the interaction between GluR2 and μ2-adaptin we constructed chimeric proteins between synaptotagmin 1 lacking its C2B domain and wild-type (pep2r) or mutant (pep2M) versions of the GluR2-derived AP-2μ binding peptide (Fig. 4A Upper). When transfected into fibroblasts synaptotagmin 1 ΔC2B neither displays detectable interaction with AP-2 nor does it become internalized. By contrast, when fibroblasts expressing synaptotagmin 1 ΔC2Bpep2r (tagged with an extracellular FLAG epitope) were subjected to an antibody feeding assay the chimeric reporter protein underwent rapid internalization in 29 ± 10% of transfected cells (SI Fig. 9B), similar to that seen for native homomeric GluR2 receptors (SI Fig. 9A). However, if two of the basic residues within the pep2r sequence that were essential for AP-2μ binding were mutated to alanines the ability of the chimeric reporter to become endocytosed was lost: internalization was detectable in only 4% of the transfected cells (Fig. 4A Lower). The synaptotagmin 1 ΔC2B reporter membrane protein itself did not undergo detectable internalization (Fig. 4A Lower), even after prolonged incubation times (data not shown). In agreement with this, we noted that upon cotransfection of a synaptotagmin 1 ΔC2B chimera harboring a functional AP-2μ binding GluR2 peptide (pep2r) soluble μ2-adaptin (157–435) was recruited to the plasmalemma (SI Fig. 10), suggesting an association of both proteins in living cells.
Fig. 4.
The GluR2-derived AP-2μ binding peptide targets a synaptotagmin 1 ΔC2B chimeric reporter protein for internalization. (A) Synaptotagmin 1 ΔC2B chimeric proteins (Upper) were expressed in Cos7 and assayed for internalization (compare SI Fig. 9B). Cells were classified visually into three phenotypic categories. Histograms (Lower) display the averaged percentage values collected from four independent experiments (n = 4; 20 cells per experiment). Error bars correspond to the SEM. Statistical significance was analyzed by the Pearson χ2 test (P < 0.01). (B) Immunoblot analysis of Cos7 cell extracts (10 μg total protein) transfected with siRNAs against a control protein (γ-BAR) or AP-2μ (μ2 siRNA) using antibodies against hsc70 or μ2-adaptin (72 h after transfection). (C) Internalization of synaptotagmin 1 ΔC2B-pep2r chimera in Cos7 cells transfected with siRNAs against a control protein or AP-2μ. Surface-exposed chimeric protein was labeled with Alexa Fluor 594 (red) under nonpermeabilizing conditions. Endocytosed anti-FLAG primary antibody was detected after Triton X-100 permeabilization of the cells followed by decoration with Alexa Fluor 488-conjugated secondary antibodies (green). (Scale bar: 15 μm.) (D) Semiquantitative analysis (see A) of FLAG-Syt1 ΔC2Bpep2r internalization in Cos7 cells depleted of AP-2μ. Histograms display the averaged percentage values collected from four independent experiments (n = 4; 20 cells per experiment). Error bars correspond to the SEM. The decrease of FLAG-Syt1 ΔC2Bpep2r endocytosis from 39 ± 11% in control transfected cells to 8 ± 3% in μ2-adaptin depleted cells is statistically significant (P < 0.01).
To test whether endocytosis of synaptotagmin 1 ΔC2Bpep2r is an AP-2-dependent process we used siRNAs directed against AP-2μ (11, 12). Transfection of Cos7 fibroblasts with the anti-μ2 siRNA but not with a control siRNA (directed against the TGN protein γ-BAR) resulted in knockdown of AP-2μ expression by >85% (Fig. 4B) and a corresponding loss of AP-2α-coated pit staining, paired with the inability of the AP-2 knockdown cells to internalize fluorescently labeled transferrin (data not shown; see also ref. 12). AP-2 knockdown cells also displayed a strongly reduced ability to endocytose the synaptotagmin 1 ΔC2Bpep2r chimera, whereas control siRNA-treated fibroblasts were unaffected (Fig. 4 C and D) (P < 0.01%). A randomized μ2 siRNA sequence had no effect on AP-2 levels (11) or on internalization (data not shown). Thus, the GluR2-derived AP-2μ binding motif is able to target a chimeric reporter protein for clathrin/AP-2-dependent internalization.
Disruption of AP-2 Binding to GluR by a Synaptotagmin 1-Derived AP-2 Binding Peptide Leads to Increased Numbers of Surface Active Receptors in Living Neurons.
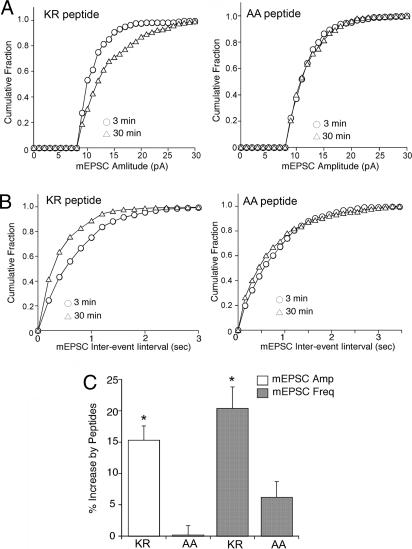
We finally analyzed the functional consequences of disrupting AP-2 recruitment to native GluRs in neurons. To this aim we carried out whole-cell patch clamp electrophysiological experiments to measure AMPA receptor-mediated miniature excitatory postsynaptic currents (mEPSC). It has been previously reported that blocking dynamin-dependent endocytosis results in an increase in the amplitude of AMPA receptor responses (13). We predicted that the Syt-1 peptide (KR), which binds AP-2 with high affinity, would block GluR internalization and similarly cause an increase in mEPSC. In agreement with this prediction, we found that dialysis with Syt-1 KR peptide (40 μg/ml) caused a significant increase in the mEPSC amplitude (Fig. 5A) and frequency (Fig. 5B). Representative mEPSC traces are illustrated in SI Fig. 11. Presumably, the increase in frequency is due to the recruitment of mEPSC previously below the threshold of detection, owing to an increased number of surface active GluRs. Increased mEPSC amplitudes (15.3 ± 2.3%, n = 7) and frequencies (20.4 ± 8.2%, n = 7) were seen only for the Syt-1 KR peptide, whereas the mutant control peptide (AA), in which two lysines had been exchanged for alanines, had little effect (mEPSC amplitude; 0.2 ± 1.5%, n = 6; mEPSC frequency: 3.4 ± 2.5%, n = 6) (Fig. 5C). No significant difference between these two peptides was observed on the mEPSC rise time or the mEPSC decay kinetics, suggesting that the effects of Syt-1 KR are not due to modulation of channel gating. These electrophysiological data thus confirm our proposal derived from biochemical studies that pre- and postsynaptic membrane proteins may use common mechanisms for clathrin/AP-2-mediated internalization.
Fig. 5.
The Syt-1 AP-2 binding peptide increases mEPSC amplitude and frequency. (A and B) Cumulative plots of the distribution of mEPSC amplitude (A) and frequency (B; shown as interevent interval) from the Syt-1 KR peptide or mutant AA control peptide-injected neurons. (C) Bar plot summary showing the different effects of Syt-1 KR and mutant AA peptides on mEPSC amplitude and frequency. (Scale bar: 50 pA and 2 sec.)
Discussion
Recent studies have provided strong evidence that AMPA receptors undergo rapid recycling in postsynaptic neurons via a clathrin- and AP-2-dependent process (2, 3, 5, 7, 14–16). In agreement with the observation that GluR2 exerts dominant effects over GluR1 with regard to AMPA receptor internalization (15), we have found that AP-2μ appears to interact much more efficiently with GluR2 or GluR3 than with GluR1 CTs. Moreover, we have mapped the GluR2 binding activity to a site within μ2 that is also responsible for its binding to inhibitory postsynaptic GABAA receptors (17) and to the presynaptic vesicle protein synaptotagmin (9, 18), suggesting a common mechanism for recognition of pre- and postsynaptic membrane cargo proteins by AP-2. This view is also supported by our observation that increasing concentrations of inositolpolyphosphates inhibit the association of GluR2 with AP-2, similar to what has been reported before for the synaptotagmin 1-AP-2 complex (10). Consistent with this we show that the atypical AP-2μ binding sequence of GluR2 is sufficient to target a synaptotagmin 1ΔC2B fusion protein chimera for AP-2-mediated internalization and, most importantly, that microinjection of a synaptotagmin 1-derived AP-2 binding peptide leads to increased surface levels of GluRs in living neurons in vivo.
AMPA receptor internalization can be triggered by different stimuli including low-frequency synaptic stimulation, ligand occupancy, or NMDA receptor activation (2, 3, 5). In addition, GluR2-containing receptors undergo constitutive cycling in and out of synapses and between endosomal compartments and the cell surface (19). The molecular determinants of these pathways have remained unexplored. Several of these mechanisms may contribute to hippocampal or cerebellar LTD. We have shown here that disruption of AP-2 binding to AMPA receptors leads to profound changes in the number of surface active GluRs and concomitant increases in the amplitude and frequency of mEPSCs in primary neurons. Our findings thus differ somewhat from the observations made by Lee et al. (7), who observed a specific inhibition of low-frequency stimulation-induced LTD in hippocampal CA1 pyramidal cells after infusion of a GluR2-derived AP-2 binding peptide. These differences are most likely caused by distinct experimental conditions (i.e., EPSCs after low-frequency stimulation vs. mEPSC measurements). However, both types of physiological readouts underscore the importance of the interaction between AMPA receptors and AP-2 for regulating the number of surface active GluRs. It is conceivable that NMDA-induced changes in the phosphorylation state of postsynaptic proteins (14, 16, 20) promote the association of AP-2μ with the atypical basic sorting signal within GluR CTs and lead to the accumulation of AMPA receptors in clathrin/AP-2-coated pits (21). The fact that the basic atypical AP-2μ binding motif is conserved between species ranging from worms (22) to mice and between different AMPA receptor subtypes suggests that homo- and heterooligomeric assembly of AMPA receptor tetramers could modulate the affinity of the complex for AP-2 and might thus regulate clathrin/AP-2-mediated receptor internalization under different physiological conditions. Other mechanisms, including phosphorylation (23, 24) or ubiquitination (14) of GluR CTs, association with HIP1 (25), etc., are likely to also contribute to the regulation of AMPA receptor internalization.
The observation that various pre- and postsynaptic membrane receptors including AMPA and GABAA receptors (17), as well as synaptotagmin family members (9) use a common mechanism for interacting with a cognate recognition site within subdomain B of μ2-adaptin (17, 18) poses the question of how exactly this is accomplished and why a distinct atypical internalization signal is used for this type of cargo proteins. One possibility could be that atypical endocytosis signals within synaptic cargo membrane proteins could contribute to their sorting into specialized endosomes. For example, presynaptic vesicle proteins are sorted away from constitutive cargo within the presynaptic terminal. Likewise, postsynaptic membrane proteins may undergo targeting to distinct populations of early endosomes (15), which are capable of segregating synaptic from constitutively internalized nonsynaptic cargo proteins, similar to what has been reported recently for other internalized ligands (26). The use of a distinct high-affinity cargo binding site within AP-2μ together with additional regulatory mechanisms, perhaps involving phosphoinositides such as PI(4,5)P2 or inositolpolyphosphates, could contribute to this.
Materials and Methods
Antibodies.
For details see SI Materials and Methods.
Peptides.
Synthetic HPLC-purified peptides used were pep2r (CKRMKVAKNPQ), pepKR (CKRLKKKKTTIKK), pepAA (CKRLKKAATTIKK), TGN38-derived tyrosine motif peptide [YQRL; (C)KVTRRPKASDYQRL] or its alanine mutant [AQRL; (C)KVTRRPKASDAQRL], and CD3γ-derived dileucine motif peptide (RQSRASDKQTLLPN).
siRNAs.
siRNAs were purchased from MWG Biotech (Ebersberg, Germany). The following sequences were used: anti-human γBAR-siRNA as control, uga cga cag cac cuc cuu att; anti-human μ2 siRNA, gug gau gcc uuu cgg guc att (see ref. 12).
Plasmid DNA and Site-Directed Mutagenesis.
DNA manipulations were carried out by standard procedures. Single, double, and triple mutants were produced by PCR using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The presence of the mutation(s) was verified by dsDNA sequencing. Plasmids encoding rat FLAG-synaptotagmin 1 (9), HA-GluR2, GST-GluR1–3 CTs (7), and the αχ, β2, μ2, and σ2 subunits of AP-2 (18) have been described before.
Affinity Purification and Peptide Competition Assays.
Fusion proteins were purified in the presence of benzonase to remove possible nucleic acid contaminations. Affinity purification from the brain was performed as described (9). Briefly, Triton X-100-extracted rat brain lysates from 12-week-old Wistar rats (≈4 mg/ml protein concentration) were incubated for 1 h at 4°C with immobilized GST or GST-GluR2 CT (50 μg each) on a rotating wheel in binding buffer (20 mM Hepes, pH 7.4/100 mM KCl/2 mM MgCl2/1% Triton X-100). Inositol phosphates or peptides were added as indicated in the figure legends. For competition assays shown in Fig. 2, peptides were preincubated with 6 μg of purified His6-μ2 (157–435) in 500 μl of binding buffer (see above) for 20 min at 4°C on a rotating wheel. The mixture was cleared by ultracentrifugation and incubated with 20 μg of GST-GluR2 wt or K844A immobilized on glutathione agarose for 90 min at 4°C on a rotating wheel.
Surface Plasmon Resonance Measurements.
Surface plasmon resonance experiments were carried out on a BIACORE X instrument (Biacore, Uppsala, Sweden) at 25°C using biotinylated pep2r (CKRMKVAKNPQ-CONH2) or glutathione immobilized on a sensor chip SA (Biacore) preimmobilized with streptavidin (see SI Materials and Methods for details) and purified μ2 (157–435). Rate constants were calculated by using BIAevaluation software 4.1 assuming a 1:1 mode of binding with drifting baseline.
Internalization Assays.
Cos7 cells transfected by using Lipofectamine 2000 (according to the manufacturer's instructions) were split 24 h after transfection onto polyL-lysine-coated glass coverslips and allowed to grow for another 24 h. Cells were starved for 45 min in OptiMEM before labeling with 2 μg/ml monoclonal anti-FLAG antibody M2 (Sigma, Henningsdorf) (45 min on ice). Antibody uptake was allowed for 30 min at 37°C. Cells were washed twice with PBS before fixation with 4% paraformaldehyde. Plasma membrane labeling was detected by staining with Alexa Fluor 594-GαM under nonpermeabilizing conditions. Excess plasma membrane labeling was blocked by applying unlabeled GαM antiserum under nonpermeabilizing conditions. Cells were then permeabilized with 0.3% Triton X-100 followed by detection of internalized antibodies using Alexa Fluor 488-GαM secondary IgG. For each condition four independent experiments (n = 4) were carried out, and 20 cells for each experiment and construct were counted. The statistical significance of different internalization levels for each condition was tested by applying a Pearson χ2 test to the contingency table.
Whole-Cell Recordings.
Whole-cell recordings of currents in cortical neurons from acute slices used standard voltage-clamp techniques (27). The whole-cell patch technique (17) was used for recordings of mEPSC. See SI Materials and Methods for details.
Miscellaneous.
For SDS/PAGE, immunoblotting, in vitro transcription/translation (Promega), and indirect immunofluorescence microscopy standard procedures were used. Knockdown experiments using siRNAs against μ2-adaptin were done as described (11, 12).
Supplementary Material
Acknowledgments
We thank Dr. Pietro De Camilli (Yale University School of Medicine, New Haven, CT) for the gift of antibodies, Dr. M. Kasim Diril (Freie Universität Berlin) for advice, and Dr. Nicolai Bissantz (University of Göttingen, Göttingen, Germany) for help with statistical analysis. This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB449 TP A11, GRK 1123, and CMPB) and the European Molecular Biology Organization (European Molecular Biology Organization Young Investigator Programme award; to V.H.).
Abbreviations
- AMPA
α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid
- CT
cytoplasmic tail
- LTD
long-term depression
- mEPSC
miniature excitatory postsynaptic currents.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611170104/DC1.
References
- 1.Fukata Y, Tzingounis AV, Trinidad JC, Fukata M, Burlingame AL, Nicoll RA, Bredt DS. J Cell Biol. 2005;169:399–404. doi: 10.1083/jcb.200501121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bredt DS, Nicoll RA. Neuron. 2003;40:361–379. doi: 10.1016/s0896-6273(03)00640-8. [DOI] [PubMed] [Google Scholar]
- 3.Malinow R, Malenka RC. Annu Rev Neurosci. 2002;25:103–126. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- 4.Perez-Otano I, Ehlers MD. Trends Neurosci. 2005;28:229–238. doi: 10.1016/j.tins.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Sheng M, Kim MJ. Science. 2002;298:776–780. doi: 10.1126/science.1075333. [DOI] [PubMed] [Google Scholar]
- 6.Praefcke GJ, Ford MG, Schmid EM, Olesen LE, Gallop JL, Peak-Chew SY, Vallis Y, Babu MM, Mills IG, McMahon HT. EMBO J. 2004;23:4371–4383. doi: 10.1038/sj.emboj.7600445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee SH, Liu L, Wang YT, Sheng M. Neuron. 2002;36:661–674. doi: 10.1016/s0896-6273(02)01024-3. [DOI] [PubMed] [Google Scholar]
- 8.Robinson MS. Trends Cell Biol. 2004;14:167–174. doi: 10.1016/j.tcb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Grass I, Thiel S, Honing S, Haucke V. J Biol Chem. 2004;279:54872–54880. doi: 10.1074/jbc.M409995200. [DOI] [PubMed] [Google Scholar]
- 10.Mizutani A, Fukuda M, Niinobe M, Mikoshiba K. Biochem Biophys Res Commun. 1997;240:128–131. doi: 10.1006/bbrc.1997.7578. [DOI] [PubMed] [Google Scholar]
- 11.Diril MK, Wienisch M, Jung N, Klingauf J, Haucke V. Dev Cell. 2006;10:233–244. doi: 10.1016/j.devcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 12.Motley A, Bright NA, Seaman MN, Robinson MS. J Cell Biol. 2003;162:909–918. doi: 10.1083/jcb.200305145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luscher C, Xia H, Beattie EC, Carroll RC, von Zastrow M, Malenka RC, Nicoll RA. Neuron. 1999;24:649–658. doi: 10.1016/s0896-6273(00)81119-8. [DOI] [PubMed] [Google Scholar]
- 14.Beattie EC, Carroll RC, Yu X, Morishita W, Yasuda H, von Zastrow M, Malenka RC. Nat Neurosci. 2000;3:1291–1300. doi: 10.1038/81823. [DOI] [PubMed] [Google Scholar]
- 15.Lee SH, Simonetta A, Sheng M. Neuron. 2004;43:221–236. doi: 10.1016/j.neuron.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 16.Lin JW, Ju W, Foster K, Lee SH, Ahmadian G, Wyszynski M, Wang YT, Sheng M. Nat Neurosci. 2000;3:1282–1290. doi: 10.1038/81814. [DOI] [PubMed] [Google Scholar]
- 17.Kittler JT, Chen G, Honing S, Bogdanov Y, McAinsh K, Arancibia-Carcamo IL, Jovanovic JN, Pangalos MN, Haucke V, Yan Z, Moss SJ. Proc Natl Acad Sci USA. 2005;102:14871–14876. doi: 10.1073/pnas.0506653102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haucke V, Wenk MR, Chapman ER, Farsad K, De Camilli P. EMBO J. 2000;19:6011–6019. doi: 10.1093/emboj/19.22.6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park M, Penick EC, Edwards JG, Kauer JA, Ehlers MD. Science. 2004;305:1972–1975. doi: 10.1126/science.1102026. [DOI] [PubMed] [Google Scholar]
- 20.Man HY, Lin JW, Ju WH, Ahmadian G, Liu L, Becker LE, Sheng M, Wang YT. Neuron. 2000;25:649–662. doi: 10.1016/s0896-6273(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 21.Racz B, Blanpied TA, Ehlers MD, Weinberg RJ. Nat Neurosci. 2004;7:917–918. doi: 10.1038/nn1303. [DOI] [PubMed] [Google Scholar]
- 22.Burbea M, Dreier L, Dittman JS, Grunwald ME, Kaplan JM. Neuron. 2002;35:107–120. doi: 10.1016/s0896-6273(02)00749-3. [DOI] [PubMed] [Google Scholar]
- 23.Ahmadian G, Ju W, Liu L, Wyszynski M, Lee SH, Dunah AW, Taghibiglou C, Wang Y, Lu J, Wong TP, et al. EMBO J. 2004;23:1040–1050. doi: 10.1038/sj.emboj.7600126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esteban JA, Shi SH, Wilson C, Nuriya M, Huganir RL, Malinow R. Nat Neurosci. 2003;6:136–143. doi: 10.1038/nn997. [DOI] [PubMed] [Google Scholar]
- 25.Metzler M, Li B, Gan L, Georgiou J, Gutekunst CA, Wang Y, Torre E, Devon RS, Oh R, Legendre-Guillemin V, et al. EMBO J. 2003;22:3254–3266. doi: 10.1093/emboj/cdg334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lakadamyali M, Rust MJ, Zhuang X. Cell. 2006;124:997–1009. doi: 10.1016/j.cell.2005.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen G, Greengard P, Yan Z. Proc Natl Acad Sci USA. 2004;101:2596–2600. doi: 10.1073/pnas.0308618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.